Biological Control of Pythium aphanidermatum, the Causal Agent of Tomato Root Rot by Two Streptomyces Root Symbionts
Abstract
1. Introduction
2. Materials and Methods
2.1. Culture Media and Preparation of the Pathogen
2.2. Pathogenicity Assessment
2.3. Soil Samples and Isolating Actinobacteria
2.4. In Vitro Screening of Actinobacteria
2.5. In Vitro Petri Plate Seedling-Bioassay
2.6. In Vitro Root Colonization Bioassay
2.7. DNA Extraction, PCR Amplification, and Electrophoresis
2.8. DNA Sequencing and Phylogenetic Analysis
2.9. Physiological Characterization
2.10. Greenhouse Evaluations
2.10.1. Preparation of Tomato Seed
2.10.2. Preparation of P. aphanidermatum Inocula
2.10.3. Soil Inoculation
2.10.4. Design of Sandwich Bed-Mixes
2.10.5. Evaluation of Plants Responses
Disease Incidence
Number of Leaves
Length of the Plants
Plant Fresh Weights
Plant Dry Weights
2.11. Statistical Analyses
3. Results
3.1. Pathogenicity Test and Damping-Off Incidence
3.2. Isolated Soil Actinobacteria and In Vitro Screening
3.3. In Vitro Petri Plate Seedling Bioassay
3.4. In Vitro Root Colonization Bioassay
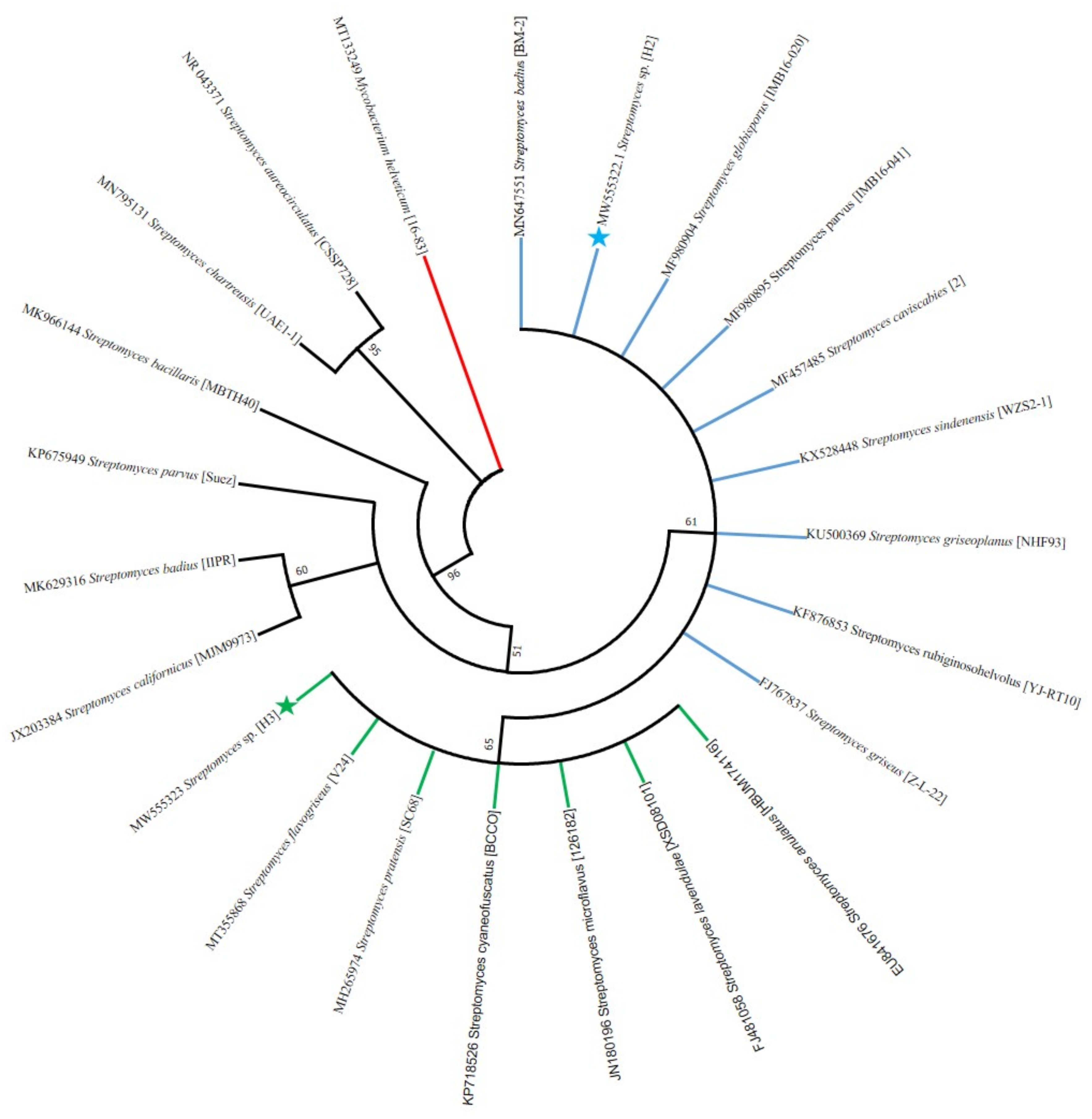
3.5. PCR Amplification of 16S rRNA Gene and Phylogeny Analysis
3.6. Physiological Characterization
3.7. Greenhouse Evaluations
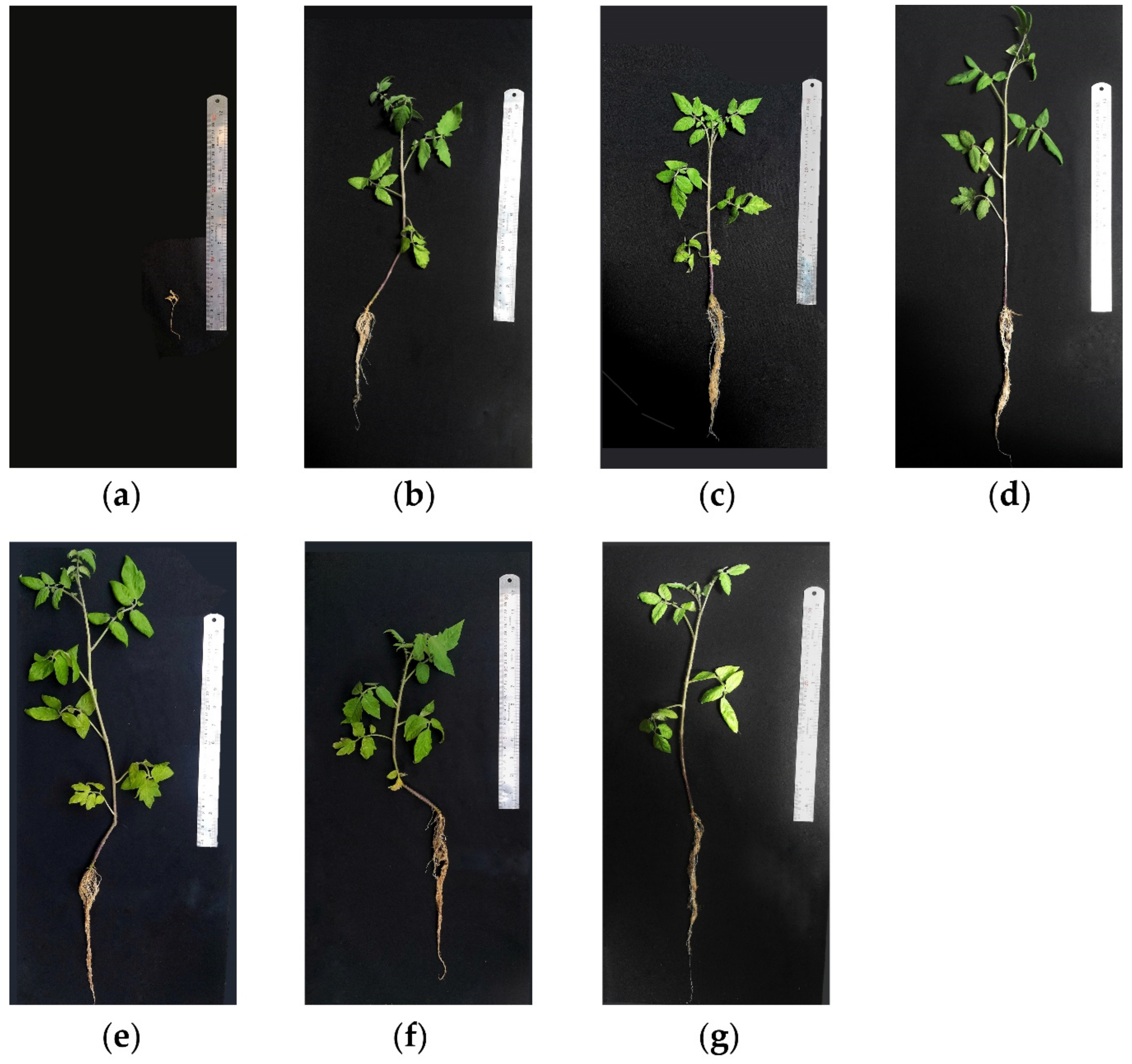
3.7.1. Biocontrol Efficacy of Symbiont Streptomyces Strains
Disease Incidence
Number of Leaves
Plants Length
Fresh and Dry Plant Weights
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAO; IFAD; UNICEF; WFP; WHO. The State of Food Security and Nutrition in the World 2020. Transforming Food Systems for Affordable Healthy Diets; FAO: Rome, Italy, 2020. [Google Scholar] [CrossRef]
- Hartley, C.; Pierce, R. The Control of Damping-Off of Coniferous Seedlings; United States Department of Agriculture: Washington, DC, USA, 1917; Volume 453.
- Lamichhane, J.; Dürr, C.; Schwanck, A.; Robin, M.; Sarthou, J.; Cellier, V.; Messéan, A.; Aubertot, J. Integrated management of damping-off diseases. A review. Agron. Sustain. Dev. 2017, 37, 10. [Google Scholar] [CrossRef]
- Al-Sheikh, H. Two pathogenic species of Pythium: P. aphanidermatum and P. diclinum from a wheat field. Saudi J. Biol. Sci. 2010, 17, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, F.; Campbell, W. Pythiums as plant pathogens. Annu. Rev. Phytopathol. 1973, 11, 77–98. [Google Scholar] [CrossRef]
- Daughtrey, M.L.; Wick, R.L.; Peterson, J.L. Compendium of Flowering Potted Plant Diseases; APS Press: St. Paul, MN, USA, 1995. [Google Scholar]
- Parveen, T.; Sharma, K. Pythium diseases, control and management strategies: A review. Int. J Plant Anim. Environ. Sci. 2015, 5, 244–257. [Google Scholar]
- Van der Plaats-Niterink, A.J. Monograph of the genus Pythium. In Studies in Mycology; Centraalbureau voor Schimmelcultures (Baarn): Baarn, The Netherlands, 1981; Volume 21, 244p. [Google Scholar]
- Abdelzaher, H.M. Occurrence of damping-off of wheat caused by Pythium diclinum tokunaga in El-Minia, Egypt and its possible control by Gliocladium roseum and Trichoderma harzianum. Arch. Phytopathol. Plant Prot. 2004, 37, 147–159. [Google Scholar] [CrossRef]
- Elshahawy, I.; Abouelnasr, H.M.; Lashin, S.M.; Darwesh, O.M. First report of Pythium aphanidermatum infecting tomato in Egypt and its control using biogenic silver nanoparticles. J. Plant Prot. Res. 2018, 58, 137–151. [Google Scholar] [CrossRef]
