Multi-Environmental Evaluation of Protein Content and Yield Stability among Tropical Soybean Genotypes Using GGE Biplot Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Description of the Locations Used in the Study
2.3. Experimental Design, Data Collection, and Analysis
3. Results
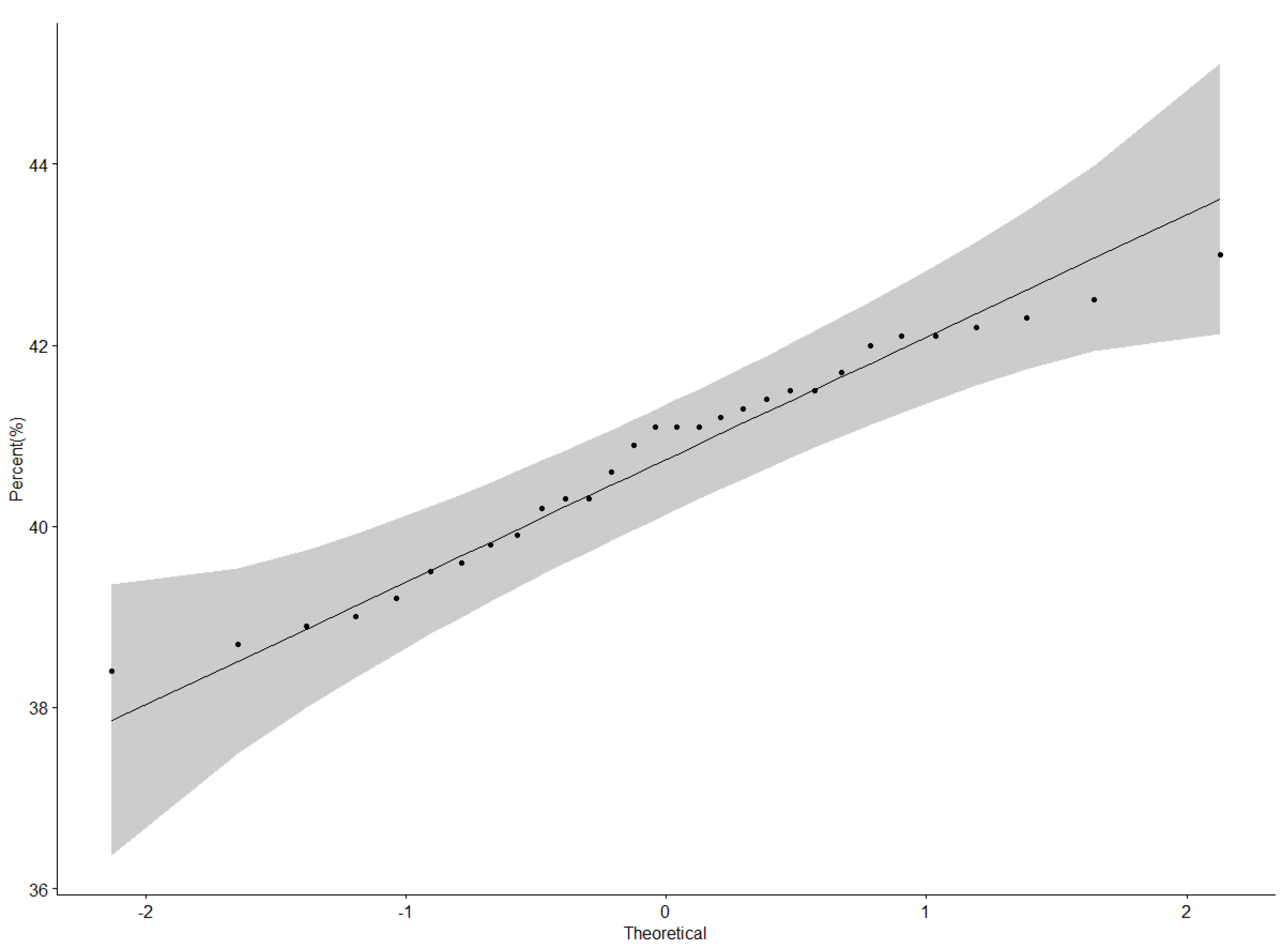
3.1. Protein Content (%) Performance of Genotypes across Locations
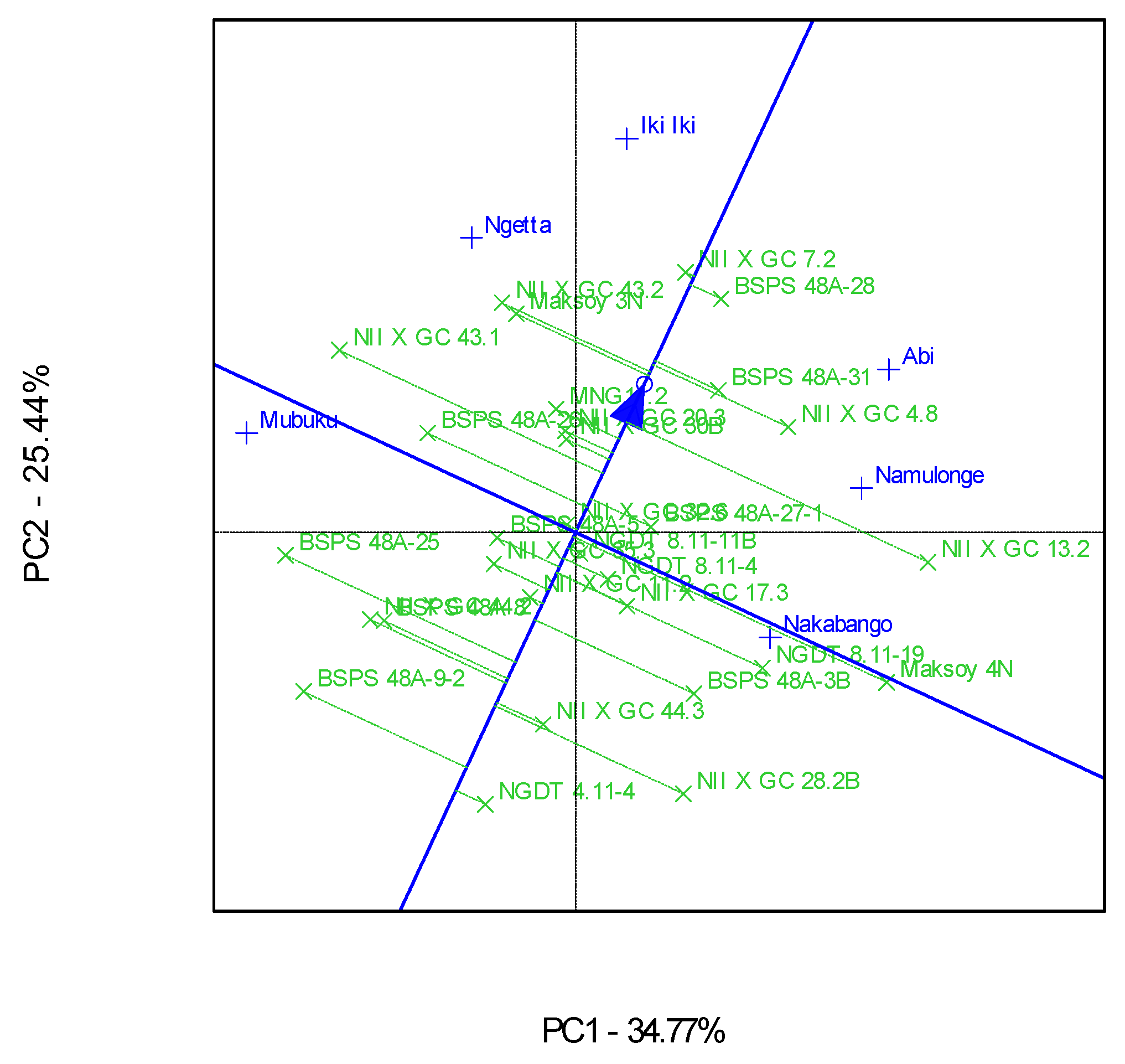
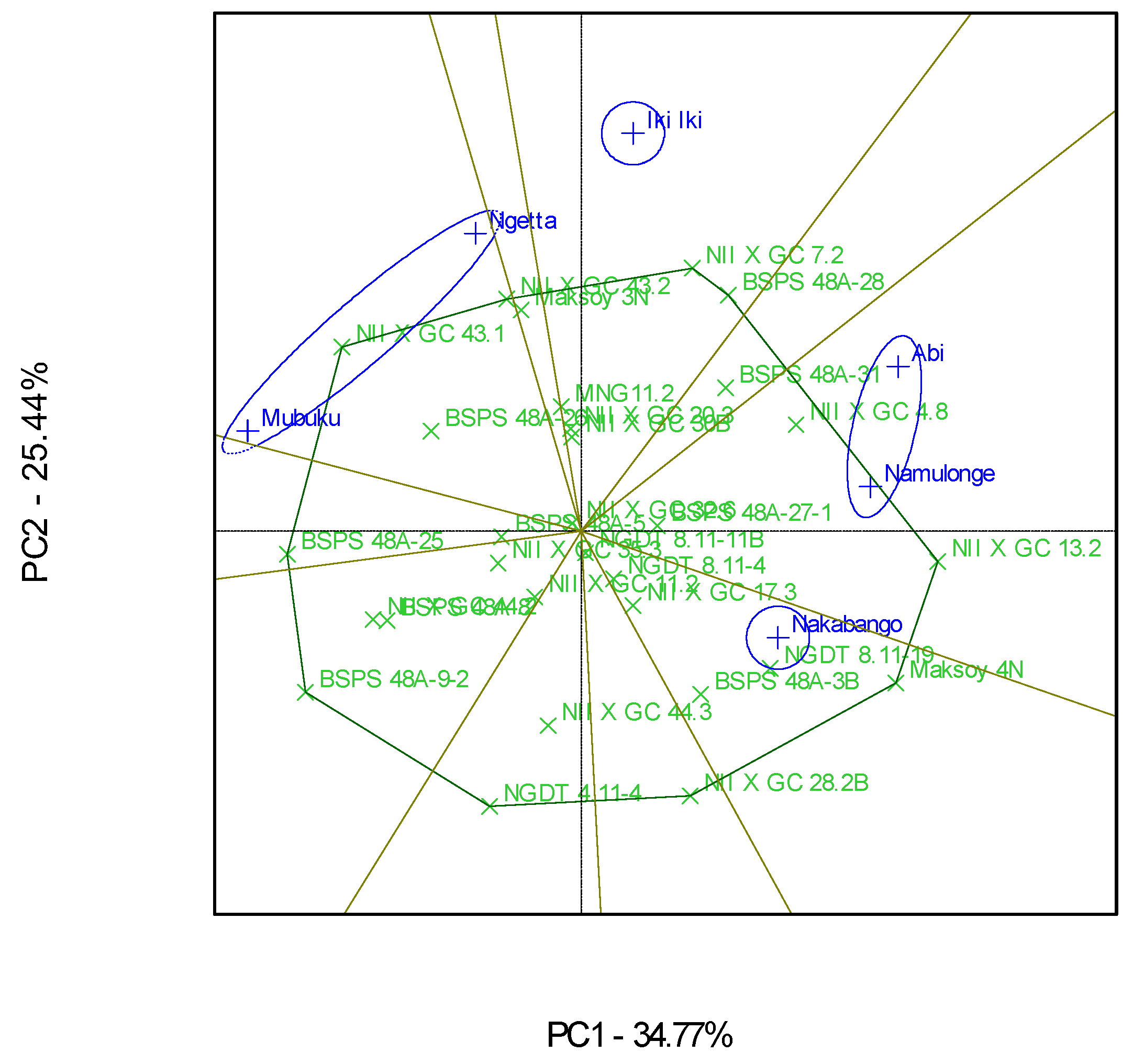
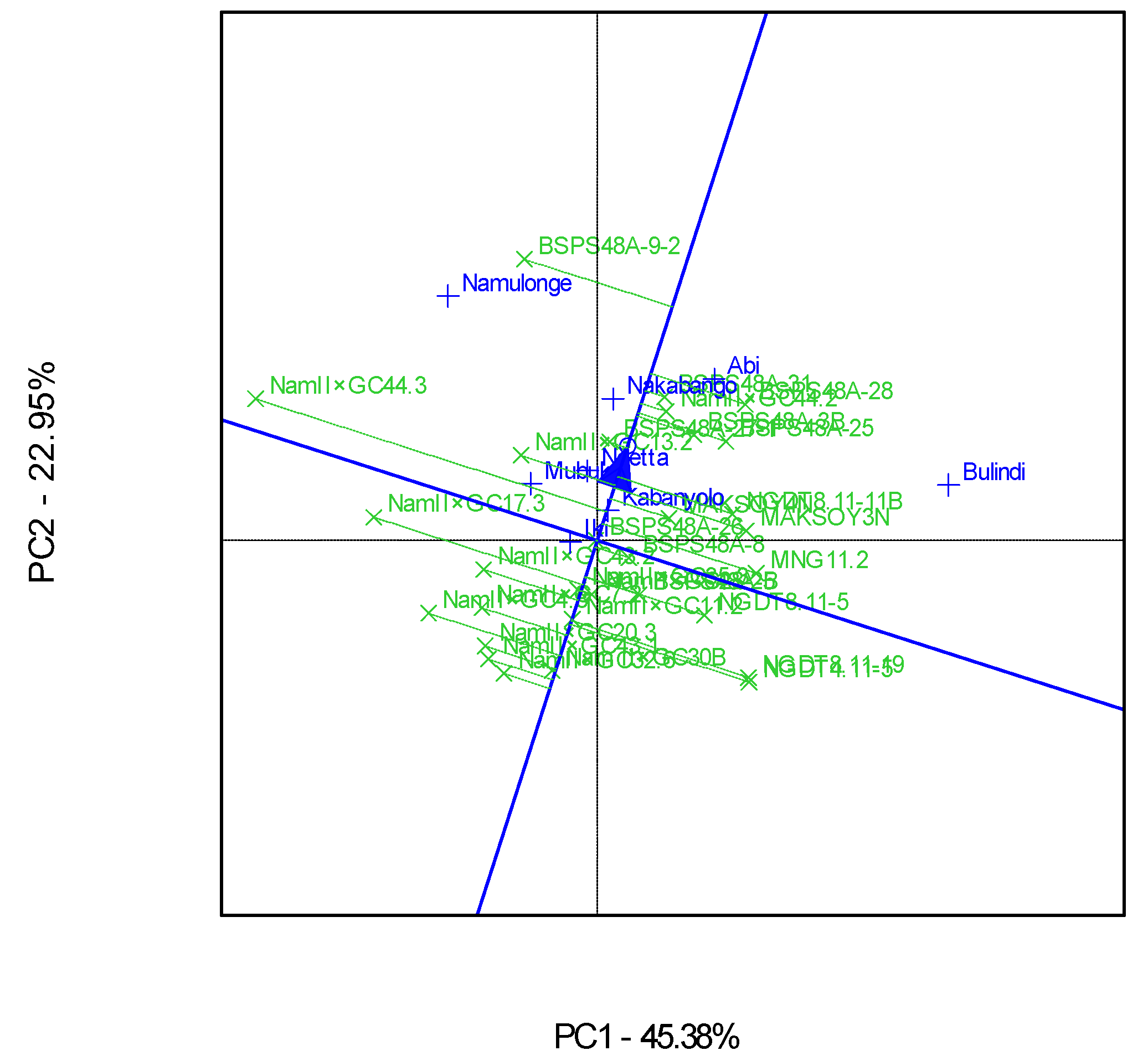
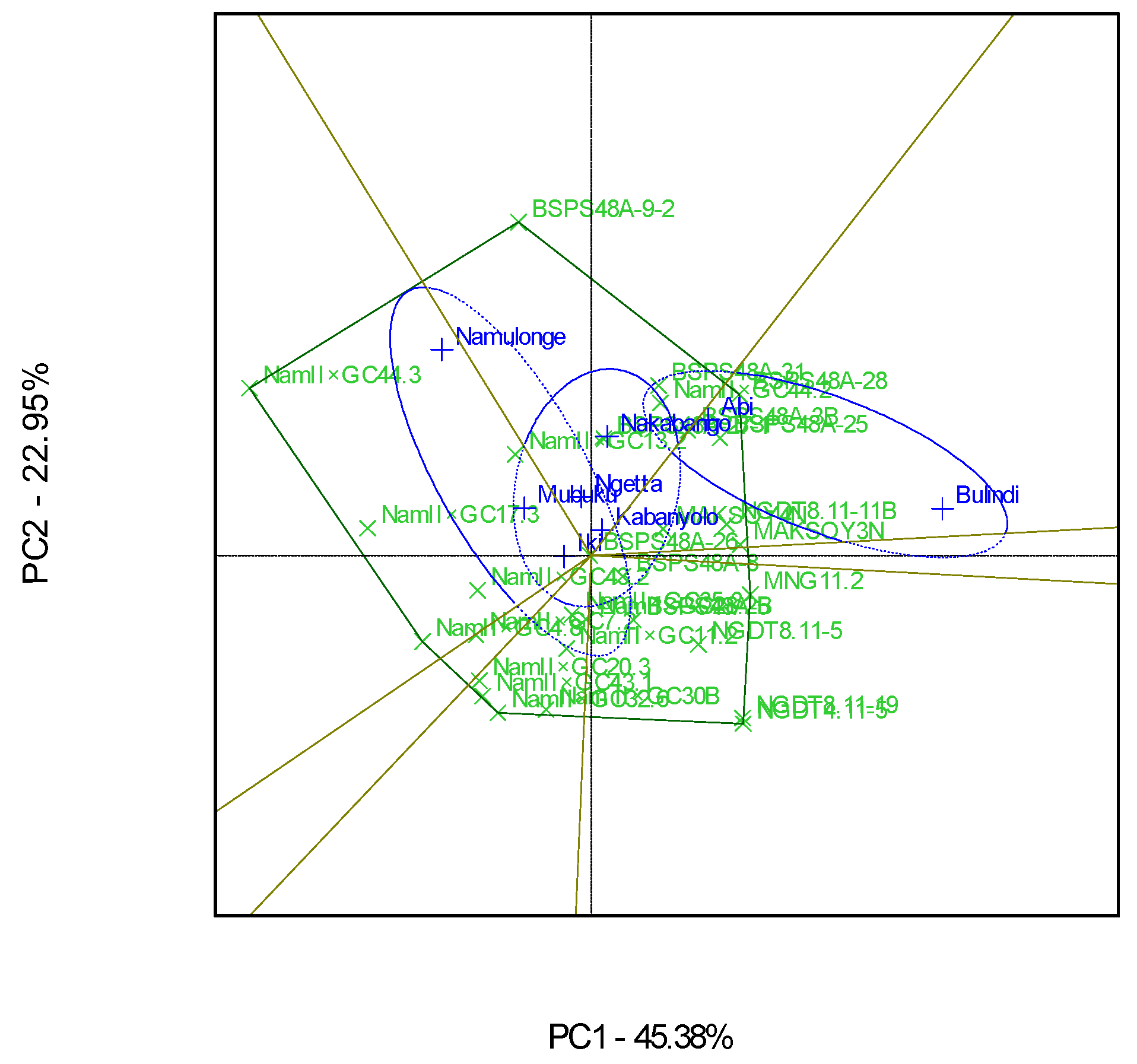
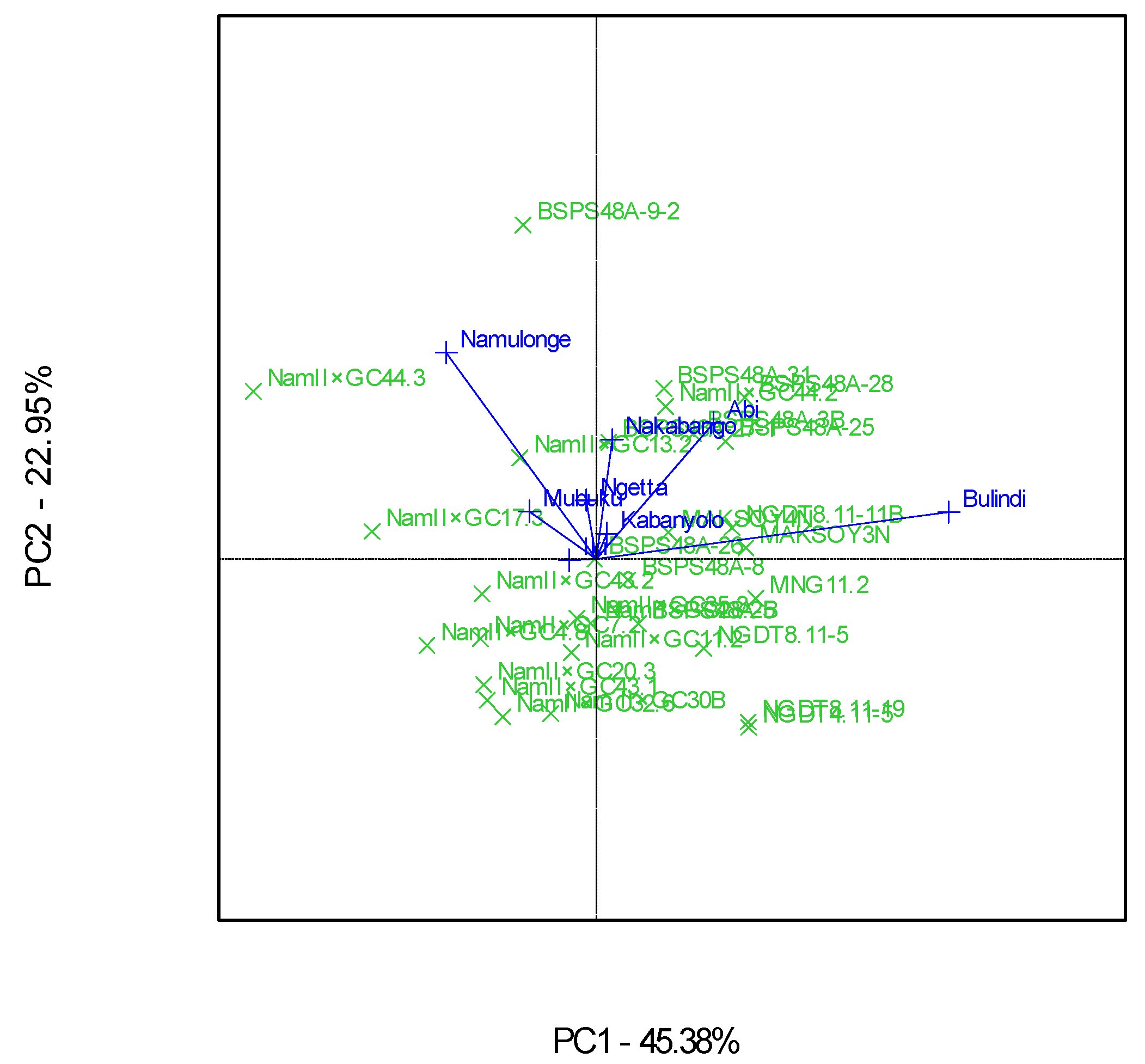
3.2. Which-Won-Where Patterns and Stability of Genotypes for Protein Content
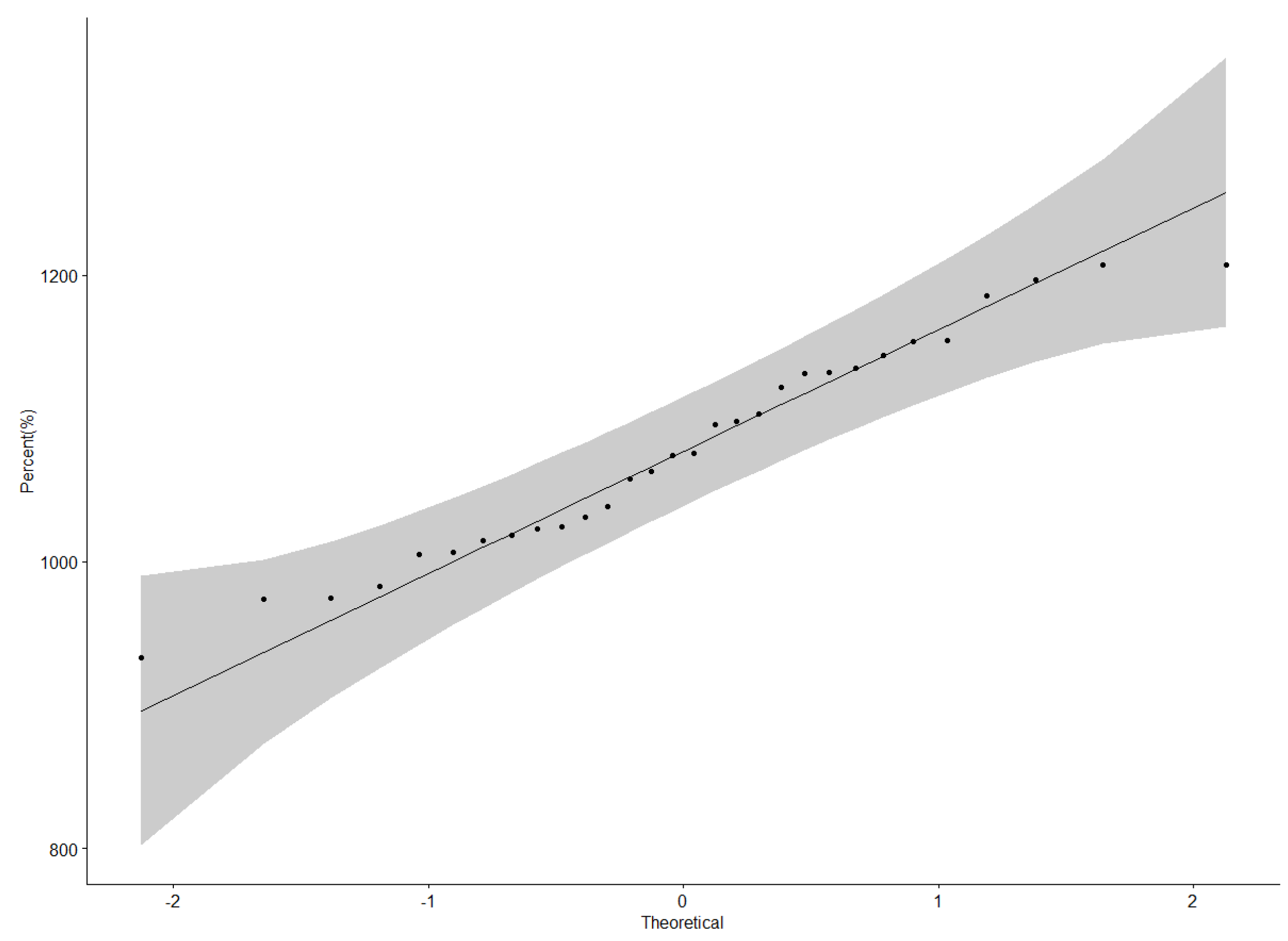
3.3. Grain Yield Performance of Genotypes across Locations
3.4. “Which-Won-Where” Patterns and Stability of Genotypes for Grain Yield
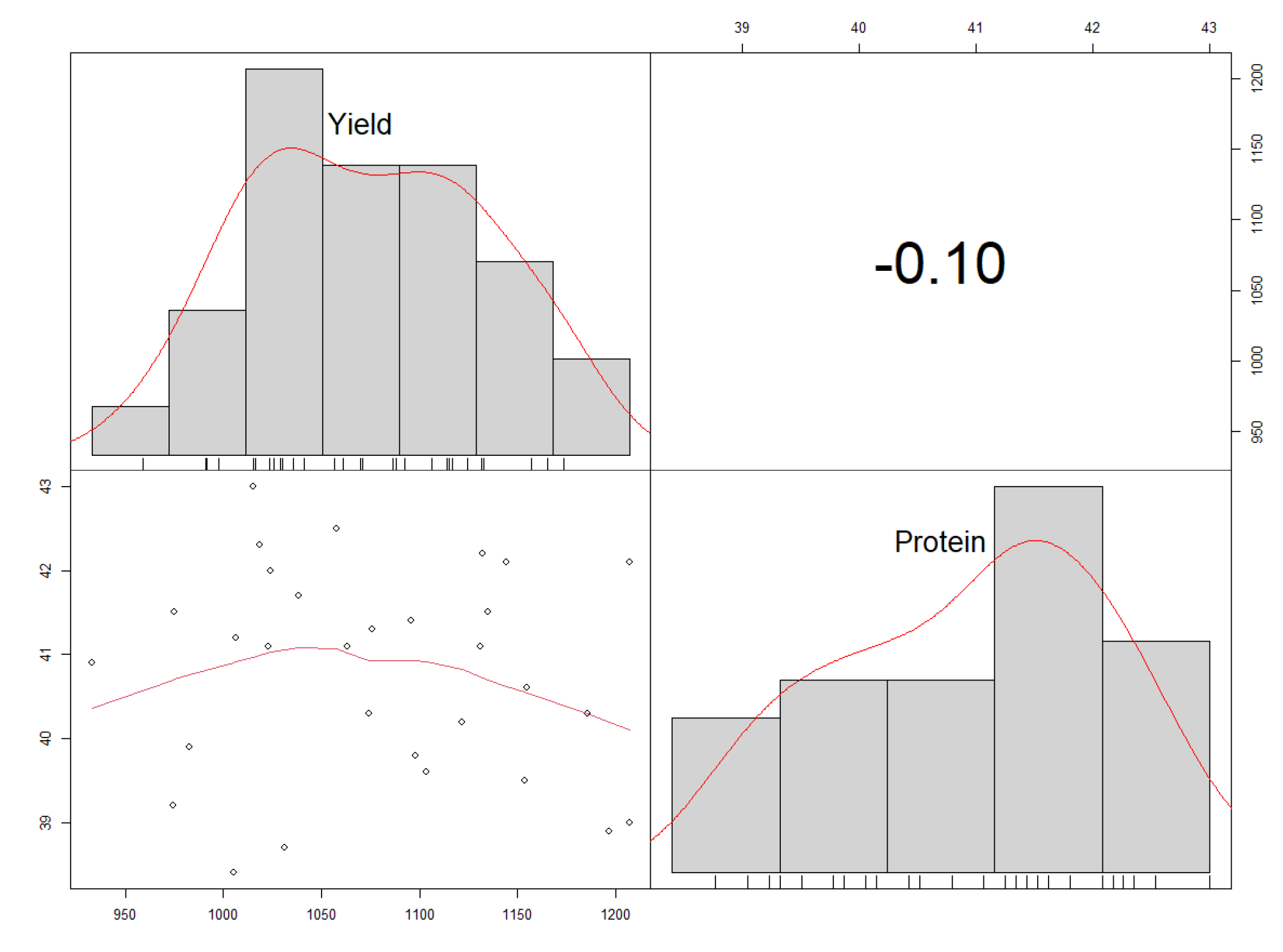
3.5. Correlation Analysis for Protein Content (%) and Yield (kg ha−1)
4. Discussion
4.1. Protein Performance and Stability
4.2. Clustering Test Environments Concerning Protein Content (%)
4.3. Yield Performance and Stability
4.4. Clustering Test Environments Concerning Yield
4.5. Correlation between Protein Content (%) and Yield (kg ha−1)
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tukamuhabwa, P.; Obua, T.; Namara, M.; Okii, D.; Kabayi, P.; Yiga, G. Soybean Research and Development in Uganda: Highlights 2002–2018; Makerere University: Kampala, Uganda, 2019. [Google Scholar]
- Agralytica, United Soybean Board Connections 2012: Soybean Market Scan. 2012. Available online: https://unitedsoybean.org/wp-content/uploads/Soybean-Market-scan-report-final.pdf (accessed on 21 June 2021).
- FAO. 2018. Available online: http://www.fao.org/faostat (accessed on 21 June 2021).
- WWF. The Growth of Soy Impacts and Solutions. 2014. Available online: http://issuu.com/wwfsoyreport/docs/wwf_soy_report_final_jan_19/1?e=10667775/6569194 (accessed on 21 June 2021).
- Singh, P.; Kumar, R.; Sabapathy, S.N.; Bawa, A.S. Functional and Edible Uses of Soy Protein Products. Compr. Rev. Food Sci. Food Saf. 2008, 7, 14–28. [Google Scholar] [CrossRef]
- Graham, P.H.; Vance, C.P. Update on Legume Utilization Legumes: Importance and Constraints to Greater Use. Plant Physiol. 2003, 131, 872–877. [Google Scholar] [CrossRef] [Green Version]
