Genome-Wide Investigation of Spliceosomal SM/LSM Genes in Wheat (Triticum aestivum L.) and Its Progenitors
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of Smith (SM)/SM-Like (SSM/SLSM) Genes in Wheat and Its Progenitors
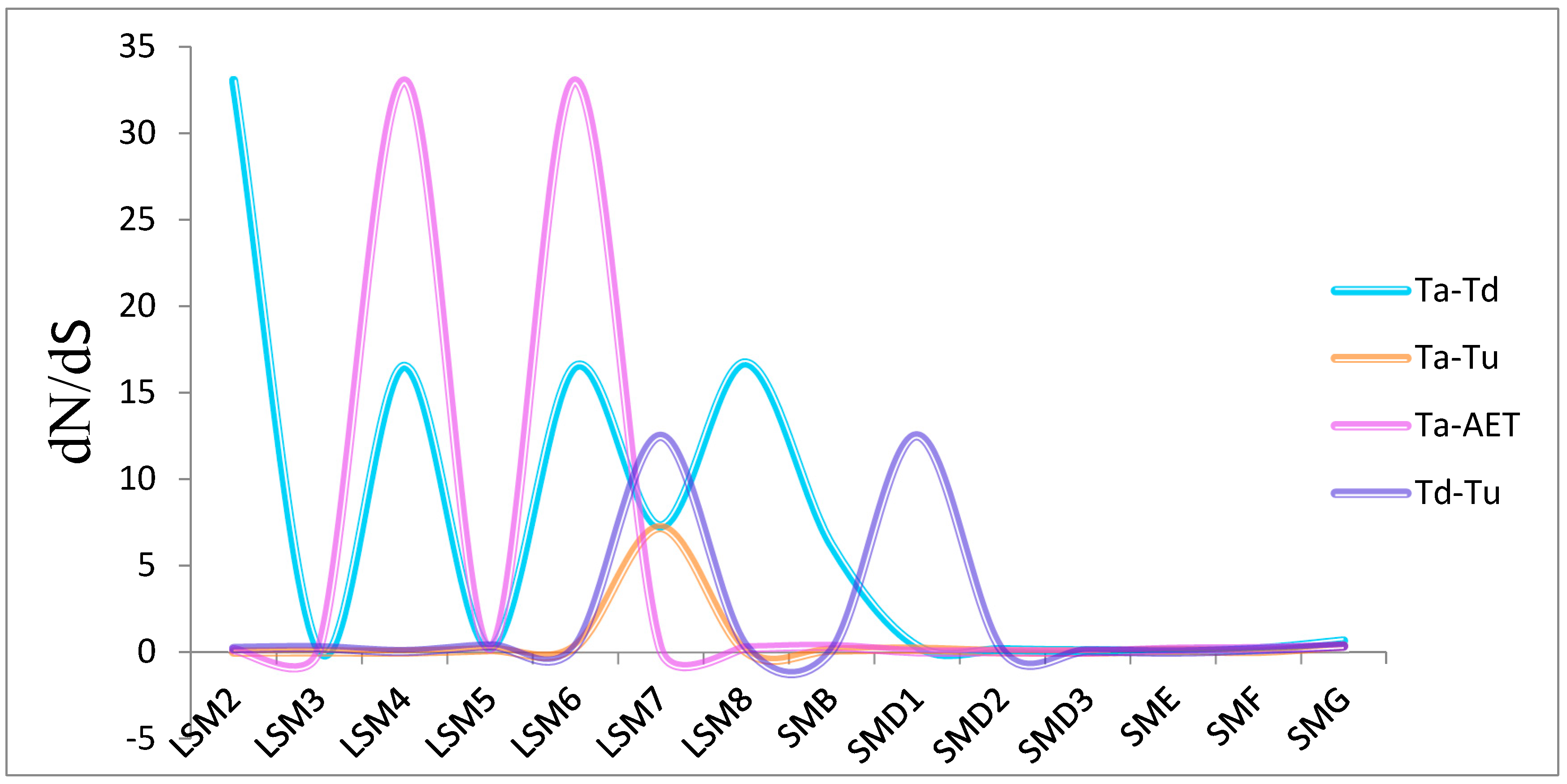
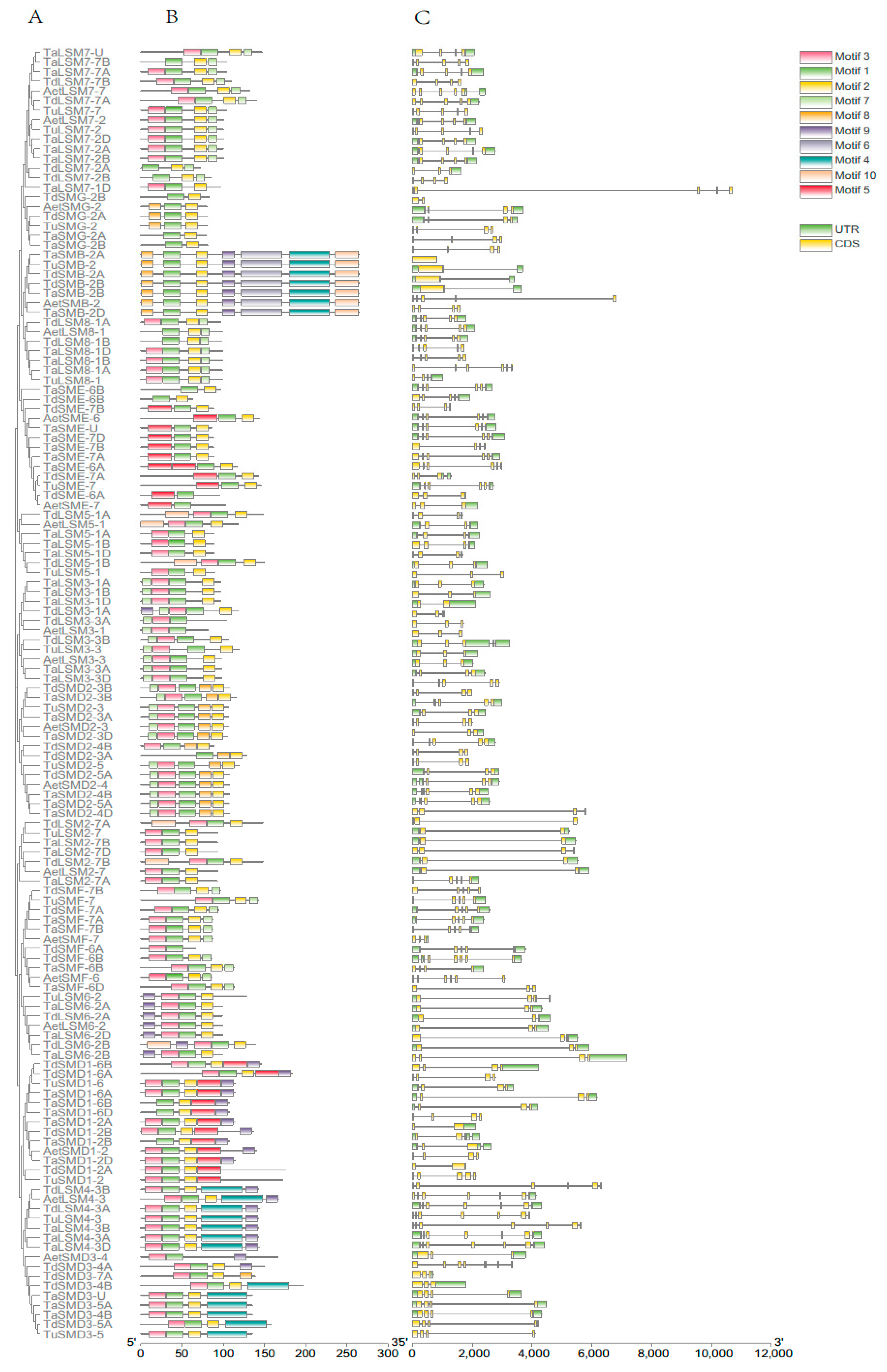
2.2. Chromosome Location, Gene Structure, Collinearity Analyses and Non-Synonymous to Synonymous (dN/dS) of SSM/SLSM Genes
2.3. Analysis of Cis-Acting Elements of SSM/SLSM Genes Promoter
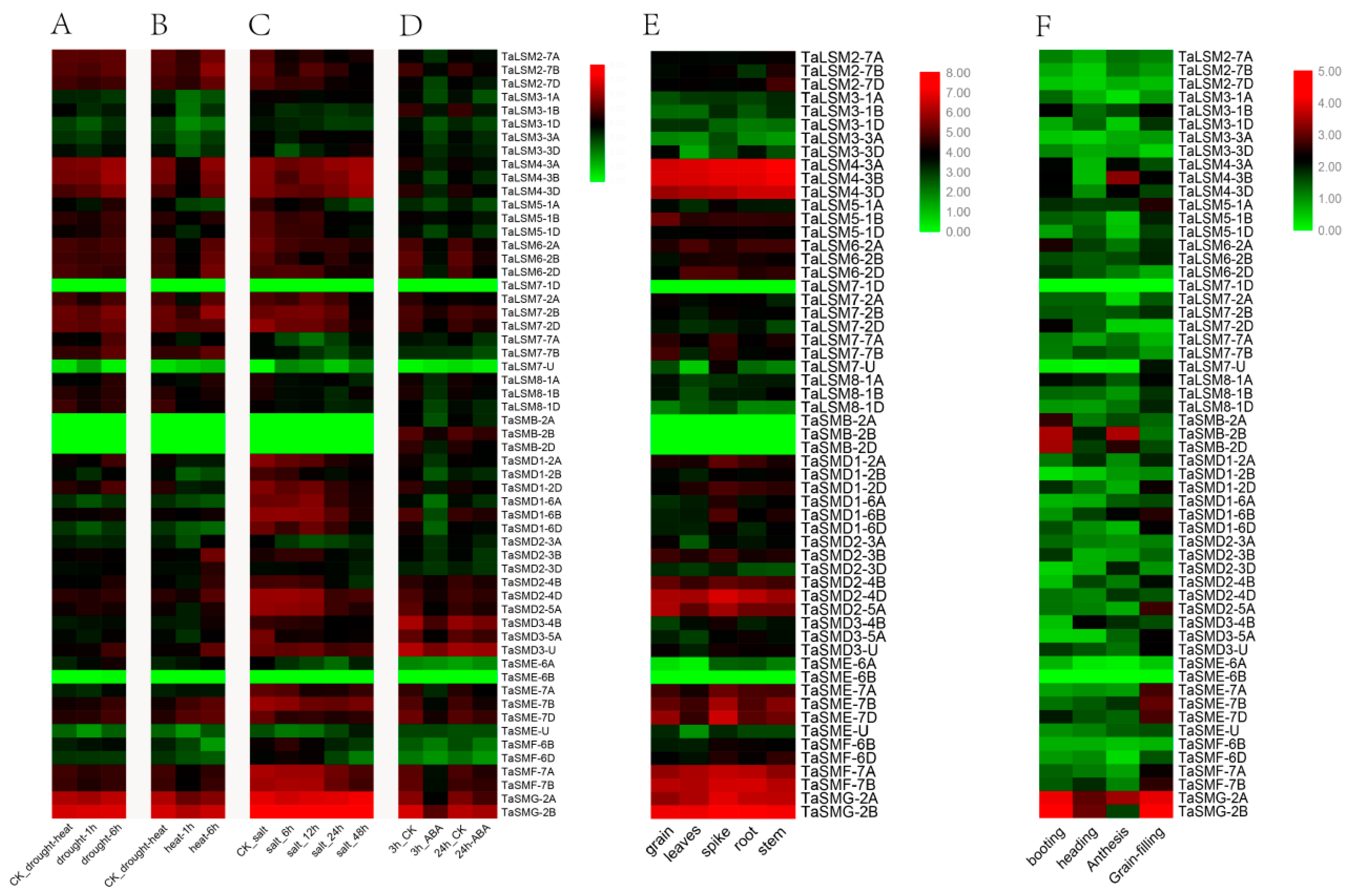
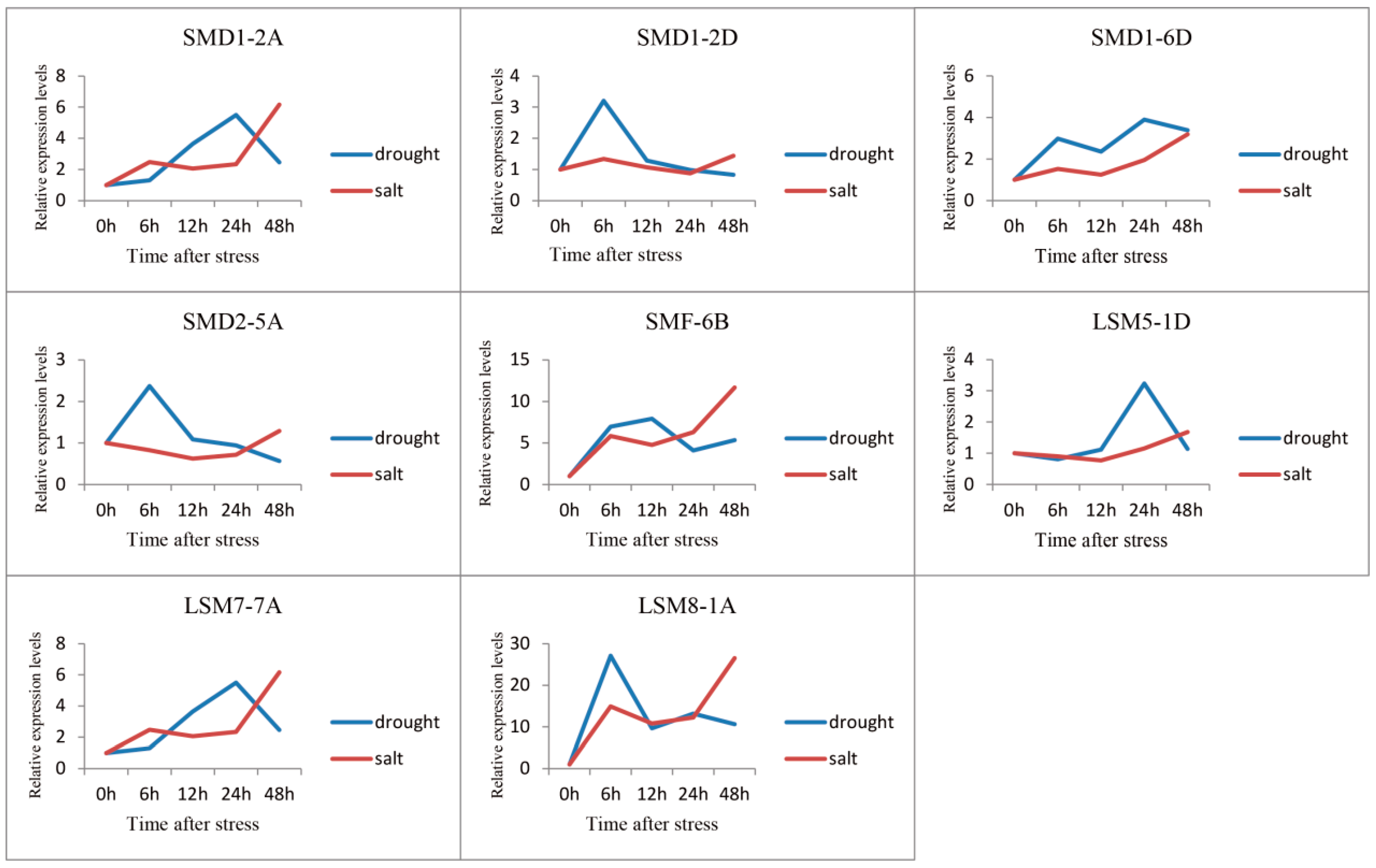
2.4. Analysis of the Specific Expression of SSM/SLSM Genes
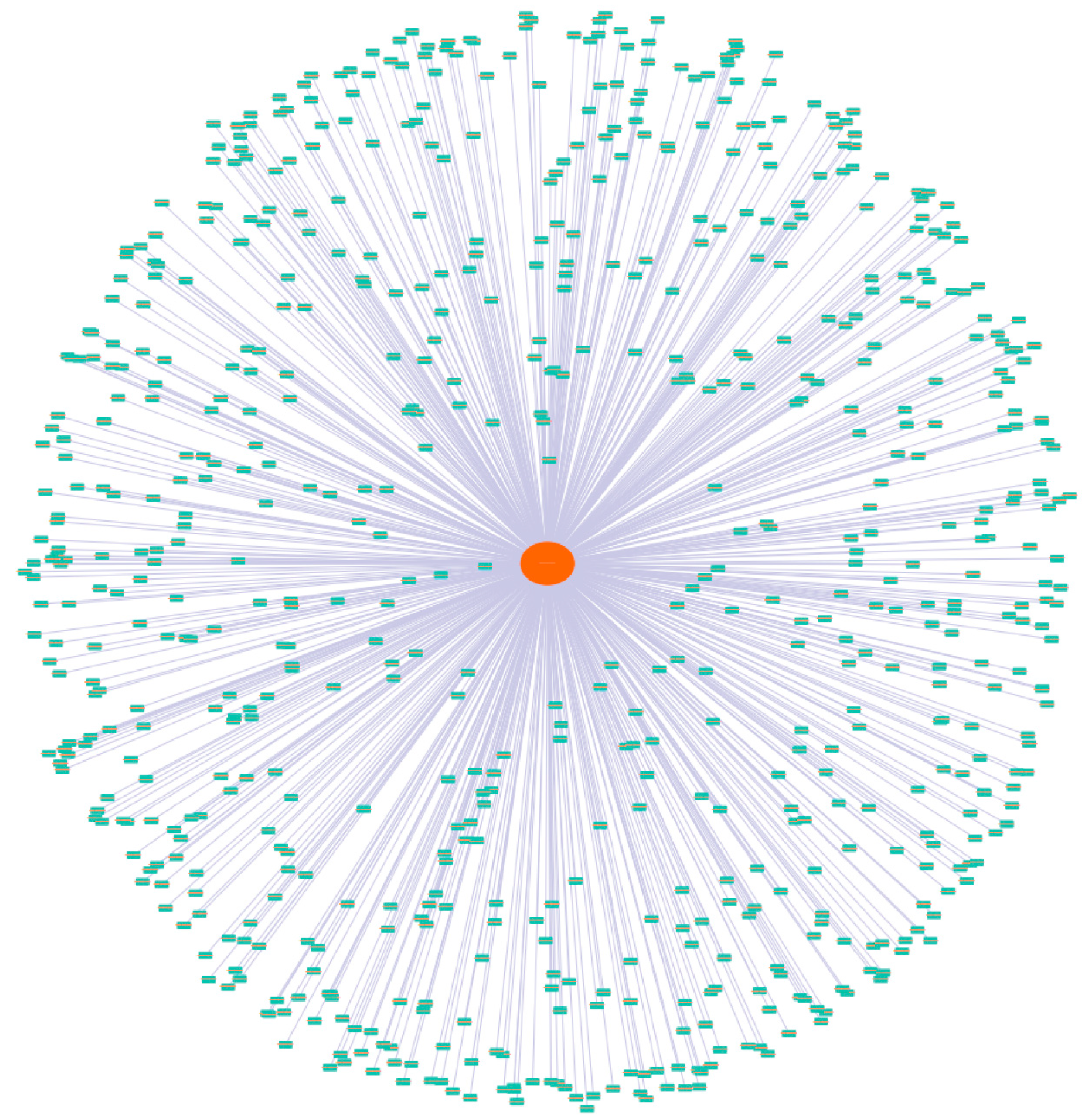
2.5. Construction of the Co-Expression Network and Functional Search of Key Genes
3. Results
3.1. Identification of SSM/SLSM Gene Members and Their Physico-Chemical Properties in Wheat and Its Progenitors
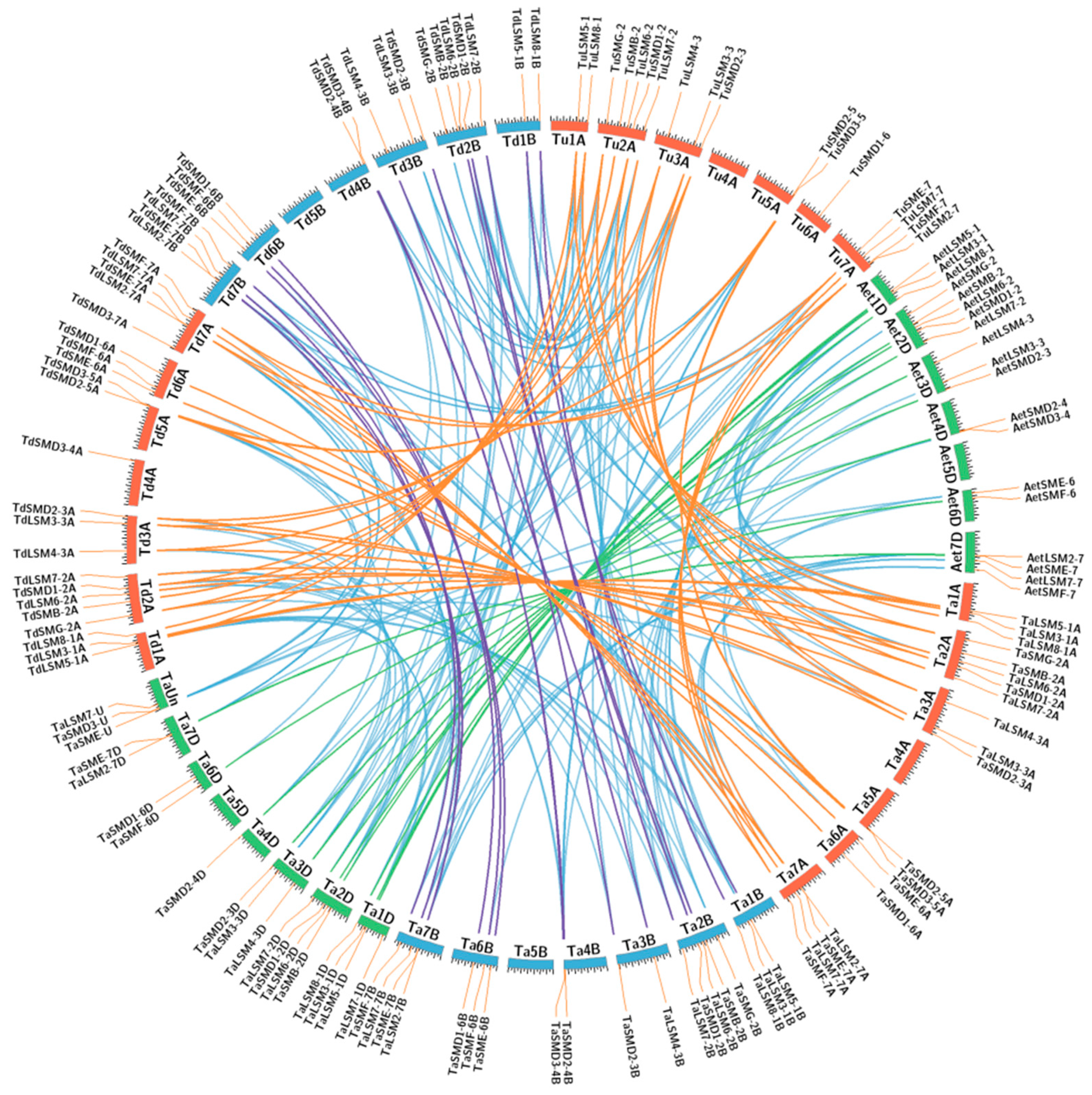
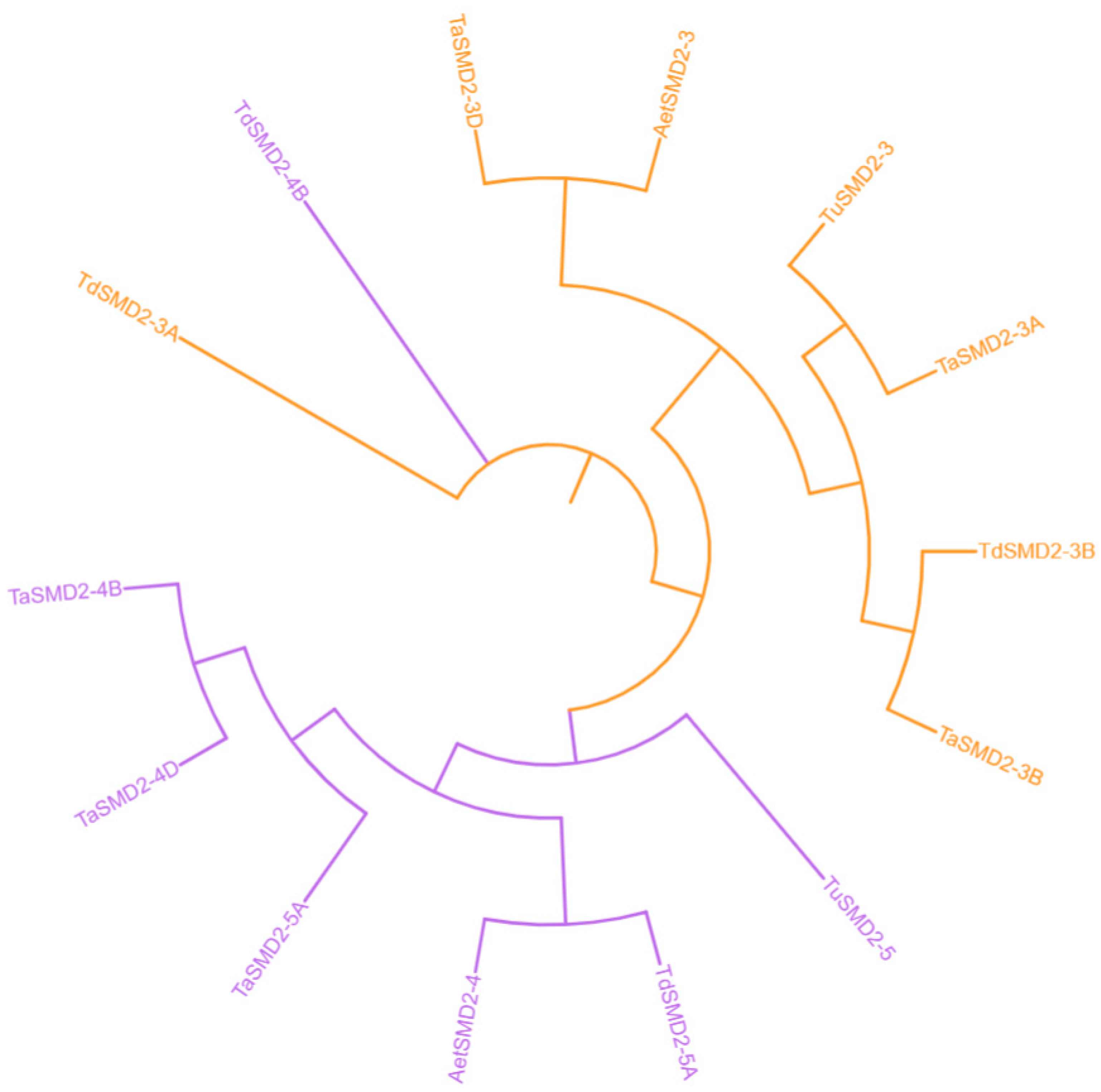
3.2. Genome Distribution and Synthetic Analysis of SSM/SLSM Genes among Bread Wheat and Its Progenitors
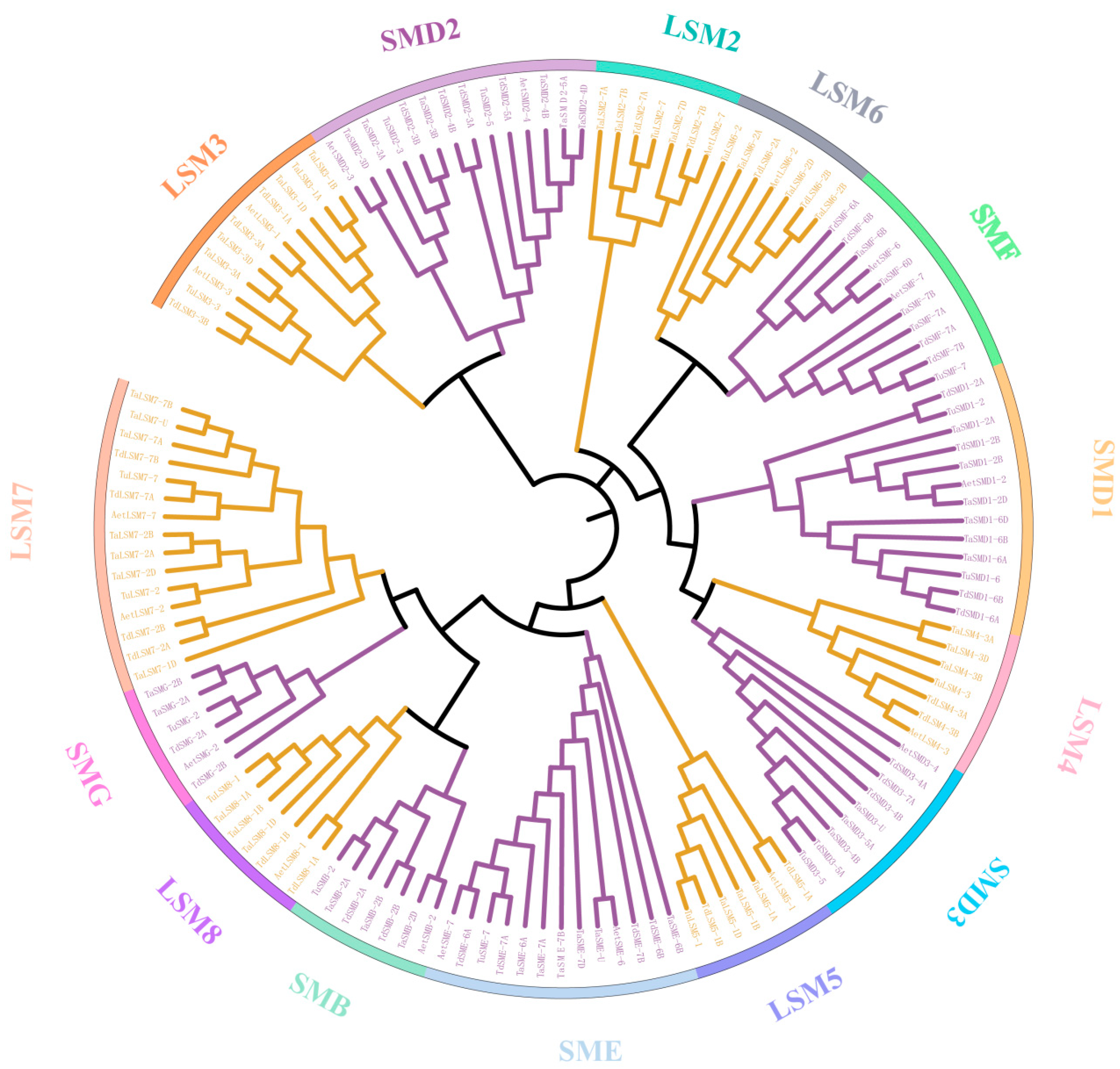
3.3. The Phylogenetic and Gene Structure of SSM/SLSM Proteins in Wheat and Its Progenitors
3.4. Analysis of Cis-Regulatory Elements in the Promoter Regions of TaSSM/SLSMs
3.5. Expression Profile Analysis of SSM/SLSM Genes in Wheat
3.6. Co-Expression Network Analysis of Wheat SSM/SLSM Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wahl, M.C.; Will, C.L.; Luhrmann, R. The spliceosome: Design principles of a dynamic RNP machine. Cell 2009, 136, 701–718. [Google Scholar] [CrossRef]
- Nilsen, T.W. The spliceosome: The most complex macromolecular machine in the cell? Bioessays 2003, 25, 1147–1149. [Google Scholar] [CrossRef]
- Reeves, W.H.; Narain, S.; Satoh, M. Henry Kunkel, Stephanie Smith, clinical immunology, and split genes. Lupus 2003, 12, 213–217. [Google Scholar] [CrossRef]
- Urlaub, H.; Raker, V.A.; Kostka, S.; Luhrmann, R. Sm protein-Sm site RNA interactions within the inner ring of the spliceosomal snRNP core structure. EMBO J. 2001, 20, 187–196. [Google Scholar] [CrossRef]
- Will, C.L.; Luhrmann, R. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 2001, 13, 290–301. [Google Scholar] [CrossRef]
- Hackl, W.; Fischer, U.; Luhrmann, R. A 69-Kd Protein That Associates Reversibly with the Sm Core Domain of Several Spliceosomal Snrnp Species. J. Cell Biol. 1994, 124, 261–272. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Parker, R. Functions of Lsm proteins in mRNA degradation and splicing. Curr. Opin. Cell Biol. 2000, 12, 346–350. [Google Scholar] [CrossRef]
- Ru, Y.; Wang, B.B.; Brendel, V. Spliceosomal Proteins in Plants. Curr. Top. Microbiol. 2008, 326, 1–15. [Google Scholar]
- Veretnik, S.; Wills, C.; Youkharibache, P.; Valas, R.E.; Bourne, P.E. Sm/Lsm genes provide a glimpse into the early evolution of the spliceosome. PLoS Comput. Biol. 2009, 5, e1000315. [Google Scholar] [CrossRef]
- Golisz, A.; Sikorski, P.J.; Kruszka, K.; Kufel, J. Arabidopsis thaliana LSM proteins function in mRNA splicing and degradation. Nucleic Acids Res. 2013, 41, 6232–6249. [Google Scholar] [CrossRef] [PubMed]