- Jones, S.; Donaldson, S.; Deacon, J. Behaviour of zoospores and zoospore cysts in relation to root infection by Pythium aphanidermatum. New Phytol. 1991, 117, 289–301. [Google Scholar] [CrossRef]
- Moorman, G.; Kang, S.; Geiser, D.M.; Kim, S. Identification and characterization of Pythium species associated with greenhouse floral crops in Pennsylvania. Plant Dis. 2002, 86, 1227–1231. [Google Scholar] [CrossRef]
- Stephens, C.; Herr, L.; Schmitthenner, A.; Powell, C. Sources of Rhizoctonia solani and Pythium spp. in the bedding plant greenhouse. Plant Dis. 1983, 67, 272–275. [Google Scholar] [CrossRef]
- Lookabaugh, E.C.; Kerns, J.P.; Cubeta, M.A.; Shew, B.B. Fitness Attributes of Pythium aphanidermatum with Dual Resistance to Mefenoxam and Fenamidone. Plant Dis. 2018, 102, 1938–1943. [Google Scholar] [CrossRef]
- Pérez, W.; Lara, J.; Forbes, G.A. Resistance to metalaxyl-M and cymoxanil in a dominant clonal lineage of Phytophthora infestans in Huánuco, Peru, an area of continuous potato production. Eur. J. Plant Pathol. 2009, 125, 87–95. [Google Scholar] [CrossRef]
- Utkhede, R.; Gupta, V. In vitro selection of strains of Phytophthora cactorum resistant to metalaxyl. J. Phytopathol. 1988, 122, 35–44. [Google Scholar] [CrossRef]
- Sabarwal, A.; Kumar, K.; Singh, R.P. Hazardous effects of chemical pesticides on human health—Cancer and other associated disorders. Environ. Toxicol. Pharmacol. 2018, 63, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Wightwick, A.; Walters, R.; Allinson, G.; Reichman, S.; Menzies, N. Environmental Risks of Fungicides Used in Horticultural Production Systems; Carisse, O., Ed.; InTech: Rijeka, Croatia, 2010; Volume 1, pp. 273–304. [Google Scholar]
- Sood, M.; Kapoor, D.; Kumar, V.; Sheteiwy, M.S.; Ramakrishnan, M.; Landi, M.; Araniti, F.; Sharma, A. Trichoderma: The “Secrets” of a Multitalented Biocontrol Agent. Plants 2020, 9, 762. [Google Scholar] [CrossRef] [PubMed]
- Kipngeno, P.; Losenge, T.; Maina, N.; Kahangi, E.; Juma, P. Efficacy of Bacillus subtilis and Trichoderma asperellum against Pythium aphanidermatum in tomatoes. Biol. Control 2015, 90, 92–95. [Google Scholar] [CrossRef]
- Sajeena, A.; Nair, D.S.; Sreepavan, K. Non-pathogenic Fusarium oxysporum as a biocontrol agent. Indian Phytopathol. 2020, 73, 177–183. [Google Scholar] [CrossRef]
- Kobayashi, D.Y.; Guglielmoni, M.; Clarke, B.B. Isolation of the chitinolytic bacteria Xanthomonas maltophilia and Serratia marcescens as biological control agents for summer patch disease of turfgrass. Soil Biol. Biochem. 1995, 27, 1479–1487. [Google Scholar] [CrossRef]
- Ben Khedher, S.; Boukedi, H.; Laarif, A.; Tounsi, S. Biosurfactant produced by Bacillus subtilis V26: A potential biological control approach for sustainable agriculture development. Org. Agric. 2020, 10, 117–124. [Google Scholar] [CrossRef]
- Wang, S.; Sun, L.; Zhang, W.; Chi, F.; Hao, X.; Bian, J.; Li, Y. Bacillus velezensis BM21, a potential and efficient biocontrol agent in control of corn stalk rot caused by Fusarium graminearum. Egypt. J. Biol. Pest Control 2020, 30, 9. [Google Scholar] [CrossRef]