- Sanginga, P.C.; Adesina, A.A.; Manyong, V.M.; Otite, O.; Dashiell, K.E. Social Impact of Soybean in Nigeria’s Southern Guinea Savanna; IITA: Ibadan, Nigeria, 1999; pp. 1–34. [Google Scholar]
- Tukamuhabwa, P.; Oloka, H.K. Soybean Research and Development in Uganda. A Case of Paradigm Shift in an African University; Makerere University: Kampala, Uganda, 2016. [Google Scholar]
- Obua, T.; Nabasirye, M.; Namara, M.; Tusiime, G.; Maphosa, M. Yield stability of tropical soybean genotypes in selected agro-ecologies in Uganda Yield stability of tropical soybean genotypes in selected agro-ecologies in Uganda. S. Afr. J. Plant Soil 2020. [Google Scholar] [CrossRef]
- Mukuze, C. Genetic Diversity and Genotype × Environment Interaction of Advanced Elite Soybean Genotypes in Uganda. Master’s Thesis, Makerere University, Kampala, Uganda, 2019. [Google Scholar]
- Tukamuhabwa, P.; Asiimwe, M.; Nabasirye, M.; Kabayi, P.; Maphosa, M. Genotype by Environment Interaction of Advanced Generation. Afr. Crop Sci. J. 2012, 20, 107–115. [Google Scholar]
- Tukamuhabwa, P.; Oloka, H.K.; Sengooba, T.; Kabayi, P. Yield stability of rust-resistant soybean lines at four mid-altitude tropical locations. Euphytica 2012, 183, 1–10. [Google Scholar] [CrossRef]
- Kumar, V.; Rani, A.; Solanki, S.; Hussain, S.M. Influence of growing environment on the biochemical composition and physical characteristics of soybean seed Influence of growing environment on the biochemical composition and physical characteristics of soybean seed. J. Food Compos. Anal. 2006. [Google Scholar] [CrossRef]
- Arslanoglu, F.; Aytac, S.; Oner, E.K. Effect of genotype and environment interaction on oil and protein content of soybean (Glycine max (L.) Merrill) seed. Afr. J. Biotechnol. 2011, 10, 18409–18417. [Google Scholar] [CrossRef]
- Ojo, D.K.; Adebisi, M.A.; Tijani, B.O. Influence of environment on protein and oil contents of soybeans seed (Glycine max (L.) Merril). Glob. J. Agric. Sci. 2002, 1, 27–32. [Google Scholar] [CrossRef]
- Piper, E.L.; Boote, K.I. Temperature and cultivar effects on soybean seed oil and protein concentrations. J. Am. Oil Chem. Soc. 1999, 76, 1233–1241. [Google Scholar] [CrossRef]