- Seraphin, B. Sm and Sm-Like Proteins Belong to a Large Family-Identification of Proteins of the U6 as Well as the U1, U2, U4 and U5 Snrnps. EMBO J. 1995, 14, 2089–2098. [Google Scholar] [CrossRef]
- Cooper, M.; Johnston, L.H.; Beggs, J.D. Identification and characterization of Uss1p (Sdb23p): A novel U6 snRNA-associated protein with significant similarity to core proteins of small nuclear ribonucleoproteins. EMBO J. 1995, 14, 2066–2075. [Google Scholar] [CrossRef]
- Hermann, H.; Fabrizio, P.; Raker, V.A.; Foulaki, K.; Hornig, H.; Brahms, H.; Luhrmann, R. snRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein-protein interactions. EMBO J. 1995, 14, 2076–2088. [Google Scholar] [CrossRef]
- Kambach, C.; Walke, S.; Young, R.; Avis, J.M.; de la Fortelle, E.; Raker, V.A.; Luhrmann, R.; Li, J.; Nagai, K. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell 1999, 96, 375–387. [Google Scholar] [CrossRef]
- Cao, J.; Shi, F.; Liu, X.; Jia, J.; Zeng, J.; Huang, G. Genome-wide identification and evolutionary analysis of Arabidopsis sm genes family. J. Biomol. Struct. Dyn. 2011, 28, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, Y.; Zhang, S.T.; Su, L.Y.; Xu, X.P.; Chen, X.H.; Lai, Z.X.; Lin, Y.L. Genome-wide identification and expression analyses of Sm genes reveal their involvement in early somatic embryogenesis in Dimocarpus longan Lour. PLoS ONE 2020, 15, e0230795. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Cao, J. Comparative genomic analysis of the Sm gene family in rice and maize. Gene 2014, 539, 238–249. [Google Scholar] [CrossRef]
- Wang, B.B.; Brendel, V. The ASRG database: Identification and survey of Arabidopsis thaliana genes involved in pre-mRNA splicing. Genome Biol. 2004, 5, R102. [Google Scholar] [CrossRef] [PubMed]
- Chen, L. Identification and Survey of Pre-mRNA Splicing-Related Proteins in 10 Plant Species. Master’s Thesis, Iowa State University, Ames, IA, USA, 2011. [Google Scholar]
- Sharpe, N.G.; Williams, D.G.; Norton, P.; Latchman, D.S. Expression of the SmB’ splicing protein in rodent cells capable of following an alternative RNA splicing pathway. FEBS Lett. 1989, 243, 132–136. [Google Scholar] [CrossRef][Green Version]
- Anne, J. Arginine methylation of SmB is required for Drosophila germ cell development. Development 2010, 137, 2819–2828. [Google Scholar] [CrossRef]
- Scruggs, B.S.; Michel, C.I.; Ory, D.S.; Schaffer, J.E. SmD3 Regulates Intronic Noncoding RNA Biogenesis. Mol. Cell. Biol. 2012, 32, 4092–4103. [Google Scholar] [CrossRef]
- Li, Z.; Putzer, B.M. Spliceosomal protein E regulates neoplastic cell growth by modulating expression of Cyclin E/CDK2 and G2/M checkpoint proteins. J. Cell. Mol. Med. 2008, 12, 2427–2438. [Google Scholar] [CrossRef]
- Mabonga, L.; Kappo, A.P. The oncogenic potential of small nuclear ribonucleoprotein polypeptide G: A comprehensive and perspective view. Am. J. Transl. Res. 2019, 11, 6702–6716. [Google Scholar]
- Perez-Santangelo, S.; Mancini, E.; Francey, L.J.; Schlaen, R.G.; Chernomoretz, A.; Hogenesch, J.B.; Yanovsky, M.J. Role for LSM genes in the regulation of circadian rhythms. Proc. Natl. Acad. Sci. USA 2014, 111, 15166–15171. [Google Scholar] [CrossRef] [PubMed]