- Mohammed, A.F.; Oloyede, A.R.; Odeseye, A.O. Biological control of bacterial wilt of tomato caused by Ralstonia solanacearum using Pseudomonas species isolated from the rhizosphere of tomato plants. Arch. Phytopathol. Plant Prot. 2020, 53, 1–16. [Google Scholar] [CrossRef]
- El-Tarabily, K.A.; Nassar, A.H.; Hardy, G.E.; Sivasithamparam, K. Plant growth promotion and biological control of Pythium aphanidermatum, a pathogen of cucumber, by endophytic actinomycetes. J. Appl. Microbiol. 2009, 106, 13–26. [Google Scholar] [CrossRef]
- Kaur, T.; Rani, R.; Manhas, R.K. Biocontrol and plant growth promoting potential of phylogenetically new Streptomyces sp. MR14 of rhizospheric origin. AMB Express 2019, 9, 125. [Google Scholar] [CrossRef]
- Newitt, J.T.; Prudence, S.M.M.; Hutchings, M.I.; Worsley, S.F. Biocontrol of Cereal Crop Diseases Using Streptomycetes. Pathogens 2019, 8, 78. [Google Scholar] [CrossRef] [PubMed]
- Olanrewaju, O.S.; Babalola, O.O. Streptomyces: Implications and interactions in plant growth promotion. Appl. Microbiol. Biotechnol. 2019, 103, 1179–1188. [Google Scholar] [CrossRef]
- Elshahawy, I.E.; El-Mohamedy, R.S. Biological control of Pythium damping-off and root-rot diseases of tomato using Trichoderma isolates employed alone or in combination. J. Plant Pathol. 2019, 101, 597–608. [Google Scholar] [CrossRef]
- Halo, B.A.; Al-Yahyai, R.A.; Al-Sadi, A.M. Aspergillus terreus Inhibits Growth and Induces Morphological Abnormalities in Pythium aphanidermatum and Suppresses Pythium-Induced Damping-Off of Cucumber. Front. Microbiol. 2018, 9, 95. [Google Scholar] [CrossRef] [PubMed]
- Al-Hussini, H.S.; Al-Rawahi, A.Y.; Al-Marhoon, A.A.; Al-Abri, S.A.; Al-Mahmooli, I.H.; Al-Sadi, A.M.; Velazhahan, R. Biological control of damping-off of tomato caused by Pythium aphanidermatum by using native antagonistic rhizobacteria isolated from Omani soil. J. Plant Pathol. 2019, 101, 315–322. [Google Scholar] [CrossRef]
- Halo, B.A.; Al-Yahyai, R.A.; Maharachchikumbura, S.S.N.; Al-Sadi, A.M. Talaromyces variabilis interferes with Pythium aphanidermatum growth and suppresses Pythium-induced damping-off of cucumbers and tomatoes. Sci. Rep. 2019, 9, 11255. [Google Scholar] [CrossRef] [PubMed]
- Beena Kanimozhi, R.; Paul Raj, R. Biological Control of Pythium Damping-off in Seedlings with Streptomyces sp. Int. J. Recent Technol. Eng. 2019, 8, 8035–8039. [Google Scholar] [CrossRef]
- Kloepper, J.; Schroth, M. Plant growth-promoting rhizobacteria and plant growth under gnotobiotic conditions. Phytopathology 1981, 71, 642–644. [Google Scholar] [CrossRef]
- Mohammadipanah, F.; Dehhaghi, M. Classification and taxonomy of Actinobacteria. In Biology and Biotechnology of Actinobacteria; Springer: Berlin/Heidelberg, Germany, 2017; pp. 51–77. [Google Scholar]
- Kämpfer, P. The family Streptomycetaceae, part I: Taxonomy. In The Prokaryotes, 3rd ed.; Springer: New York, NY, USA, 2006; pp. 538–604. [Google Scholar]
- Manteca, Á.; Yagüe, P. Streptomyces as a source of antimicrobials: Novel approaches to activate cryptic secondary metabolite pathways. In Antimicrobials, Antibiotic Resistance, Antibiofilm Strategies and Activity Methods; IntechOpen: Rijeka, Croatia, 2019; p. 119. [Google Scholar]
- Euzéby, J. “Genus Streptomyces”. List of Prokaryotic Names with Standing in Nomenclature. 2008. Available online: http://www.bacterio.cict.fr/s/streptomycesa.html (accessed on 19 April 2021).