- Mulugeta, A.; Kidane, S.; Abadi, S.; Fisha, Z. GGE biplots to analyze soybean multi-environment yield trial data in north Western Ethiopia. J. Plant Breed. Crop Sci. 2013, 5, 245–254. [Google Scholar] [CrossRef] [Green Version]
- Adie, M.M.; Krisnawati, A.; Gatut-Wahyu, A.S. Assessment of Genotype × Environment Interactions for Black Soybean Yield using Ammi and GGE Biplot. Int. J. Agric. Innov. Res. 2014, 2, 673–678. [Google Scholar]
- Cheelo, P.; Lungu, D.; Mwala, M. GGE Biplot Analysis for Identification of Ideal Soybean [Glycine max L. Merrill] Test and Production Locations in Zambia. J. Exp. Agric. Int. 2017, 15, 1–15. [Google Scholar] [CrossRef]
- Anthony, P.; Malzer, G.; Sparrow, S.; Zhang, M. Soybean Yield and Quality in Relation to Soil Properties. Agron. J. 2012. [Google Scholar] [CrossRef]
- Yin, X.; Vyn, T.J. Relationships of Isoflavone, Oil, and Protein in Seed with Yield of Soybean. Agron. J. 2005, 97, 1314–1321. [Google Scholar] [CrossRef] [Green Version]
- Assefa, Y.; Purcell, L.C.; Salmeron, M.; Naeve, S.; Casteel, S.N.; Kovács, P.; Archontoulis, S.; Licht, M.; Below, F.; Kandel, H.; et al. Assessing Variation in US Soybean Seed Composition (Protein and Oil). Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Filho, O.M.; Sediyama, C.S.; Piovesan, N. Grain yield and seed quality of soybean selected for high protein content Grain yield and seed quality of soybean selected for high protein content. Pesqui. Agropecuária Bras. 2004, 39, 445–450. [Google Scholar] [CrossRef]
- Helms, T.C.; Orf, J.H. Protein, oil, and yield of soybean lines selected for increased protein. Crop Sci. 1998, 38, 707–711. [Google Scholar] [CrossRef]
- Wilcox, J.R.; Shibles, R.M. Interrelationships among seed quality attributes in soybean. Crop Sci. 2001, 41, 11–14. [Google Scholar] [CrossRef]
- Rotundo, L.; Westgate, M.E. Meta-analysis of environmental effects on soybean seed composition Field Crops Research Meta-analysis of environmental effects on soybean seed composition. Field Crops Res. 2009. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Yan, W. GGE Biplot—A Windows application for graphical analysis of multi-environment trial data and other types of two-way data. Agron. J. 2001, 93, 1111–1118. [Google Scholar] [CrossRef] [Green Version]