- Huertas, R.; Catala, R.; Jimenez-Gomez, J.M.; Castellano, M.M.; Crevillen, P.; Pineiro, M.; Jarillo, J.A.; Salinas, J. Arabidopsis SME1 Regulates Plant Development and Response to Abiotic Stress by Determining Spliceosome Activity Specificity. Plant Cell 2019, 31, 537–554. [Google Scholar] [CrossRef]
- Cui, P.; Zhang, S.; Ding, F.; Ali, S.; Xiong, L. Dynamic regulation of genome-wide pre-mRNA splicing and stress tolerance by the Sm-like protein LSm5 in Arabidopsis. Genome Biol. 2014, 15, R1. [Google Scholar] [CrossRef]
- Zhang, Z.L.; Zhang, S.P.; Zhang, Y.; Wang, X.; Li, D.; Li, Q.L.; Yue, M.H.; Li, Q.; Zhang, Y.E.; Xu, Y.Y.; et al. Arabidopsis Floral Initiator SKB1 Confers High Salt Tolerance by Regulating Transcription and Pre-mRNA Splicing through Altering Histone H4R3 and Small Nuclear Ribonucleoprotein LSM4 Methylation. Plant Cell 2011, 23, 396–411. [Google Scholar] [CrossRef]
- Perea-Resa, C.; Carrasco-Lopez, C.; Catala, R.; Tureckova, V.; Novak, O.; Zhang, W.P.; Sieburth, L.; Jimenez-Gomez, J.M.; Salinas, J. The LSM1-7 Complex Differentially Regulates Arabidopsis Tolerance to Abiotic Stress Conditions by Promoting Selective mRNA Decapping. Plant Cell 2016, 28, 505–520. [Google Scholar] [CrossRef]
- Carrasco-Lopez, C.; Hernandez-Verdeja, T.; Perea-Resa, C.; Abia, D.; Catala, R.; Salinas, J. Environment-dependent regulation of spliceosome activity by the LSM2-8 complex in Arabidopsis. Nucleic Acids Res. 2017, 45, 7416–7431. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, Y. Evolution of Polyploid Triticum Wheats under Cultivation: The Role of Domestication, Natural Hybridization and Allopolyploid Speciation in their Diversification. Plant Cell Physiol. 2011, 52, 750–764. [Google Scholar] [CrossRef] [PubMed]
- Feldman, M.; Levy, A.A. Genome Evolution Due to Allopolyploidization in Wheat. Genetics 2012, 192, 763–774. [Google Scholar] [CrossRef]
- Pfeifer, M.; Kugler, K.G.; Sandve, S.R.; Zhan, B.; Rudi, H.; Hvidsten, T.R.; International Wheat Genome Sequencing Consortium; Mayer, K.F.; Olsen, O.A. Genome interplay in the grain transcriptome of hexaploid bread wheat. Science 2014, 345, 1250091. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Pasam, R.; Shi, F.; Kant, S.; Keeble-Gagnere, G.; Kay, P.; Forrest, K.; Fritz, A.; Hucl, P.; Wiebe, K.; et al. Exome sequencing highlights the role of wild-relative introgression in shaping the adaptive landscape of the wheat genome. Nat. Genet. 2019, 51, 896–904. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Zhang, Z.; Xiao, J.F.; Wu, J.Y.; Zhang, H.Y.; Liu, G.M.; Wang, X.M.; Dai, L. ParaAT: A parallel tool for constructing multiple protein-coding DNA alignments. Biochem. Biophys. Res. Commun. 2012, 419, 779–781. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Zhang, Y.; Cui, H.; Liu, J.; Wu, Y.; Cheng, Y.; Xu, H.; Huang, X.; Li, S.; Zhou, A.; et al. WEGO 2.0: A web tool for analyzing and plotting GO annotations, 2018 update. Nucleic Acids Res. 2018, 46, W71–W75. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Chen, S.; Luo, Y.; Wang, G.; Feng, C.; Li, H. Genome-wide identification of expansin genes in Brachypodium distachyon and functional characterization of BdEXPA27. Plant Sci. 2020, 296, 110490. [Google Scholar] [CrossRef]