- Waksman, S.A.; Henrici, A.T. The nomenclature and classification of the actinomycetes. J. Bacteriol. 1943, 46, 337. [Google Scholar] [CrossRef]
- Bentley, S.D.; Chater, K.F.; Cerdeño-Tárraga, A.-M.; Challis, G.L.; Thomson, N.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3 (2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Seipke, R.F.; Kaltenpoth, M.; Hutchings, M.I. Streptomyces as symbionts: An emerging and widespread theme? FEMS Microbiol. Rev. 2012, 36, 862–876. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Omura, S. Isolation of new actinomycete strains for the screening of new bioactive compounds. J. Gen. Appl. Microbiol. 2003, 49, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Sabido, E.; Tenebro, C.; Suarez, A.; Ong, S.; Trono, D.; Amago, D.; Evangelista, J.J.; Reynoso, A.; Villalobos, I.; Alit, L.; et al. Marine Sediment-Derived Streptomyces Strain Produces Angucycline Antibiotics against Multidrug-Resistant Staphylococcus aureus Harboring SCCmec Type 1 Gene. J. Mar. Sci. Eng. 2020, 8, 734. [Google Scholar] [CrossRef]
- Demain, A.L. Small bugs, big business: The economic power of the microbe. Biotechnol. Adv. 2000, 18, 499–514. [Google Scholar] [CrossRef]
- Watve, M.G.; Tickoo, R.; Jog, M.M.; Bhole, B.D. How many antibiotics are produced by the genus Streptomyces? Arch. Microbiol. 2001, 176, 386–390. [Google Scholar] [CrossRef] [PubMed]
- Benizri, E.; Baudoin, E.; Guckert, A. Root colonization by inoculated plant growth-promoting rhizobacteria. Biocontrol Sci. Technol. 2001, 11, 557–574. [Google Scholar] [CrossRef]
- Sylvia, D.M.; Fuhrmann, J.J.; Hartel, P.G.; Zuberer, D.A. Principles and Applications of Soil Microbiology; Pearson Prentice Hall: Upper Saddle River, NJ, USA, 2005. [Google Scholar]
- Loliama, B.; Morinagab, T.; Chaiyanana, S. Biocontrol of Pythium aphanidermatum by the cellulolytic actinomycetes Streptomyces rubrolavendulae S4. Sci. Asia 2013, 39, 584–590. [Google Scholar] [CrossRef]
- Costa, F.G.; Zucchi, T.D.; de Melo, I.S. Biological control of phytopathogenic fungi by endophytic actinomycetes isolated from maize (Zea mays L.). Braz. Arch. Biol. Technol. 2013, 56, 948–955. [Google Scholar] [CrossRef]
- El-Tarabily, K.A.; Hardy, G.E.S.J.; Sivasithamparam, K. Performance of three endophytic actinomycetes in relation to plant growth promotion and biological control of Pythium aphanidermatum, a pathogen of cucumber under commercial field production conditions in the United Arab Emirates. Eur. J. Plant Pathol. 2010, 128, 527–539. [Google Scholar] [CrossRef]
- Abreo, E.; Vaz-Jauri, P.; Nuñez, L.; Stewart, S.; Mattos, N.; Dini, B.; Altier, N. Pathogenicity of Pythium spp. obtained from agricultural soils and symptomatic legume seedlings in Uruguay. Australas. Plant Dis. Notes 2017, 12, 35. [Google Scholar] [CrossRef]
- Küster, E.; Williams, S. Selection of media for isolation of streptomycetes. Nature 1964, 202, 928–929. [Google Scholar] [CrossRef]