- Yan, W.; Rajcan, I. Biplot analysis of test sites and trait relations of soybean in Ontario. Crop Sci. 2002, 42, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Simko, V.; Wei, T. R Package ‘Corrplot’: Visualization of a Correlation Matrix. (Version 0.88). 2021. Available online: https://github.com/taiyun/corrplot (accessed on 21 June 2021).
- Yan, W.; Kang, M.S.; Ma, B.; Woods, S.; Cornelius, P.L. GGE biplot vs. AMMI analysis of genotype-by-environment data. Crop Sci. 2007, 47, 643–655. [Google Scholar] [CrossRef]
- Tukamuhabwa, P.; Obua, T. Soybean Production Guide in Uganda; Makerere University Agricultural Research Institute: Kabanyolo, Uganda, 2015. [Google Scholar]
- Gurmu, F.; Mohammed, H.; Alemaw, G. Genotype X environment interactions and stability of soybean for grain yield and nutrition quality. Afr. Crop Sci. J. 2010, 17. [Google Scholar] [CrossRef]
- Hampango, M. Stability Analysis for Oil and Protein Content in Soybean (Glycine max. L. merrill) Seed across Varying Environments. Master’s Thesis, University of Zambia, Lusaka, Zambia, 2016. [Google Scholar]
- Becker, H.C.; Leon, J. Stability Analysis in Plant Breeding. Plant Breed. 1988, 23, 1–23. [Google Scholar] [CrossRef]
- Lin, C.S.; Binns, M.R.; Lefkovitch, L.P. Stability Analysis: Where Do We Stand? Crop Sci. 1986, 26, 894–900. [Google Scholar] [CrossRef] [Green Version]
- Obua, T. Soybean Rust Diversity and Adaptation of Elite Soybean Lines to the Ugandan Environment. Master’s Thesis, Makerere University, Kampala, Uganda, 2013. [Google Scholar]
- Yan, W.; Kang, M.S. YGGE Biplot Analysis: A Graphical Tool for Breeders, Geneticists, and Agronomists; CRC Press: Boca Raton, FL, USA, 2003. [Google Scholar]
- Cober, E.R.; Voldeng, H.D. Developing high-protein, high-yield soybean populations and lines. Crop Sci. 2000, 40, 39–42. [Google Scholar] [CrossRef]

| Genotype | Pedigree | Comment |
|---|---|---|
| BSPS 48A-28 | GC0038-29 × Duiker | Advanced line |
| BSPS 48A-9-2 | GC0038-29 × Duiker | Advanced line |
| Nam 2 × GC 44.2 | Nam 2 × GC0038-29 | Advanced line |
| BSPS 48A-25 | GC0038-29 × Duiker | Advanced line |
| BSPS 48A-27-1 | GC0038-29 × Duiker | Advanced line |
| BSPS 48A-3B | GC0038-29 × Duiker | Advanced line |