- Ding, F.; Cui, P.; Wang, Z.; Zhang, S.; Ali, S.; Xiong, L. Genome-wide analysis of alternative splicing of pre-mRNA under salt stress in Arabidopsis. BMC Genom. 2014, 15, 431. [Google Scholar] [CrossRef]
- Feng, J.; Li, J.; Gao, Z.; Lu, Y.; Yu, J.; Zheng, Q.; Yan, S.; Zhang, W.; He, H.; Ma, L.; et al. SKIP Confers Osmotic Tolerance during Salt Stress by Controlling Alternative Gene Splicing in Arabidopsis. Mol. Plant 2015, 8, 1038–1052. [Google Scholar] [CrossRef]
- Zhan, X.Q.; Qian, B.L.; Cao, F.Q.; Wu, W.W.; Yang, L.; Guan, Q.M.; Gu, X.B.; Wang, P.C.; Okusolubo, T.A.; Dunn, S.L.; et al. An Arabidopsis PWI and RRM motif-containing protein is critical for pre-mRNA splicing and ABA responses. Nat. Commun. 2015, 6, 8139. [Google Scholar] [CrossRef]
- Thatcher, S.R.; Danilevskaya, O.N.; Meng, X.; Beatty, M.; Zastrow-Hayes, G.; Harris, C.; Van Allen, B.; Habben, J.; Li, B.L. Genome-Wide Analysis of Alternative Splicing during Development and Drought Stress in Maize. Plant Physiol. 2016, 170, 586–599. [Google Scholar] [CrossRef]
- Jiang, J.F.; Liu, X.N.; Liu, C.H.; Liu, G.T.; Li, S.H.; Wang, L.J. Integrating Omics and Alternative Splicing Reveals Insights into Grape Response to High Temperature. Plant Physiol. 2017, 173, 1502–1518. [Google Scholar] [CrossRef]
- Keller, M.; Hu, Y.J.; Mesihovic, A.; Fragkostefanakis, S.; Schleiff, E.; Simm, S. Alternative splicing in tomato pollen in response to heat stress(aEuro). DNA Res. 2017, 24, 205–217. [Google Scholar] [CrossRef]
- Zhu, F.Y.; Chen, M.X.; Ye, N.H.; Shi, L.; Ma, K.L.; Yang, J.F.; Cao, Y.Y.; Zhang, Y.J.; Yoshida, T.; Fernie, A.R.; et al. Proteogenomic analysis reveals alternative splicing and translation as part of the abscisic acid response in Arabidopsis seedlings. Plant J. 2017, 91, 518–533. [Google Scholar] [CrossRef]
- Forment, J.; Naranjo, M.A.; Roldan, M.; Serrano, R.; Vicente, O. Expression of Arabidopsis SR-like splicing proteins confers salt tolerance to yeast and transgenic plants. Plant J. 2002, 30, 511–519. [Google Scholar] [CrossRef]
- Shaked, H.; Kashkush, K.; Ozkan, H.; Feldman, M.; Levy, A.A. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 2001, 13, 1749–1759. [Google Scholar] [CrossRef]
- Szadkowski, E.; Eber, F.; Huteau, V.; Lode, M.; Huneau, C.; Belcram, H.; Coriton, O.; Manzanares-Dauleux, M.J.; Delourme, R.; King, G.J.; et al. The first meiosis of resynthesized Brassica napus, a genome blender. New Phytol. 2010, 186, 102–112. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Y.; Su, H.; Guo, X.; Han, F. Centromere structure and function analysis in wheat-rye translocation lines. Plant J. 2017, 91, 199–207. [Google Scholar] [CrossRef]
- Zhou, C.; Dong, Z.; Zhang, T.; Wu, J.; Yu, S.; Zeng, Q.; Han, D.; Tong, W. Genome-Scale Analysis of Homologous Genes among Subgenomes of Bread Wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2020, 21, 3015. [Google Scholar] [CrossRef]
- Chen, Y.; Song, W.; Xie, X.; Wang, Z.; Guan, P.; Peng, H.; Jiao, Y.; Ni, Z.; Sun, Q.; Guo, W. A Collinearity-Incorporating Homology Inference Strategy for Connecting Emerging Assemblies in the Triticeae Tribe as a Pilot Practice in the Plant Pangenomic Era. Mol. Plant 2020, 13, 1694–1708. [Google Scholar] [CrossRef] [PubMed]