- Pepper, I.L.; Gerba, C.P.; Brendecke, J.W. Environmental Microbiology: A Laboratory Manual; Elsevier Science Press: London, UK, 2005. [Google Scholar]
- Sanders, E.R. Aseptic laboratory techniques: Plating methods. J. Vis. Exp. 2012, e3064. [Google Scholar] [CrossRef] [PubMed]
- Shahidi Bonjar, G.H.; Fooladi, M.H.; Mahdavi, M.J.; Shahghasi, A. Broadspectrim, a novel antibacterial from Streptomyces sp. Biotechnology 2004, 3, 126–130. [Google Scholar] [CrossRef]
- El-Tarabily, K.; Soliman, M.; Nassar, A.; Al-Hassani, H.; Sivasithamparam, K.; McKenna, F.; Hardy, G.S.J. Biological control of Sclerotinia minor using a chitinolytic bacterium and actinomycetes. Plant Pathol. 2000, 49, 573–583. [Google Scholar] [CrossRef]
- Zhang, Y.-P.; Uyemoto, J.; Kirkpatrick, B. A small-scale procedure for extracting nucleic acids from woody plants infected with various phytopathogens for PCR assay. J. Virol. Methods 1998, 71, 45–50. [Google Scholar] [CrossRef]
- Cook, A.E.; Meyers, P.R. Rapid identification of filamentous actinomycetes to the genus level using genus-specific 16S rRNA gene restriction fragment patterns. Int. J. Syst. Evol. Microbiol. 2003, 53, 1907–1915. [Google Scholar] [CrossRef]
- Huang, X.; Madan, A. CAP3: A DNA sequence assembly program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Campanella, J.J.; Bitincka, L.; Smalley, J. MatGAT: An application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinform. 2003, 4, 29. [Google Scholar] [CrossRef]
- Palaniyandi, S.; Damodharan, K.; Yang, S.; Suh, J. Streptomyces sp. strain PGPA39 alleviates salt stress and promotes growth of ‘Micro Tom’ tomato plants. J. Appl. Microbiol. 2014, 117, 766–773. [Google Scholar] [CrossRef]
- Nonomura, H. Key for classification and identification of 458 species of the streptomycetes included in ISP. J. Ferment. Technol. 1974, 52, 78–92. [Google Scholar]
- Kathiravan, P.; Sabarinathan, R.; Subbaiya, R.; Masilamani Selvam, M. Investigation on sugar cane field Actinomycetes of Erode District. Res. J. Pharm. Biol. Chem. Sci. 2016, 7, 1145–1154. [Google Scholar]
- Hankin, L.; Anagnostakis, S.L. The use of solid media for detection of enzyme production by fungi. Mycologia 1975, 67, 597–607. [Google Scholar] [CrossRef]
- Alström, S.; Burns, R.G. Cyanide production by rhizobacteria as a possible mechanism of plant growth inhibition. Biol. Fertil. Soils 1989, 7, 232–238. [Google Scholar] [CrossRef]
- Gao, H.; Li, P.; Xu, X.; Zeng, Q.; Guan, W. Research on volatile organic compounds from Bacillus subtilis CF-3: Biocontrol effects on fruit fungal pathogens and dynamic changes during fermentation. Front. Microbiol. 2018, 9, 456. [Google Scholar] [CrossRef] [PubMed]