| Nam 2 × GC 13.2 | Nam 2 × GC0038-29 | Advanced line |
| MAKSOY 4N | GC0038-29 × Duiker | Check Variety |
| BSPS 48A-31 | GC0038-29 × Duiker | Advanced line |
| MAKSOY 3N | GC0038-29 × Duiker | Check Variety |
| NGDT 8.11-11B | Nam 2 × GC0038-29 | Advanced line |
| BSPS 48A-8 | GC0038-29 × Duiker | Advanced line |
| BSPS 48A-26 | GC0038-29 × Duiker | Advanced line |
| MNG 11.2 | Nam 2 × GC0038-29 | Advanced line |
| Nam 2 × GC 35.3 | Nam 2 × GC0038-29 | Advanced line |
| Nam 2 × GC 17.3 | Nam 2 × GC0038-29 | Advanced line |
| Nam 2 × GC 44.3 | Nam 2 × GC0038-29 | Advanced line |
| Nam 2 × GC 43.2 | Nam 2 × GC0038-29 | Advanced line |
| BSPS 48A-5 | GC0038-29 × Duiker | Advanced line |
| Nam 2 × GC 28.2B | Nam 2 × GC0038-29 | Advanced line |
| Nam 2 × GC 11.2 | Nam 2 × GC0038-29 | Advanced line |
| NGDT 8.11-4 | Nam 2 × GC0038-29 | Advanced line |
| Nam 2 × GC 7.2 | Nam 2 × GC0038-29 | Advanced line |
| Nam 2 × GC 20.3 | Nam 2 × GC0038-29 | Advanced line |
| Nam 2 × GC 4.8 | Nam 2 × GC0038-29 | Advanced line |
| NGDT 8.11-19 | Nam 2 × GC0038-29 | Advanced line |
| NGDT 4.11-5 | Nam 2 × GC0038-29 | Advanced line |
| Nam 2 × GC 30B | Nam 2 × GC0038-29 | Advanced line |
| Nam 2 × GC 32.6 | Nam 2 × GC0038-29 | Advanced line |
| Nam 2 × GC 43.1 | Nam 2 × GC0038-29 | Advanced line |
| Location | Coordinates | Region | Altitude (masl) | Mean Annual Temperature (°C) | Mean Annual Rainfall (mm) |
|---|---|---|---|---|---|
| Namulonge | 0°32′ N/32°37′ E | Central | 1160 | 22.6 | 1400 |
| Nakabango | 0°29′ N/33°14′ E | Eastern | 1210 | 22.8 | 1400 |
| Iki-Iki | 1°06′ N/34°00′ E | Eastern | 1156 | 24.7 | 1200 |
| Ngetta | 2°17′ N/32°56′ E | Northern | 1103 | 24.7 | 1200 |
| Mubuku | 0°13′ N/30°08′ E | Western | 1007 | 27.8 | 750 |
| Kabanyolo | 0°28′ N/32°36′ E | Central | 1180 | 21.4 | 1234 |
| Bulindi | 1°41′ N/31°42′ E | Mid-West | 1122 | 22.9 | 1355 |
| Abi | 3°04′ N/30°56′ E | West Nile | 1214 | 22.9 | 1404 |
| SOV | d.f. | s.s. | m.s. | v.r. | F pr. |
|---|---|---|---|---|---|
| Genotype | 29 | 1557.65 | 53.71 | 3.50 | <0.001 |
| Location | 5 | 298.79 | 74.70 | 4.87 | <0.001 |
| Genotype × Location | 94 | 3256.39 | 34.64 | 2.26 | <0.001 |
| Reps | 2 | 7.81 | 3.91 | 0.25 | 0.775 |
| Residual | 413 | 6335.51 | 15.34 | ||