- Spielman, S.J.; Wilke, C.O. The Relationship between dN/dS and Scaled Selection Coefficients. Mol. Biol. Evol. 2015, 32, 1097–1108. [Google Scholar] [CrossRef] [PubMed]
- Mugal, C.F.; Wolf, J.B.; Kaj, I. Why time matters: Codon evolution and the temporal dynamics of dN/dS. Mol. Biol. Evol. 2014, 31, 212–231. [Google Scholar] [CrossRef] [PubMed]
- Ratnakumar, A.; Mousset, S.; Glemin, S.; Berglund, J.; Galtier, N.; Duret, L.; Webster, M.T. Detecting positive selection within genomes: The problem of biased gene conversion. Philos. Trans. R. Soc. B 2010, 365, 2571–2580. [Google Scholar] [CrossRef]
- Yang, Z.; Nielsen, R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 2000, 17, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Swaraz, A.M.; Park, Y.D.; Hur, Y. Knock-out mutations of Arabidopsis SmD3-b induce pleotropic phenotypes through altered transcript splicing. Plant Sci. 2011, 180, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Elvira-Matelot, E.; Bardou, F.; Ariel, F.; Jauvion, V.; Bouteiller, N.; Le Masson, I.; Cao, J.; Crespi, M.D.; Vaucheret, H. The Nuclear Ribonucleoprotein SmD1 Interplays with Splicing, RNA Quality Control, and Posttranscriptional Gene Silencing in Arabidopsis. Plant Cell 2016, 28, 426–438. [Google Scholar] [CrossRef] [PubMed]
- Mejias, J.; Bazin, J.; Truong, N.M.; Chen, Y.; Marteu, N.; Bouteiller, N.; Sawa, S.; Crespi, M.D.; Vaucheret, H.; Abad, P.; et al. The root-knot nematode effector MiEFF18 interacts with the plant core spliceosomal protein SmD1 required for giant cell formation. New Phytol. 2020. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | AA | MW (Da) | PI | GRAVY | Gene Id | |
|---|---|---|---|---|---|---|
| LSM2 | TuLSM2-7 | 148 | 16,537.77 | 8.45 | −0.511 | TuG1812G0700005606.01.P01 |
| TdLSM2-7B | 148 | 16,319.52 | 7.43 | −0.326 | TRIDC7BG033290.2 | |
| TdLSM2-7A | 93 | 10,737.36 | 6.82 | −0.199 | TRIDC7AG042160.1 | |
| TaLSM2-7D | 93 | 10,737.36 | 6.82 | −0.199 | TraesCS7D02G297400.1 | |
| TaLSM2-7B | 93 | 10,737.36 | 6.82 | −0.199 | TraesCS7B02G202200.1 | |
| TaLSM2-7A | 93 | 10,737.36 | 6.82 | −0.199 | TraesCS7A02G302100.1 | |
| AetLSM2-7 | 93 | 10,737.36 | 6.82 | −0.199 | AET7Gv20754100.2 | |
| LSM3 | TuLSM3-3 | 118 | 13,347.21 | 5.06 | −0.311 | TuG1812G0300004141.01.P01 |
| TdLSM3-3B | 106 | 11,936.61 | 4.69 | −0.105 | TRIDC3BG060910.5 | |
| TdLSM3-3A | 104 | 12,178.97 | 5.14 | −0.15 | TRIDC3AG054250.1 | |
| TdLSM3-1A | 98 | 11,279.92 | 4.76 | −0.182 | TRIDC1AG040710.1 | |
| TaLSM3-3D | 82 | 96,07.99 | 4.57 | −0.26 | TraesCS3D02G371500.1 | |
| TaLSM3-3A | 97 | 11,196.83 | 4.76 | −0.215 | TraesCS3A02G378200.1 | |
| TaLSM3-1D | 97 | 11,196.83 | 4.76 | −0.215 | TraesCS1D02G273400.1 | |
| TaLSM3-1B | 97 | 11,196.83 | 4.76 | −0.215 | TraesCS1B02G283200.1 | |
| TaLSM3-1A | 98 | 11,279.92 | 4.76 | −0.182 | TraesCS1A02G273400.1 | |
| AetLSM3-3 | 98 | 11,279.92 | 4.76 | −0.182 | AET3Gv20844000.2 | |
| AetLSM3-1 | 119 | 13,735.82 | 4.8 | −0.05 | AET1Gv20658000.5 | |
| LSM4 | TuLSM4-3 | 143 | 15,472.57 | 9.96 | −0.914 | TuG1812G0300001955.01.P01 |
| TdLSM4-3B | 143 | 15,469.51 | 9.96 | −0.952 | TRIDC3BG028150.3 | |
| TdLSM4-3A | 143 | 15,472.57 | 9.96 | −0.914 | TRIDC3AG024440.1 | |
