Studying the Genetic Diversity of Yam Bean Using a New Draft Genome Assembly
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. DNA Extraction and Sequencing
2.3. Data Generation and Validation
2.4. Annotation
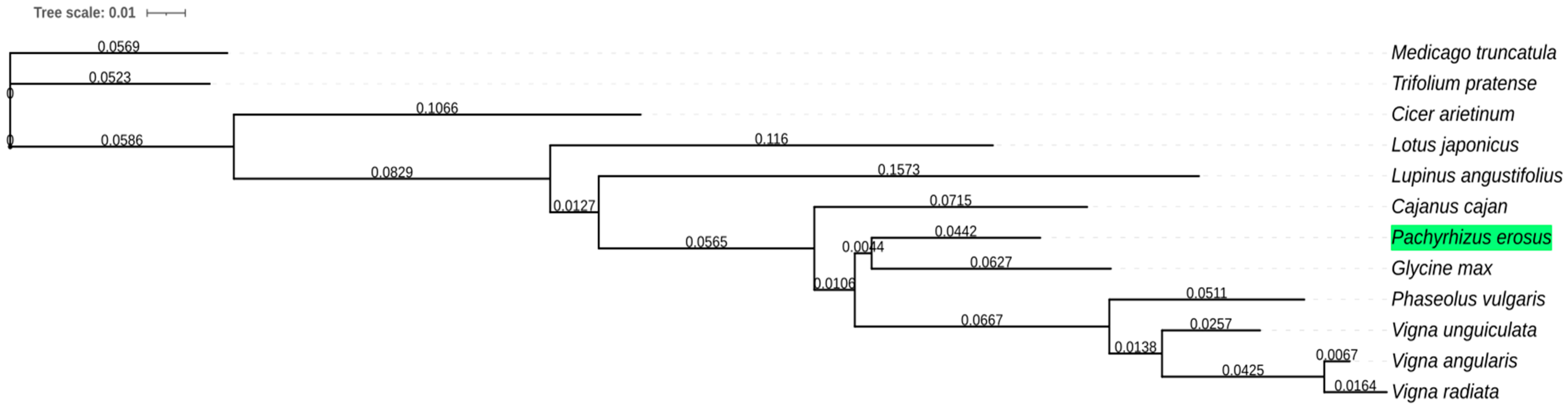
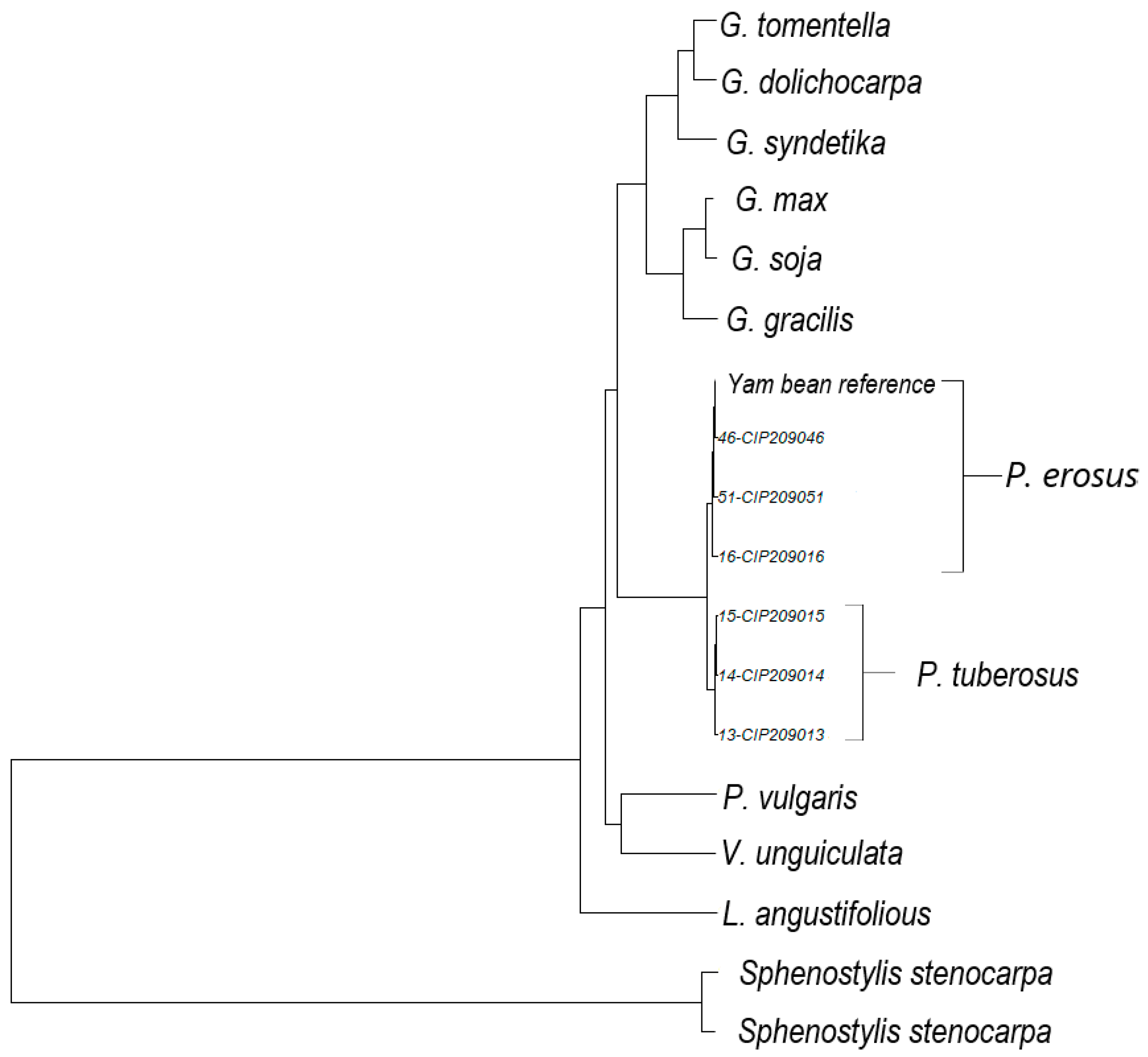
2.5. Phylogenetic Gene Trees
3. Results and Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gosálvez, M. Carcinogenesis with the insecticide rotenone. Life Sci. 1983, 32, 809–816. [Google Scholar] [CrossRef]
- Lee, J.; Hymowitz, T. A Molecular Phylogenetic Study of the Subtribe Glycininae (Leguminosae) Derived from the Chlo-roplast DNA rps16 Intron Sequences. Am. J. Bot. 2001, 88, 2064–2073. [Google Scholar] [CrossRef] [PubMed]
- National Research Council. Lost Crops of Africa; Volume II: Vegetables; National Academies Press: Washington, DC, USA, 2006. [Google Scholar]
- Agaba, R.; Tukamuhabwa, P.; Rubaihayo, P.; Mwanga, R.; Ssenyonjo, A.; Ndirigwe, J.; Tumwegamire, S.; Grüneberg, W. Heritability, combining ability and inheritance of storage root dry matter in yam beans. Afr. Crop. Sci. J. 2017, 25, 83. [Google Scholar] [CrossRef]
- Sørensen, M. Yam Bean (Pachyrhizus DC.). Promoting the Conservation and Use of Underutilized and Neglected Crops; Institute of Plant Genetics and Crop Plant Research: Gatersleben, Germany; International Plant Genetic Resources Institute: Rome, Italy, 1996. [Google Scholar]
- Grüneberg, W.; Freynhagen-Leopold, P.; Delgado-Váquez, O. A new yam bean (Pachyrhizus spp.) interspecific hybrid. Genet. Resour. Crop. Evol. 2003, 50, 757–766. [Google Scholar] [CrossRef]
- Sørensen, M.; Døygaard, S.; Estrella, J.E.; Kvist, L.P.; Nielsen, P.E. Status of the South American tuberous legume Pachyrhizus tuberosus (Lam.) Spreng: Field observations, taxonomic analysis, linguistic studies and agronomic data on the diversity of the South American Pachyrhizus tuberosus (Lam.) Spreng. complex with special reference to the identification of two new cultivar groups from Ecuador and Peru. Biodivers. Conserv. 1997, 6, 1581–1625. [Google Scholar] [CrossRef]
- Sørensen, M. A taxonomic revision of the genus Pachyrhizus (Fabaceae-Phaseoleae). Nord. J. Bot. 1988, 8, 167–192. [Google Scholar] [CrossRef]