| Total | 542 | 11,456.15 | 21.14 |
| Genotype | Abi | Iki-Iki | Mubuku | Nakabango | Namulonge | Ngetta | Mean |
|---|---|---|---|---|---|---|---|
| BSPS 48A-25 | 34.6 | 45.2 | 39.7 | 39.2 | 43.1 | 40.3 | |
| BSPS 48A-26 | 39.5 | 43.3 | 42.6 | 36.4 | 37.1 | 40.1 | 39.8 |
| BSPS 48A-27-1 | 40.3 | 40.9 | 39.3 | - | 41.7 | 40.6 | 40.6 |
| BSPS 48A-28 | - | 45.1 | 40.1 | 42.3 | 41.0 | - | 42.1 |
| BSPS 48A-31 | 43.2 | 42.4 | 39.8 | 44.7 | 41.2 | 42.0 | 42.2 |
| BSPS 48A-3B | 42.0 | 37.5 | 36.5 | 41.7 | 39.8 | 39.6 | 39.5 |
| BSPS 48A-5 | 40.2 | 40.0 | 45.2 | 44.4 | 38.9 | 41.7 | |
| BSPS 48A-8 | 38.3 | 41.0 | 43.8 | 40.9 | 35.1 | 38.6 | 39.6 |
| BSPS 48A-9-2 | - | 37.1 | - | 42.2 | 35.8 | 41.0 | 39.0 |
| Maksoy 3N | - | 45.5 | 43.6 | 38.5 | 37.8 | 40.2 | 41.1 |
| Maksoy 4N | 42.4 | 39.5 | 36.1 | 49.2 | 43.4 | 38.7 | 41.5 |
| MNG11.2 | 41.2 | 42.3 | - | 40.6 | - | 41.5 | 41.4 |
| NGDT 4.11-5 | 36.7 | 38.3 | 43.0 | 45.5 | 38.4 | 37.7 | 39.9 |
| NGDT 8.11-11B | 42.0 | 41.3 | 41.4 | 39.6 | 38.4 | 38.5 | 40.2 |
| NGDT 8.11-19 | 41.1 | 39.4 | 34.9 | - | - | - | 38.4 |
| NGDT 8.11-4 | 41.7 | 41.4 | 41.9 | 44.7 | 38.1 | 38.7 | 41.1 |
| NII X GC 11.2 | 43.9 | 40.5 | 46.5 | 48.6 | 34.2 | 38.5 | 42.0 |
| NII X GC 13.2 | 44.7 | 39.7 | 36.3 | 47.0 | 44.7 | 40.2 | 42.1 |
| NII X GC 17.3 | 39.2 | 39.4 | - | - | 42.1 | 40.4 | 40.3 |
| NII X GC 20.3 | 41.9 | 43.7 | 48.5 | 37.8 | 43.1 | 43.0 | |
| NII X GC 28.2B | 40.8 | 38.8 | 35.9 | 42.2 | 38.0 | 36.6 | 38.7 |
| NII X GC 30B | 43.2 | 42.6 | 42.7 | 43.8 | 36.3 | 40.4 | 41.5 |
| NII X GC 32.6 | 41.3 | 42.3 | - | 35.0 | - | 38.3 | 39.2 |
| NII X GC 35.3 | 40.5 | 40.2 | 46.3 | 42.2 | 39.5 | 39.4 | 41.3 |
| NII X GC 4.8 | - | 43.5 | 37.3 | 43.0 | 40.9 | - | 41.2 |
| NII X GC 43.1 | - | 46.5 | 36.5 | 39.6 | - | 40.9 | |
| NII X GC 43.2 | 40.7 | 43.5 | - | 42.4 | - | 43.2 | 42.5 |
| NII X GC 44.2 | - | 38.9 | 41.2 | 38.3 | 35.3 | 40.7 | 38.9 |
| NII X GC 44.3 | 40.5 | 38.2 | 42.9 | 45.3 | - | 38.4 | 41.1 |
| NII X GC 7.2 | 44.1 | 46.1 | 40.4 | 43.4 | 38.7 | 41.1 | 42.3 |
| Mean | 41.0 | 41.1 | 41.3 | 42.5 | 38.9 | 40.0 | 40.8 |
| LSD | 3.1 | 0.0 | 3.5 | 5.3 | 3.0 | 1.4 | 4.5 |
| CV | 5.6 | 12.3 | 8.2 | 10.9 | 11.9 | 7.2 | 9.2 |
| SOV | d.f. | s.s. | F Value | F pr. |