| TaLSM4-3D | 143 | 15,472.57 | 9.96 | −0.914 | TraesCS3D02G181600.1 | |
| TaLSM4-3B | 143 | 15,472.57 | 9.96 | −0.914 | TraesCS3B02G205600.2 | |
| TaLSM4-3A | 143 | 15,472.57 | 9.96 | −0.914 | TraesCS3A02G175900.1 | |
| AetLSM4-3 | 167 | 18,150.39 | 10.27 | −0.975 | AET3Gv20400700.6 | |
| LSM5 | TuLSM5-1 | 148 | 16,232.73 | 9.33 | −0.264 | TuG1812G0100002831.01.P01 |
| TdLSM5-1B | 150 | 16,187.58 | 8.74 | −0.29 | TRIDC1BG041610.1 | |
| TdLSM5-1A | 118 | 12,956.97 | 7.93 | −0.098 | TRIDC1AG036620.3 | |
| TaLSM5-1D | 88 | 9590.03 | 4.42 | 0 | TraesCS1D02G242900.1 | |
| TaLSM5-1B | 89 | 9753.21 | 4.42 | −0.015 | TraesCS1B02G254400.1 | |
| TaLSM5-1A | 89 | 9753.21 | 4.42 | −0.015 | TraesCS1A02G242900.1 | |
| AetLSM5-1 | 89 | 9753.21 | 4.42 | −0.015 | AET1Gv20591900.1 | |
| LSM6 | TuLSM6-2 | 128 | 13,818.85 | 9.72 | −0.241 | TuG1812G0200003237.01.P02 |
| TdLSM6-2B | 139 | 14,806.67 | 9.86 | −0.623 | TRIDC2BG044080.1 | |
| TdLSM6-2A | 99 | 10,360.7 | 9.13 | −0.362 | TRIDC2AG041140.2 | |
| TaLSM6-2D | 99 | 10,350.66 | 9.13 | −0.354 | TraesCS2D02G283200.1 | |
| TaLSM6-2B | 99 | 10,350.66 | 9.13 | −0.354 | TraesCS2B02G301300.1 | |
| TaLSM6-2A | 99 | 10,374.72 | 9.13 | −0.361 | TraesCS2A02G284300.1 | |
| AetLSM6-2 | 99 | 10,350.66 | 9.13 | −0.354 | AET2Gv20643200.1 | |
| LSM7 | TuLSM7-7 | 104 | 11,432.98 | 4.97 | −0.421 | TuG1812G0700004021.01.P01 |
| TuLSM7-2 | 100 | 10,675.18 | 4.87 | −0.183 | TuG1812G0200005747.01.P01 | |
| TdLSM7-7B | 110 | 12,203.98 | 4.74 | −0.201 | TRIDC7BG042390.2 | |
| TdLSM7-7A | 140 | 15,455.53 | 8.84 | −0.652 | TRIDC7AG051520.1 | |
| TdLSM7-2B | 85 | 9209.38 | 4.62 | −0.184 | TRIDC2BG080800.1 | |
| TdLSM7-2A | 72 | 7675.66 | 4.27 | −0.101 | TRIDC2AG074130.1 | |
| TaLSM7-U | 147 | 15,822.08 | 5.61 | −0.196 | TraesCSU02G104500.2 | |
| TaLSM7-7B | 104 | undefined | undefined | −0.286 | TraesCS7B02G259600.1 | |
| TaLSM7-7A | 104 | 11,447.01 | 4.97 | −0.418 | TraesCS7A02G368900.1 | |
| TaLSM7-2D | 100 | 10,675.18 | 4.87 | −0.183 | TraesCS2D02G531700.1 | |
| TaLSM7-2B | 100 | 10,675.18 | 4.87 | −0.183 | TraesCS2B02G559200.1 | |
| TaLSM7-2A | 100 | 10,675.18 | 4.87 | −0.183 | TraesCS2A02G528800.1 | |
| TaLSM7-1D | 97 | 10,593.4 | 8.31 | 0.218 | TraesCS1D02G062600.1 | |
| AetLSM7-7 | 132 | 14,456.32 | 7.16 | −0.572 | AET7Gv20882600.3 | |
| AetLSM7-2 | 100 | 10,675.18 | 4.87 | −0.183 | AET2Gv21167900.2 | |
| LSM8 | TuLSM8-1 | 99 | 10,756.24 | 4.55 | 0.044 | TuG1812G0100004839.01.P01 |
| TdLSM8-1B | 98 | 10,642.14 | 4.55 | 0.077 | TRIDC1BG074340.1 | |
| TdLSM8-1A | 97 | 10,770.39 | 5.63 | −0.115 | TRIDC1AG065090.2 | |
| TaLSM8-1D | 99 | 10,756.24 | 4.55 | 0.044 | TraesCS1D02G451800.1 | |
| TaLSM8-1B | 99 | 10,756.24 | 4.55 | 0.044 | TraesCS1B02G478200.1 | |
| TaLSM8-1A | 99 | 10,770.27 | 4.55 | 0.047 | TraesCS1A02G443700.1 | |
| AetLSM8-1 | 99 | 11,084.79 | 4.66 | 0.205 | AET1Gv21045900.3 | |
| SMB | TuSMB-2 | 265 | 27,936.58 | 11.3 | −0.689 | TuG1812G0200002708.01.P02 |
| TdSMB-2B | 265 | 27,965.58 | 11.3 | −0.708 | TRIDC2BG038770.1 | |
| TdSMB-2A | 265 | 27,936.58 | 11.3 | −0.689 | TRIDC2AG034250.1 | |
| TaSMB-2D | 265 | 28,038.72 | 11.3 | −0.665 | TraesCS2D02G248300.1.cds1 | |
| TaSMB-2B | 265 | 27,965.58 | 11.3 | −0.708 | TraesCS2B02G270200.1.cds1 | |