- Engelmann, J. Molecular Systematics of the Neotropical Tuberous Legume ‘Pachyrhizus’ Rich. ex DC, the Yam Bean. In Environmental and Evolutionary Biology; University of St Andrews: St Andrews, UK; ProQuest Dissertations Publishing: Morrisville, NC, USA, 1998. [Google Scholar]
- Delêtre, M.; Soengas, B.; Utge, J.; Lambourdière, J.; Sørensen, M. Microsatellite Markers for the Yam Bean Pachyrhizus (Fabaceae). Appl. Plant Sci. 2013, 1, 1200551. [Google Scholar] [CrossRef] [PubMed]
- Santayana, M.; Rossel, G.; Núñez, J.; Sørensen, M.; Delêtre, M.; Robles, R.; Fernández, V.; Grüneberg, W.J.; Heider, B. Molecu-lar characterization of cultivated species of the genus Pachyrhizus Rich. ex DC. by AFLP markers: Calling for more data. Trop. Plant Biol. 2014, 7, 121–132. [Google Scholar] [CrossRef]
- Zanklan, A.S.; Becker, H.C.; Sørensen, M.; Pawelzik, E.; Grüneberg, W.J. Genetic diversity in cultivated yam bean (Pachyrhizus spp.) evaluated through multivariate analysis of morphological and agronomic traits. Genet. Resour. Crop. Evol. 2017, 65, 811–843. [Google Scholar] [CrossRef]
- Zimin, A.V.; Marçais, G.; Puiu, D.; Roberts, M.; Salzberg, S.L.; Yorke, J.A. The MaSuRCA genome assembler. Bioinformatics 2013, 29, 2669–2677. [Google Scholar] [CrossRef]
- Marçais, G.; Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 2011, 27, 764–770. [Google Scholar] [CrossRef]
- Rahman, A.; Hallgrímsdóttir, I.; Eisen, M.; Pachter, L. Association mapping from sequencing reads using k-mers. eLife 2018, 7, e32920. [Google Scholar] [CrossRef]
- Vurture, G.; Sedlazeck, F.J.; Nattestad, M.; Underwood, C.J.; Fang, H.; Gurtowski, J.; Schatz, M.C. GenomeScope: Fast refer-ence-free genome profiling from short reads. Bioinformatics 2017, 33, 2202–2204. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, R.M.; Seppey, M.; Simão, F.A.; Manni, M.; Ioannidis, P.; Klioutchnikov, G.; Kriventseva, E.V.; Zdobnov, E. BUSCO Applications from Quality Assessments to Gene Prediction and Phylogenomics. Mol. Biol. Evol. 2018, 35, 543–548. [Google Scholar] [CrossRef]
- Hoff, K.J.; Stanke, M. Predicting Genes in Single Genomes with AUGUSTUS. Curr. Protoc. Bioinform. 2018, 65, e57. [Google Scholar] [CrossRef]