|---|---|---|---|---|
| Genotype | 29 | 19,731,420 | 4.3729 | 1.07 × 10−13 |
| Location | 7 | 1.38 × 108 | 126.761 | <2.2 × 10−16 |
| Season | 5 | 1.44 × 108 | 184.5312 | <2.2 × 10−16 |
| Rep | 2 | 3,397,014 | 10.9162 | 1.92 × 10−05 |
| Genotype × Location | 203 | 53,562,730 | 1.6958 | 2.00 × 10−08 |
| Genotype × Season | 145 | 21,094,874 | 0.935 | 0.6958 |
| Genotype × Location × Season | 660 | 3.96 × 108 | 3.8605 | <2.2 × 10−16 |
| Residuals | 2094 | 3.26 × 108 |
| Genotype | Abi | Bulindi | Iki-Iki | Kabanyolo | Mubuku | Nakabango | Namulonge | Ngetta | Mean |
|---|---|---|---|---|---|---|---|---|---|
| BSPS48A-25 | 1180 | 1674 | 810 | 1024 | 1302 | 1487 | 842 | 1166 | 1186 |
| BSPS48A-26 | 802 | 1393 | 660 | 1002 | 1348 | 1391 | 830 | 1357 | 1098 |
| BSPS48A-27-1 | 1013 | 1384 | 669 | 1020 | 1372 | 1628 | 892 | 1260 | 1155 |
| BSPS48A-28 | 1316 | 1709 | 778 | 939 | 1389 | 1496 | 843 | 1187 | 1207 |
| BSPS48A-31 | 790 | 1798 | 580 | 895 | 1259 | 1401 | 1336 | 1000 | 1132 |
| BSPS48A-3B | 999 | 1728 | 657 | 936 | 1359 | 1361 | 1048 | 1143 | 1154 |
| BSPS48A-5 | 646 | 1548 | 610 | 973 | 1192 | 1365 | 838 | 1135 | 1038 |
| BSPS48A-8 | 596 | 1610 | 761 | 972 | 1524 | 1483 | 865 | 1017 | 1104 |
| BSPS48A-9-2 | 1144 | 1346 | 638 | 936 | 1400 | 1479 | 1509 | 1205 | 1207 |
| MAKSOY3N | 945 | 1797 | 662 | 1062 | 1335 | 1276 | 783 | 1190 | 1131 |
| MAKSOY4N | 858 | 1590 | 735 | 1030 | 1316 | 1453 | 807 | 1289 | 1135 |
| MNG11.2 | 837 | 1814 | 661 | 996 | 1306 | 1390 | 697 | 1067 | 1096 |
| NamII × GC30B | 637 | 1282 | 767 | 837 | 1256 | 1228 | 777 | 1015 | 975 |
| NamII × GC11.2 | 655 | 1438 | 667 | 806 | 1463 | 1038 | 900 | 1227 | 1024 |
| NamII × GC13.2 | 892 | 1215 | 649 | 1023 | 1564 | 1536 | 960 | 1316 | 1144 |
| NamII × GC17.3 | 638 | 901 | 828 | 958 | 1528 | 1460 | 1096 | 1187 | 1074 |
| NamII × GC20.3 | 619 | 1130 | 863 | 948 | 1414 | 1253 | 822 | 1072 | 1015 |
| NamII × GC28.2B | 861 | 1310 | 705 | 950 | 1177 | 1320 | 782 | 1145 | 1031 |
| NamII × GC32.6 | 530 | 1180 | 682 | 950 | 1271 | 1206 | 816 | 1160 | 974 |
| NamII × GC35.3 | 613 | 1465 | 697 | 969 | 1648 | 1294 | 849 | 1074 | 1076 |
| NamII × GC4.8 | 493 | 1009 | 732 | 1008 | 1312 | 1409 | 944 | 1145 | 1006 |
| NamII × GC43.1 | 931 | 951 | 649 | 807 | 1157 | 1148 | 751 | 1070 | 933 |