| TaSMB-2A | 265 | 27,936.58 | 11.3 | −0.689 | TraesCS2A02G243500.1.cds1 | |
| AetSMB-2 | 265 | 28,038.72 | 11.3 | −0.665 | AET2Gv20552800.1 | |
| SMD1 | TuSMD1-6 | 114 | 12,727.99 | 11.23 | −0.538 | TuG1812G0600002404.01.P01 |
| TuSMD1-2 | 172 | 18,940.35 | 8.55 | 0.271 | TuG1812G0200003790.01.P03 | |
| TdSMD1-6B | 146 | 16,050.8 | 11.05 | −0.471 | TRIDC6BG036850.1 | |
| TdSMD1-6A | 184 | 20,193.36 | 11.29 | −0.507 | TRIDC6AG032030.1 | |
| TdSMD1-2B | 136 | 15,176.65 | 10.72 | −0.59 | TRIDC2BG050890.3 | |
| TdSMD1-2A | 175 | 19,362.8 | 9.8 | 0.126 | TRIDC2AG048420.3 | |
| TaSMD1-6D | 107 | 11,809.78 | 11.02 | −0.645 | TraesCS6D02G188600.1 | |
| TaSMD1-6B | 107 | 11,809.78 | 11.02 | −0.645 | TraesCS6B02G230500.1 | |
| TaSMD1-6A | 114 | 12,727.99 | 11.23 | −0.538 | TraesCS6A02G207000.1 | |
| TaSMD1-2D | 114 | 12,722.1 | 11.23 | −0.642 | TraesCS2D02G331500.1 | |
| TaSMD1-2B | 107 | 11,821.72 | 11.02 | −0.721 | TraesCS2B02G350800.1 | |
| TaSMD1-2A | 114 | 12,703.42 | 11.21 | −0.641 | TraesCS2A02G331400.1 | |
| AetSMD1-2 | 140 | 15,650.33 | 10.74 | −0.487 | AET2Gv20748800.2 | |
| SMD2 | TuSMD2-5 | 119 | 13,325.73 | 9.81 | −0.354 | TuG1812G0500005377.01.P02 |
| TuSMD2-3 | 106 | 12,085.23 | 9.95 | −0.477 | TuG1812G0300005644.01.P01 | |
| TdSMD2-5A | 107 | 12,224.42 | 9.95 | −0.457 | TRIDC5AG075140.1 | |
| TdSMD2-4B | 89 | 10,219.24 | 10.82 | −0.343 | TRIDC4BG060470.1 | |
| TdSMD2-3B | 107 | 12,156.31 | 9.95 | −0.456 | TRIDC3BG084500.1 | |
| TdSMD2-3A | 129 | 14,372.68 | 10.24 | −0.729 | TRIDC3AG073200.5 | |
| TaSMD2-5A | 107 | 12,224.42 | 9.95 | −0.457 | TraesCS5A02G529800.2 | |
| TaSMD2-4D | 107 | 12,224.42 | 9.95 | −0.457 | TraesCS4D02G354800.2 | |
| TaSMD2-4B | 107 | 12,224.42 | 9.95 | −0.457 | TraesCS4B02G361800.1 | |
| TaSMD2-3D | 105 | 11,985.11 | 9.88 | −0.422 | TraesCS3D02G524800.1 | |
| TaSMD2-3B | 115 | 13,068.47 | 10.07 | −0.365 | TraesCS3B02G584900.1 | |
| TaSMD2-3A | 106 | 12,085.23 | 9.95 | −0.477 | TraesCS3A02G517300.1 | |
| AetSMD2-4 | 107 | 12,224.42 | 9.95 | −0.457 | AET4Gv20832900.1 | |
| AetSMD2-3 | 106 | 12,113.28 | 9.95 | −0.455 | AET3Gv21214400.2 | |
| SMD3 | TuSMD3-5 | 135 | 14,505.94 | 11.06 | −0.23 | TuG1812G0500005669.01.P01 |
| TdSMD3-7A | 139 | 15,484.09 | 9.96 | −0.266 | TRIDC7AG005000.1 | |
| TdSMD3-5A | 158 | 17,171.04 | 11.5 | −0.385 | TRIDC5AG078210.2 | |
| TdSMD3-4B | 196 | 22,029.53 | 10.43 | −0.37 | TRIDC4BG064290.3 | |
| TdSMD3-4A | 150 | Undefined | undefined | −0.135 | TRIDC4AG065300.1 | |
| TaSMD3-U | 135 | 14,505.94 | 11.06 | −0.23 | TraesCSU02G033900.1 | |
| TaSMD3-5A | 135 | 14,505.94 | 11.06 | −0.23 | TraesCS5A02G554700.1 | |
| TaSMD3-4B | 135 | 14,505.94 | 11.06 | −0.23 | TraesCS4B02G392700.1 | |
| AetSMD3-4 | 166 | 18,806.14 | 8.73 | −0.654 | AET4Gv20876200.2 | |
| SME | TuSME-7 | 146 | 16,460.43 | 10.74 | −0.116 | TuG1812G0700003308.01.P01 |
| TdSME-7B | 88 | 10,359.28 | 9.89 | −0.265 | TRIDC7BG033630.1 | |
| TdSME-7A | 143 | 16,103.99 | 10.75 | −0.093 | TRIDC7AG042500.1 | |
| TdSME-6B | 63 | 7409.69 | 8.3 | −0.335 | TRIDC6BG015620.1 | |
| TdSME-6A | 95 | 11,020.96 | 9.58 | −0.052 | TRIDC6AG011490.2 | |
| TaSME-U | 86 | 10,185.06 | 9.52 | −0.141 | TraesCSU02G017200.1 | |