- Slater, G.S.C.; Birney, E. Automated generation of heuristics for biological sequence comparison. BMC Bioinform. 2005, 6, 31. [Google Scholar] [CrossRef]
- Emms, D.; Kelly, S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves or-thogroup inference accuracy. Genome Biol. 2015, 16, 157. [Google Scholar] [CrossRef]
- Suzek, B.E.; Wang, Y.; Huang, H.; McGarvey, P.B.; Wu, C.H. The UniProt Consortium UniRef clusters: A comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 2015, 31, 926–932. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Dash, S.; Campbell, J.D.; Cannon, E.K.S.; Cleary, A.M.; Huang, W.; Kalberer, S.R.; Karingula, V.; Rice, A.G.; Singh, J.; Umale, P.E.; et al. Legume information system (LegumeInfo.org): A key component of a set of federated data resources for the legume family. Nucleic Acids Res. 2016, 44, D1181–D1188. [Google Scholar] [CrossRef]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Leinonen, R.; Sugawara, H.; Shumway, M.; on behalf of the International Nucleotide Sequence Database Collaboration. The Sequence Read Archive. Nucleic Acids Res. 2010, 39, D19–D21. [Google Scholar] [CrossRef] [PubMed]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef]
- Schliep, K.P. phangorn: Phylogenetic analysis in R. Bioinformatics 2011, 27, 592–593. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Levine, D.; Shen, J.; Gogarten, S.M.; Laurie, C.; Weir, B.S. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 2012, 28, 3326–3328. [Google Scholar] [CrossRef]
- Pati, K.; Zhang, F.; Batley, J. First report of genome size and ploidy of the underutilized leguminous tuber crop Yam Bean (Pachyrhizus erosus and P. tuberosus) by flow cytometry. Plant Genet. Resour. 2019, 17, 456–459. [Google Scholar] [CrossRef]
- Kreplak, J.; Madoui, M.-A.; Cápal, P.; Novák, P.; Labadie, K.; Aubert, G.; Bayer, P.E.; Gali, K.K.; Syme, R.A.; Main, D.; et al. A reference genome for pea provides insight into legume genome evolution. Nat. Genet. 2019, 51, 1411–1422. [Google Scholar] [CrossRef]
- Xie, M.; Chung, C.Y.-L.; Li, M.-W.; Wong, F.-L.; Wang, X.; Liu, A.; Wang, Z.; Leung, A.K.-Y.; Wong, T.-H.; Tong, S.-W.; et al. A reference-grade wild soybean genome. Nat. Commun. 2019, 10, 1216. [Google Scholar] [CrossRef]