| NamII × GC43.2 | 783 | 1086 | 729 | 891 | 1572 | 1417 | 819 | 1166 | 1058 |
| NamII × GC44.2 | 903 | 1652 | 749 | 912 | 1411 | 1649 | 976 | 1325 | 1197 |
| NamII × GC44.3 | 772 | 743 | 771 | 917 | 1483 | 1080 | 1701 | 1039 | 1063 |
| NamII × GC7.2 | 651 | 1124 | 707 | 904 | 1520 | 1375 | 852 | 1015 | 1019 |
| NGDT4.11-5 | 1052 | 1641 | 635 | 884 | 1143 | 1014 | 547 | 945 | 983 |
| NGDT8.11-11B | 952 | 1779 | 681 | 954 | 1339 | 1325 | 848 | 1095 | 1122 |
| NGDT8.11-19 | 849 | 1763 | 674 | 898 | 1260 | 1051 | 626 | 922 | 1005 |
| NGDT8.11-5 | 705 | 1733 | 660 | 821 | 1185 | 1245 | 814 | 1018 | 1023 |
| Mean | 935 | 1598 | 687 | 981 | 1345 | 1438 | 963 | 1177 | 1141 |
| LSD | 504.37 | 523.26 | 413.13 | 249.23 | 328.40 | 314.36 | 485.84 | 503.19 | |
| CV | 37.20 | 22.44 | 71.99 | 39.62 | 36.18 | 35.07 | 80.40 | 60.83 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Obua, T.; Sserumaga, J.P.; Awio, B.; Nganga, F.; Odong, T.L.; Tukamuhabwa, P.; Tusiime, G.; Mukasa, S.B.; Nabasirye, M. Multi-Environmental Evaluation of Protein Content and Yield Stability among Tropical Soybean Genotypes Using GGE Biplot Analysis. Agronomy 2021, 11, 1265. https://doi.org/10.3390/agronomy11071265
Obua T, Sserumaga JP, Awio B, Nganga F, Odong TL, Tukamuhabwa P, Tusiime G, Mukasa SB, Nabasirye M. Multi-Environmental Evaluation of Protein Content and Yield Stability among Tropical Soybean Genotypes Using GGE Biplot Analysis. Agronomy. 2021; 11(7):1265. https://doi.org/10.3390/agronomy11071265
Chicago/Turabian StyleObua, Tonny, Julius Pyton Sserumaga, Bruno Awio, Fredrick Nganga, Thomas L. Odong, Phinehas Tukamuhabwa, Geoffrey Tusiime, Settumba B. Mukasa, and Margaret Nabasirye. 2021. "Multi-Environmental Evaluation of Protein Content and Yield Stability among Tropical Soybean Genotypes Using GGE Biplot Analysis" Agronomy 11, no. 7: 1265. https://doi.org/10.3390/agronomy11071265
APA StyleObua, T., Sserumaga, J. P., Awio, B., Nganga, F., Odong, T. L., Tukamuhabwa, P., Tusiime, G., Mukasa, S. B., & Nabasirye, M. (2021). Multi-Environmental Evaluation of Protein Content and Yield Stability among Tropical Soybean Genotypes Using GGE Biplot Analysis. Agronomy, 11(7), 1265. https://doi.org/10.3390/agronomy11071265