| TaSME-7D | 88 | 10,359.28 | 9.89 | −0.265 | TraesCS7D02G299900.1 | |
| TaSME-7B | 88 | 10,359.28 | 9.89 | −0.265 | TraesCS7B02G204700.1 | |
| TaSME-7A | 88 | 10,359.28 | 9.89 | −0.265 | TraesCS7A02G304400.1 | |
| TaSME-6B | 97 | 11,483.56 | 5.37 | −0.069 | TraesCS6B02G111500.1 | |
| TaSME-6A | 117 | 13,453.83 | 10.26 | −0.306 | TraesCS6A02G089100.2 | |
| AetSME-7 | 103 | 12,280.32 | 9.2 | −0.403 | AET7Gv20759600.3 | |
| AetSME-6 | 143 | 16,406.26 | 11.53 | −0.49 | AET6Gv20214400.2 | |
| SMF | TuSMF-7 | 143 | 15,245.44 | 7.23 | −0.272 | TuG1812G0700005332.01.P01 |
| TdSMF-7B | 97 | 10,571.1 | 4.69 | −0.205 | TRIDC7BG063710.1 | |
| TdSMF-7A | 94 | 10,270.78 | 4.5 | −0.151 | TRIDC7AG069290.2 | |
| TdSMF-6B | 86 | 9664.09 | 4.42 | −0.127 | TRIDC6BG026950.1 | |
| TdSMF-6A | 66 | 7491.65 | 8.24 | −0.191 | TRIDC6AG021120.1 | |
| TaSMF-7B | 87 | 9673.16 | 4.5 | −0.116 | TraesCS7B02G401200.1 | |
| TaSMF-7A | 87 | 9673.16 | 4.5 | −0.116 | TraesCS7A02G496000.1 | |
| TaSMF-6D | 113 | 12,580.39 | 4.43 | −0.068 | TraesCS6D02G141900.1 | |
| TaSMF-6B | 113 | 12,612.47 | 4.56 | −0.05 | TraesCS6B02G180100.1 | |
| AetSMF-7 | 87 | 9768.23 | 4.69 | −0.167 | AET7Gv21208500.4 | |
| AetSMF-6 | 86 | 9632.97 | 4.42 | −0.217 | AET6Gv20390000.6 | |
| SMG | TuSMG-2 | 80 | 8933.39 | 8.06 | −0.226 | TuG1812G0200001854.01.P01 |
| TdSMG-2B | 83 | 9151.47 | 4.37 | 0.2 | TRIDC2BG026270.2 | |
| TdSMG-2A | 80 | 8933.39 | 8.06 | −0.226 | TRIDC2AG021760.1 | |
| TaSMG-2B | 81 | 9121.44 | 6.55 | −0.249 | TraesCS2B02G198200.1 | |
| TaSMG-2A | 79 | 8818.08 | 5.67 | −0.234 | TraesCS2A02G171800.1 | |
| AetSMG-2 | 80 | 8933.39 | 8.06 | −0.226 | AET2Gv20357300.1 | |
| Gene Name | Wheat Minimum Similarity (%) | Wheat Maximum Similarity (%) |
|---|---|---|
| LSM2 | 98.582 | 99.291 |
| LSM3 | 83.274 | 99.66 |
| LSM4 | 96.065 | 97.454 |
| LSM5 | 97.407 | 99.259 |
| LSM6 | 94.667 | 97.333 |
| LSM7 | 77.5 | 99.01 |
| LSM8 | 96.333 | 99.333 |
| SMB | 97.619 | 98.246 |
| SMD1 | 87.126 | 99.383 |
| SMD2 | 90.034 | 99.074 |
| SMD3 | 97.304 | 97.794 |
| SME | 85.992 | 100 |
| SMF | 97.661 | 97.727 |
| SMG | 95.935 | 95.935 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, R.; Su, N.; Pan, W.; Bao, Q.; Li, Z.; Nie, X.; Tong, W.; Song, W. Genome-Wide Investigation of Spliceosomal SM/LSM Genes in Wheat (Triticum aestivum L.) and Its Progenitors. Agronomy 2021, 11, 1429. https://doi.org/10.3390/agronomy11071429
Gao R, Su N, Pan W, Bao Q, Li Z, Nie X, Tong W, Song W. Genome-Wide Investigation of Spliceosomal SM/LSM Genes in Wheat (Triticum aestivum L.) and Its Progenitors. Agronomy. 2021; 11(7):1429. https://doi.org/10.3390/agronomy11071429
Chicago/Turabian StyleGao, Ruiting, Ning Su, Wenqiu Pan, Qiaoyu Bao, Zhen Li, Xiaojun Nie, Wei Tong, and Weining Song. 2021. "Genome-Wide Investigation of Spliceosomal SM/LSM Genes in Wheat (Triticum aestivum L.) and Its Progenitors" Agronomy 11, no. 7: 1429. https://doi.org/10.3390/agronomy11071429
APA StyleGao, R., Su, N., Pan, W., Bao, Q., Li, Z., Nie, X., Tong, W., & Song, W. (2021). Genome-Wide Investigation of Spliceosomal SM/LSM Genes in Wheat (Triticum aestivum L.) and Its Progenitors. Agronomy, 11(7), 1429. https://doi.org/10.3390/agronomy11071429