- Chen, H.; Zeng, Y.; Yang, Y.; Huang, L.; Tang, B.; Zhang, H.; Hao, F.; Liu, W.; Li, Y.; Liu, Y.; et al. Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the auto-tetraploid cultivated alfalfa. Nat. Commun. 2020, 11, 2494. [Google Scholar] [CrossRef]
- Chang, Y.; Liu, H.; Liu, M.; Liao, X.; Sahu, S.K.; Fu, Y.; Song, B.; Cheng, S.; Kariba, R.; Muthemba, S.; et al. The draft genomes of five agriculturally important African orphan crops. GigaScience 2019, 8, 152. [Google Scholar] [CrossRef]
- Oyebanji, O.; Zhang, R.; Chen, S.-Y.; Yi, T.-S. New Insights into the Plastome Evolution of the Millettioid/Phaseoloid Clade (Papilionoideae, Leguminosae). Front. Plant Sci. 2020, 11, 151. [Google Scholar] [CrossRef]
- Doyle, J.; Chappill, J.; Bailey, C.; Kajita, T.; Herendeen, P.; Bruneau, A. Advances in Legume Systematics, Part 9; Royal Botanic Gardens Kew: Richmond, UK, 2000; pp. 1–20. [Google Scholar]
- Lackey, J.A. A revised classification of the tribe Phaseoleae (Leguminosae: Papilionoideae), and its relation to canavanine distribution. Bot. J. Linn. Soc. 1977, 74, 163–178. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. Chloroplast DNA Phylogeny of the Papilionoid Legume Tribe Phaseoleae. Syst. Bot. 1993, 18, 309–327. [Google Scholar] [CrossRef]
- Polhill, R. Classification of the Leguminosae. In Phytochemical Dictionary of the Leguminosae; Harborne, J.B., Ed.; Chapman & Hall: Cambridge, UK, 1994; Volume 1, pp. XXXV–XLVIII. [Google Scholar]
- Choi, I.S.; Choi, B.H. The distinct plastid genome structure of Maackia fauriei (Fabaceae: Papilionoideae) and its system-atic implications for genistoids and tribe Sophoreae. PLoS ONE 2017, 12, e0173766. [Google Scholar] [CrossRef]
- Tapia, C.; Sørensen, M. Morphological characterization of the genetic variation existing in a Neotropical collection of yam bean, Pachyrhizus tuberosus (Lam.) Spreng. Genet. Resour. Crop. Evol. 2003, 50, 681–692. [Google Scholar] [CrossRef]
- Estrella, E.; Phillips, S.; Abbott, R.; Gillies, A.; Sørensen, M. Genetic Variation and Relationships in Agronomically Important Species of Yam Bean (Pachyrhizus DC.) Based on RAPD Markers. In kke Angivet, Proceedings of the 2. International Symposium on Tuberous Legumes, Celaya, Guanajuato, Mexico, 5–8 August 1996; MacKeenzie: Dublin, UK, 1998; pp. 43–59. [Google Scholar]