- Jorjandi, M.; Bonjar, G.S.; Baghizadeh, A.; Sirchi, G.S.; Massumi, H.; Baniasadi, F.; Aghighi, S.; Farokhi, P.R. Biocontrol of Botrytis allii Munn the causal agent of neck rot, the post harvest disease in onion, by use of a new Iranian isolate of Streptomyces. Am. J. Agric. Biol. Sci. 2009, 4, 72–78. [Google Scholar] [CrossRef][Green Version]
- Kyuchukova, M.A.; Büttner, C.; Gabler, J.; Bar-Yosef, B.; Grosch, R.; Kläring, H.-P. Evaluation of a method for quantification of Pythium aphanidermatum in cucumber roots at different temperatures and inoculum densities. J. Plant Dis. Prot. 2006, 113, 113–119. [Google Scholar] [CrossRef]
- Zheng, X.; Wang, J.; Chen, Z.; Zhang, H.; Wang, Z.; Zhu, Y.; Liu, B. A Streptomyces sp. strain: Isolation, identification, and potential as a biocontrol agent against soilborne diseases of tomato plants. Biol. Control 2019, 136, 104004. [Google Scholar] [CrossRef]
- Martinuz, A.; Schouten, A.; Sikora, R. Systemically induced resistance and microbial competitive exclusion: Implications on biological control. Phytopathology 2012, 102, 260–266. [Google Scholar] [CrossRef]
- Parke, J.L. Root colonization by indigenous and introduced microorganisms. In The Rhizosphere and Plant Growth; Keister, D.L., Cregan, P.B., Eds.; Beltsville Symposia in Agricultural Research; Springer: Dordrecht, The Netherlands, 1991; Volume 4, pp. 33–42. [Google Scholar]
- Evangelista-Martínez, Z.; Contreras-Leal, E.A.; Corona-Pedraza, L.F.; Gastélum-Martínez, É. Biocontrol potential of Streptomyces sp. CACIS-1.5CA against phytopathogenic fungi causing postharvest fruit diseases. Egypt. J. Biol. Pest Control 2020, 30, 117. [Google Scholar] [CrossRef]
- Getha, K.; Vikineswary, S. Antagonistic effects of Streptomyces violaceusniger strain G10 on Fusarium oxysporum f. sp. cubense race 4: Indirect evidence for the role of antibiosis in the antagonistic process. J. Ind. Microbiol. Biotechnol. 2002, 28, 303–310. [Google Scholar] [PubMed]
- Zhao, J.; Xue, Q.; Niu, G.; Xue, L.; Shen, G.; Du, J. Extracellular enzyme production and fungal mycelia degradation of antagonistic Streptomyces induced by fungal mycelia preparation of cucurbit plant pathogens. Ann. Microbiol. 2013, 63, 809–812. [Google Scholar] [CrossRef]
- Viaene, T.; Langendries, S.; Beirinckx, S.; Maes, M.; Goormachtig, S. Streptomyces as a plant’s best friend? FEMS Microbiol. Ecol. 2016, 92, fiw119. [Google Scholar] [CrossRef] [PubMed]
- Ait Barka, E.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.-P.; Clément, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef]
- Huddleston, A.S.; Cresswell, N.; Neves, M.; Beringer, J.E.; Baumberg, S.; Thomas, D.I.; Wellington, E. Molecular detection of streptomycin-producing streptomycetes in Brazilian soils. Appl. Environ. Microbiol. 1997, 63, 1288–1297. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Zheng, W.; Rong, X.; Huang, Y. A multilocus phylogeny of the Streptomyces griseus 16S rRNA gene clade: Use of multilocus sequence analysis for streptomycete systematics. Int. J. Syst. Evol. Microbiol. 2008, 58, 149–159. [Google Scholar] [CrossRef]