- Andiku, C.; Tukamuhabwa, P.; Ssebuliba, J.M.; Talwana, H.; Tumwegamire, S.; Grüneberg, W.J. Evaluation of the American Yam Bean (Pachyrhizus spp.) for Storage Root Yield Across Varying Eco-geographic Conditions in Uganda. J. Agric. Sci. 2019, 11, 100. [Google Scholar] [CrossRef]
- Jean, N.; Patrick, R.; Phenihas, T.; Rolland, A.; Placide, R.; Robert, M.O.M.; Silver, T.; Vestine, K.; Evrard, K.; Grüneberg, W.J. Evaluation of Performance of Introduced Yam Bean (Pachyrhizus spp.) in Three Agro-Ecological Zones of Rwanda. Trop. Plant Biol. 2017, 10, 97–109. [Google Scholar] [CrossRef]

| No. | Accession | Origin | Pachyrhizus Species | Total Gbp | Sequence Coverage |
|---|---|---|---|---|---|
| 13 | Pt-CIP-209013 | Peru | P. tuberosus | 16.51 | 35.89× |
| 14 | Pt-CIP-209014 | Peru | P. tuberosus | 12.19 | 26.49× |
| 15 | Pt-CIP-209015 | Peru | P. tuberosus | 16.79 | 36.50× |
| 16 | Pe-CIP-209016 | Guatemala | P. erosus | 21.06 | 45.78× |
| 46 | Pe-CIP-209046 | Cartago Costa Rica | P. erosus | 16.05 | 34.89× |
| 51 | Pe-CIP-209051 | Mexico | P. erosus | 19.18 | 41.69× |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tay Fernandez, C.G.; Pati, K.; Severn-Ellis, A.A.; Batley, J.; Edwards, D. Studying the Genetic Diversity of Yam Bean Using a New Draft Genome Assembly. Agronomy 2021, 11, 953. https://doi.org/10.3390/agronomy11050953
Tay Fernandez CG, Pati K, Severn-Ellis AA, Batley J, Edwards D. Studying the Genetic Diversity of Yam Bean Using a New Draft Genome Assembly. Agronomy. 2021; 11(5):953. https://doi.org/10.3390/agronomy11050953
Chicago/Turabian StyleTay Fernandez, Cassandria G., Kalidas Pati, Anita A. Severn-Ellis, Jacqueline Batley, and David Edwards. 2021. "Studying the Genetic Diversity of Yam Bean Using a New Draft Genome Assembly" Agronomy 11, no. 5: 953. https://doi.org/10.3390/agronomy11050953
APA StyleTay Fernandez, C. G., Pati, K., Severn-Ellis, A. A., Batley, J., & Edwards, D. (2021). Studying the Genetic Diversity of Yam Bean Using a New Draft Genome Assembly. Agronomy, 11(5), 953. https://doi.org/10.3390/agronomy11050953