- Gopalakrishnan, S.; Pande, S.; Sharma, M.; Humayun, P.; Kiran, B.K.; Sandeep, D.; Vidya, M.S.; Deepthi, K.; Rupela, O. Evaluation of actinomycete isolates obtained from herbal vermicompost for the biological control of Fusarium wilt of chickpea. Crop Prot. 2011, 30, 1070–1078. [Google Scholar] [CrossRef]
- Shahidi Bonjar, G.; Barkhordar, B.; Pakgohar, N.; Aghighi, S.; Biglary, S.; Rashid Farrokhi, P.; Aminaii, M.; Mahdavi, M.; Aghelizadeh, A. Biological control of Phytophthora drechsleri Tucker, the causal agent of pistachio gummosis, under greenhouse conditions by use of actinomycetes. Plant Pathol. 2006, 5, 20–23. [Google Scholar] [CrossRef]
- Yang, Y.; Wu, Z.-M.; Li, K.-T. The peculiar physiological responses of Rhizoctonia solani under the antagonistic interaction coupled by a novel antifungalmycin N2 from Streptomyces sp. N2. Arch. Microbiol. 2019, 201, 787–794. [Google Scholar] [CrossRef]
- Cao, P.; Liu, C.; Sun, P.; Fu, X.; Wang, S.; Wu, F.; Wang, X. An endophytic Streptomyces sp. strain DHV3-2 from diseased root as a potential biocontrol agent against Verticillium dahliae and growth elicitor in tomato (Solanum lycopersicum). Antonie Leeuwenhoek 2016, 109, 1573–1582. [Google Scholar] [CrossRef] [PubMed]
- Hamdali, H.; Hafidi, M.; Virolle, M.J.; Ouhdouch, Y. Growth promotion and protection against damping-off of wheat by two rock phosphate solubilizing actinomycetes in a P-deficient soil under greenhouse conditions. Appl. Soil Ecol. 2008, 40, 510–517. [Google Scholar] [CrossRef]
- Goudjal, Y.; Toumatia, O.; Yekkour, A.; Sabaou, N.; Mathieu, F.; Zitouni, A. Biocontrol of Rhizoctonia solani damping-off and promotion of tomato plant growth by endophytic actinomycetes isolated from native plants of Algerian Sahara. Microbiol. Res. 2014, 169, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Loqman, S.; Barka, E.A.; Clément, C.; Ouhdouch, Y. Antagonistic actinomycetes from Moroccan soil to control the grapevine gray mold. World J. Microb. Biotechnol. 2009, 25, 81–91. [Google Scholar] [CrossRef]
- Esmaeel, Q.; Jacquard, C.; Sanchez, L.; Clément, C.; Barka, E.A. The mode of action of plant associated Burkholderia against grey mould disease in grapevine revealed through traits and genomic analyses. Sci. Rep. 2020, 10, 19393. [Google Scholar] [CrossRef]
- Michel-Aceves, A.; Díaz-Nájera, J.; Ariza-Flores, R.; Otero-Sánchez, M.; Escobar-Martínez, R.; Avendaño-Arrazate, C. Control Alternatives for Damping-Off in Tomato Seedling Production. Phyton 2019, 88, 325–333. [Google Scholar] [CrossRef]
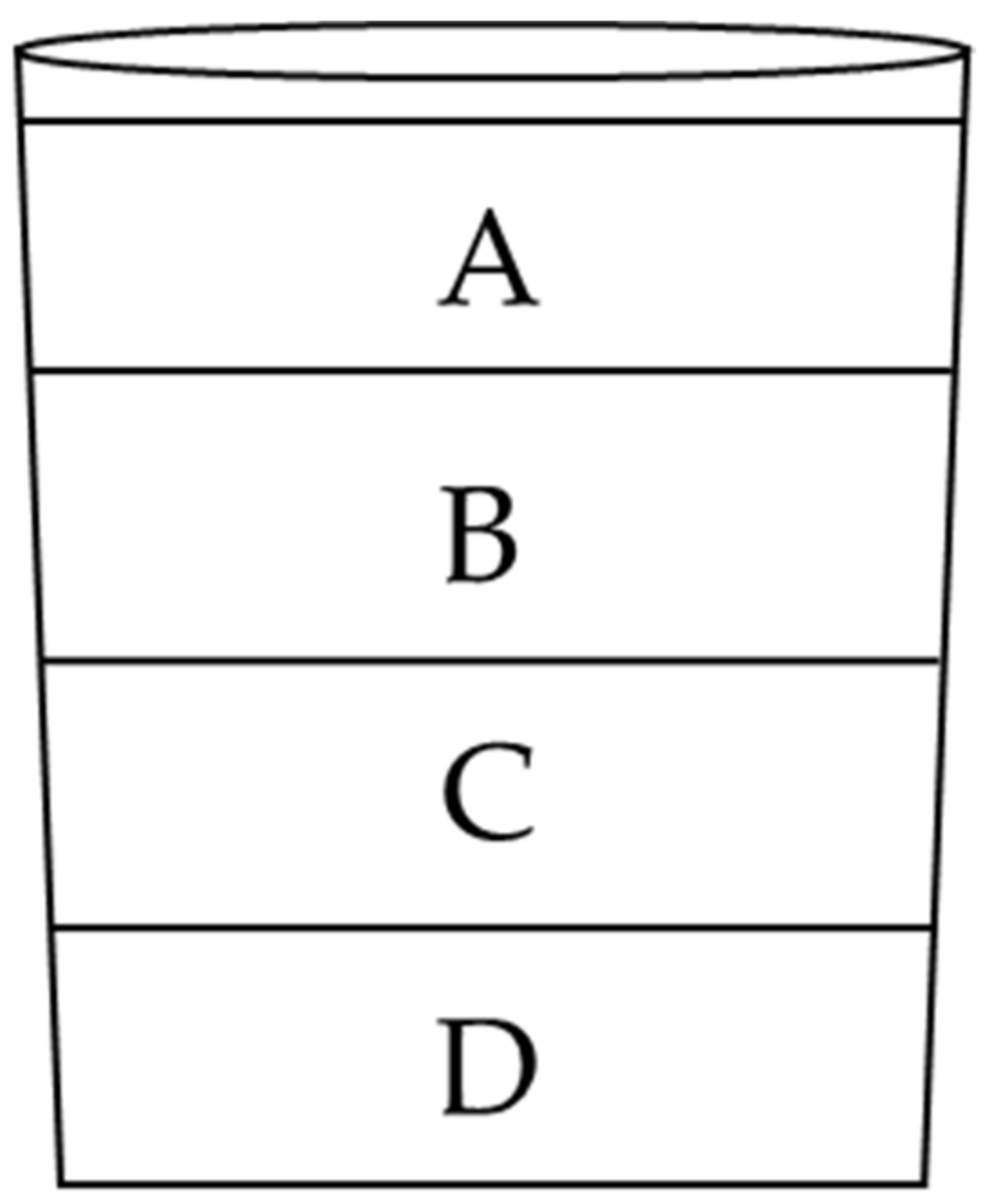
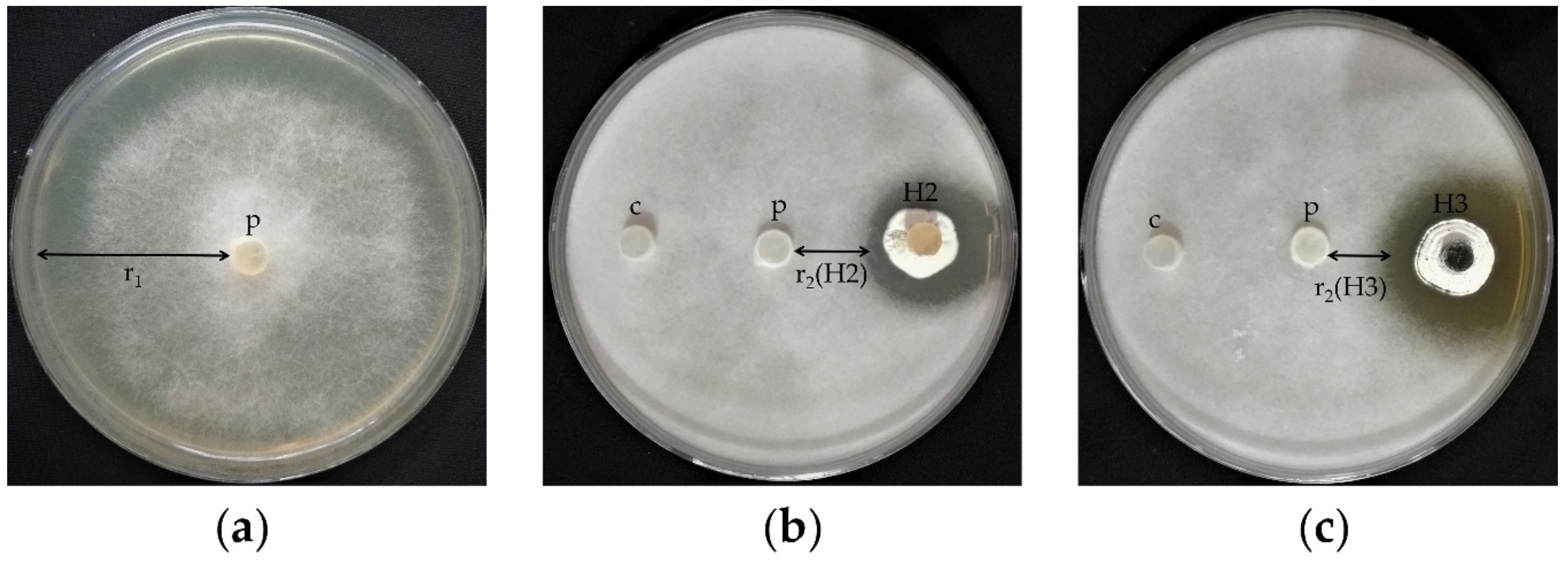

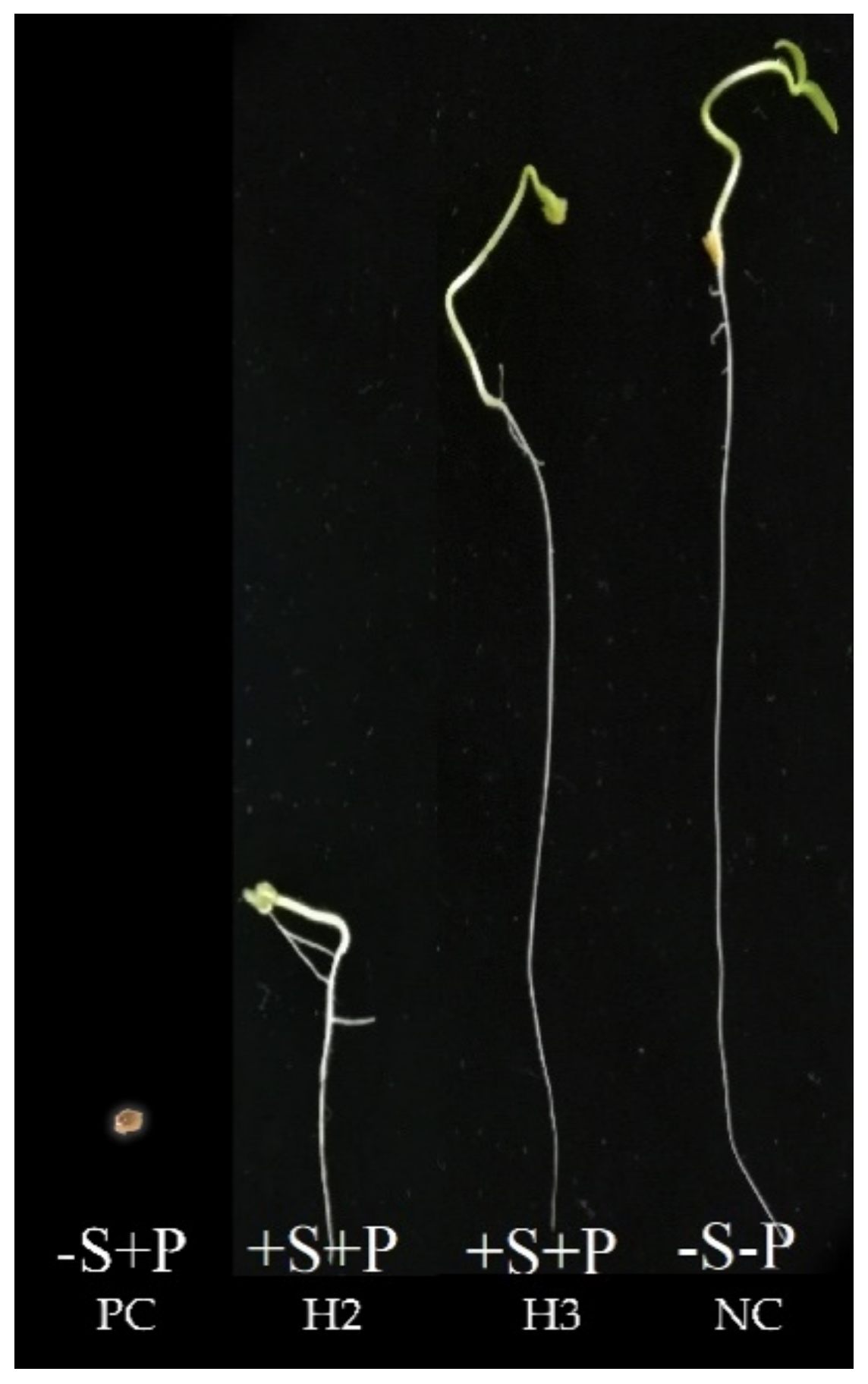
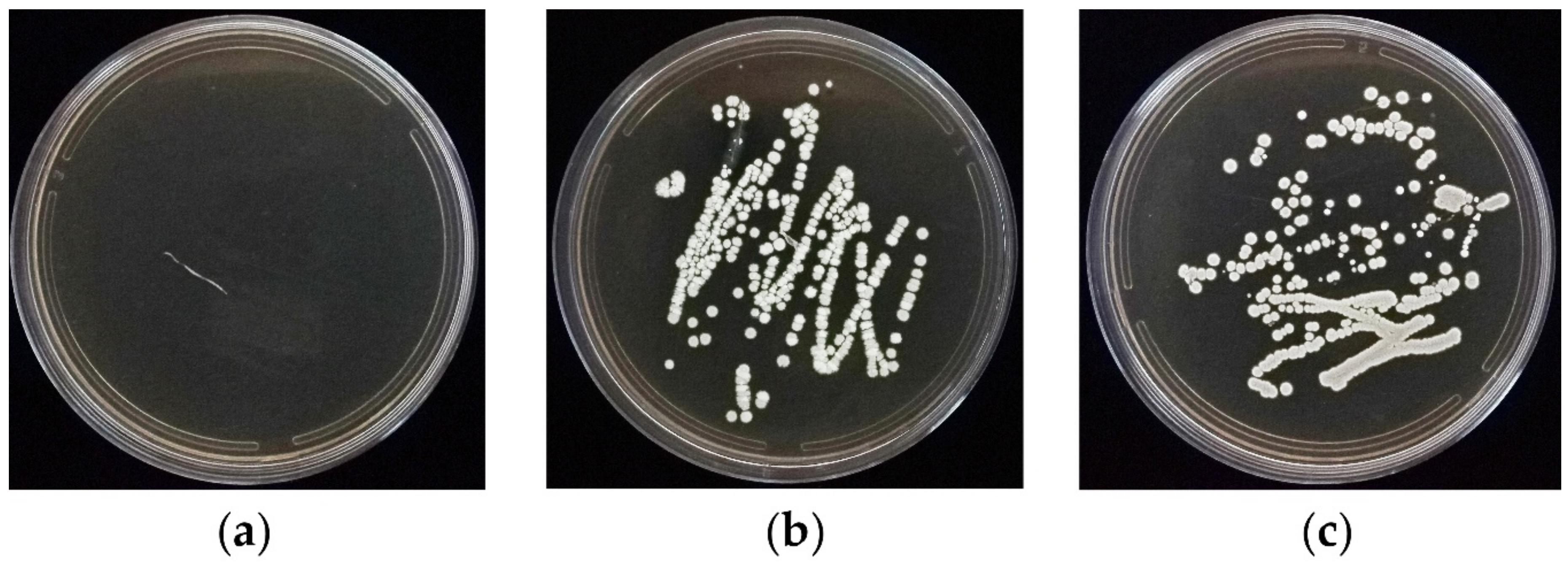

| Treatment No. | Treatment | Treatment Description |
|---|---|---|
| 1 | Positive control | Millet seed Pythium aphanidermatum inoculum (0.5% w/w) |
| 2 | P. aphanidermatum + H2 | 10 mL 108 CFU spore suspension of H2 + Millet seed Pythium aphanidermatum inoculum (0.5% w/w) |
| 3 | P. aphanidermatum + H3 | 10 mL 108 CFU spore suspension of H3 + Millet seed Pythium aphanidermatum inoculum (0.5% w/w) |
| 4 | P. aphanidermatum + Metalaxyl (chemical fungicides) | Millet seed Pythium aphanidermatum inoculum (0.5% w/w) + Metalaxyl (1%) |
| 5 | Symbiont Streptomyces isolate H2 | 10 mL 108 CFU spore suspension of H2 kg−1 sterilized soil |
| 6 | Symbiont Streptomyces isolate H3 | 10 mL 108 CFU spore suspension of H3 kg−1 sterilized soil |
| 7 | Negative control | 10 mL sterile water kg−1 sterilized soil |
| Bioassay | H2 | H3 |
|---|---|---|
| Catalase production | + | + |
| Gelatinase production | + | + |
| Amylase production | + | + |
| Citrate utilization | + | + |
| HCN 1 production | − | − |
| VOC 2 production | − | − |
| Deactivated by chloroform | + | − |
| Treatment No. | Treatment | %DI 1 | %APC 2 | %RDI 3 | MNL 4 | MPL 5 (cm) | MFSW 6 (g) | MDSW 7 (g) |
|---|---|---|---|---|---|---|---|---|
| 1 | P. aphanidermatum8 | 47 a | 100% | 0% | 1.9 c | 8 c | 0.8 c | 0.1 e |
| 2 | P. aphanidermatum + H2 | 31.3 ab | 66.6% | 33.4% | 4.4 b | 42.3 ab | 3.0 b | 0.3 cd |
| 3 | P. aphanidermatum + H3 | 21 b | 44.7% | 55.3% | 4.9 ab | 43.3 ab | 2.6 b | 0.2 de |
| 4 | P. aphanidermatum + Metalaxyl | 0 b | 0% | 100% | 5 ab | 40.4 ab | 2.6 b | 0.3 cd |
| 5 | Streptomyces strains H2 | 0 b | 0% | 100% | 6.8 a | 43.8 ab | 5.6 a | 0.6 a |
| 6 | Streptomyces strains H3 | 0 b | 0% | 100% | 6 ab | 46 a | 3.9 b | 0.5 a |
| 7 | NC 9 | 0 b | 0% | 100% | 5.5 ab | 40 ab | 3.4 b | 0.4 bc |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hassanisaadi, M.; Shahidi Bonjar, G.H.; Hosseinipour, A.; Abdolshahi, R.; Ait Barka, E.; Saadoun, I. Biological Control of Pythium aphanidermatum, the Causal Agent of Tomato Root Rot by Two Streptomyces Root Symbionts. Agronomy 2021, 11, 846. https://doi.org/10.3390/agronomy11050846
Hassanisaadi M, Shahidi Bonjar GH, Hosseinipour A, Abdolshahi R, Ait Barka E, Saadoun I. Biological Control of Pythium aphanidermatum, the Causal Agent of Tomato Root Rot by Two Streptomyces Root Symbionts. Agronomy. 2021; 11(5):846. https://doi.org/10.3390/agronomy11050846
Chicago/Turabian StyleHassanisaadi, Mohadeseh, Gholam Hosein Shahidi Bonjar, Akbar Hosseinipour, Roohollah Abdolshahi, Essaid Ait Barka, and Ismail Saadoun. 2021. "Biological Control of Pythium aphanidermatum, the Causal Agent of Tomato Root Rot by Two Streptomyces Root Symbionts" Agronomy 11, no. 5: 846. https://doi.org/10.3390/agronomy11050846
APA StyleHassanisaadi, M., Shahidi Bonjar, G. H., Hosseinipour, A., Abdolshahi, R., Ait Barka, E., & Saadoun, I. (2021). Biological Control of Pythium aphanidermatum, the Causal Agent of Tomato Root Rot by Two Streptomyces Root Symbionts. Agronomy, 11(5), 846. https://doi.org/10.3390/agronomy11050846







