Optimizing Biocontrol Activity of Paenibacillus xylanexedens for Management of Hairy Root Disease in Tomato Grown in Hydroponic Greenhouses
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Preparation of Bacterial Inocula
2.2. Greenhouse Experiments
2.3. Quantification of Rhizogenic Agrobacterium and Paenibacillus in Root Samples
2.4. Statistical Analysis
3. Results
3.1. Parameters Influencing the Biocontrol Activity of Paenibacillus xylanexedens
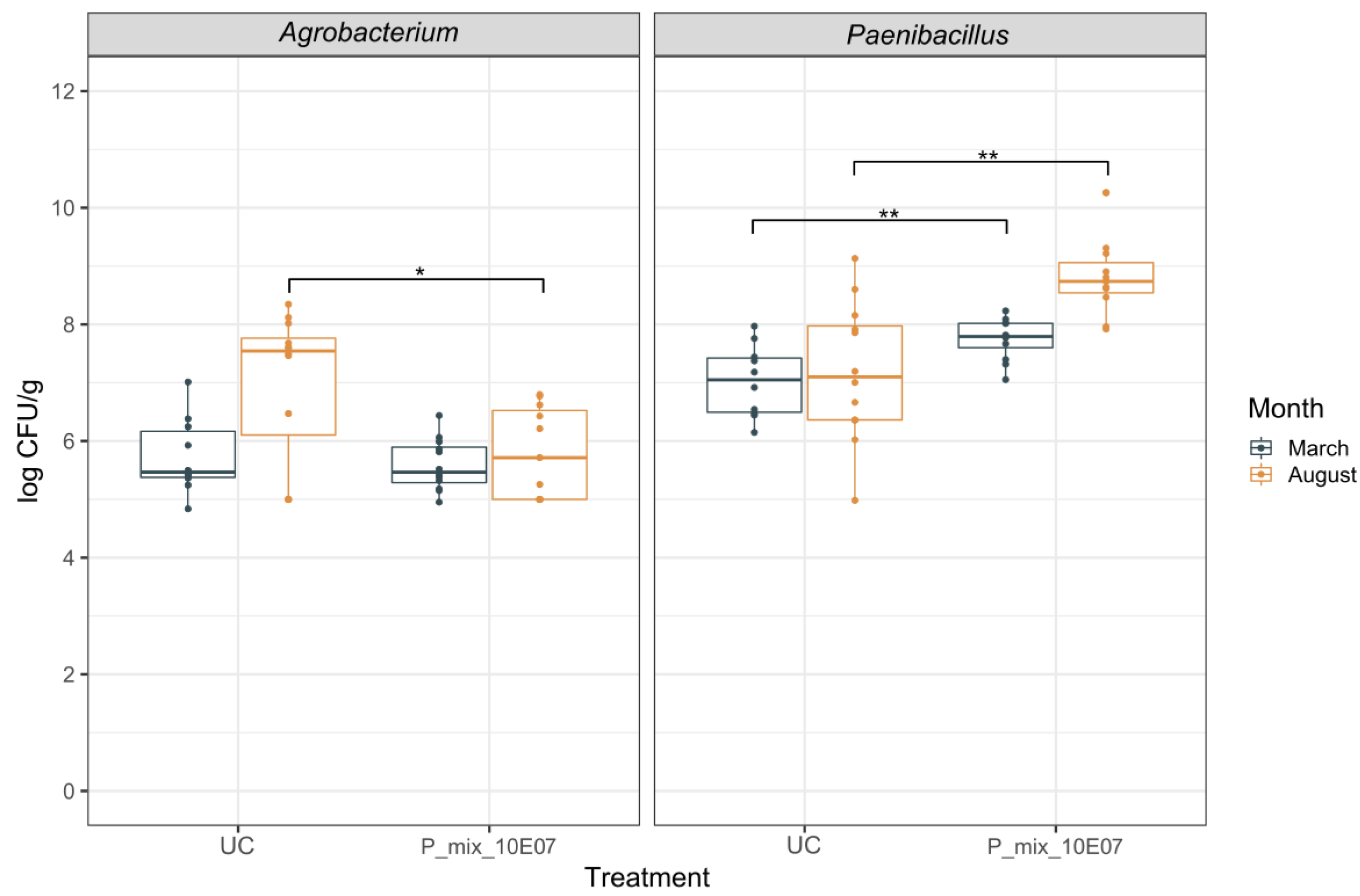
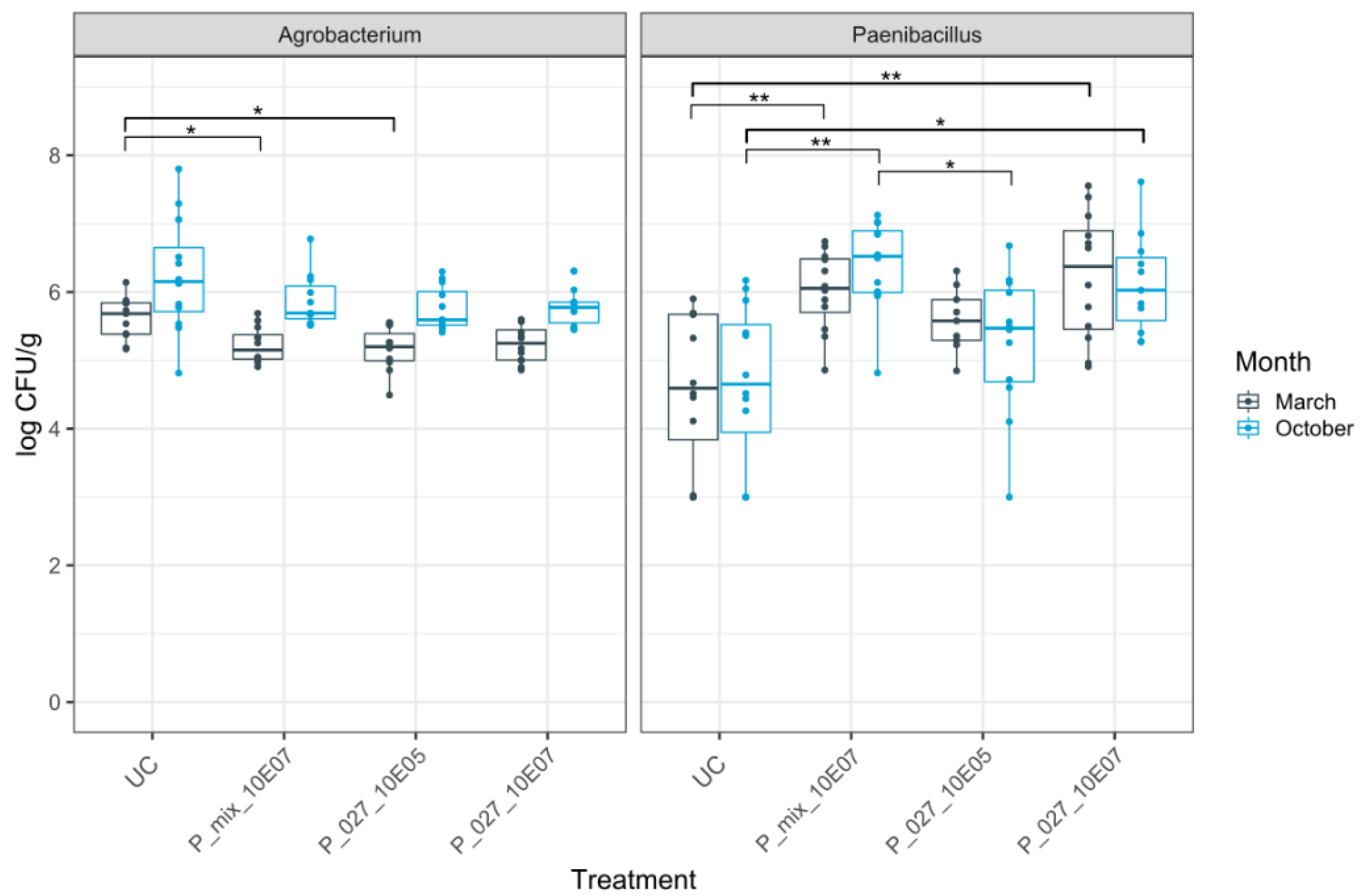
3.2. Dynamics of Rhizogenic Agrobacterium and Paenibacillus Populations in Infested and Non-Infested Rockwool Substrate
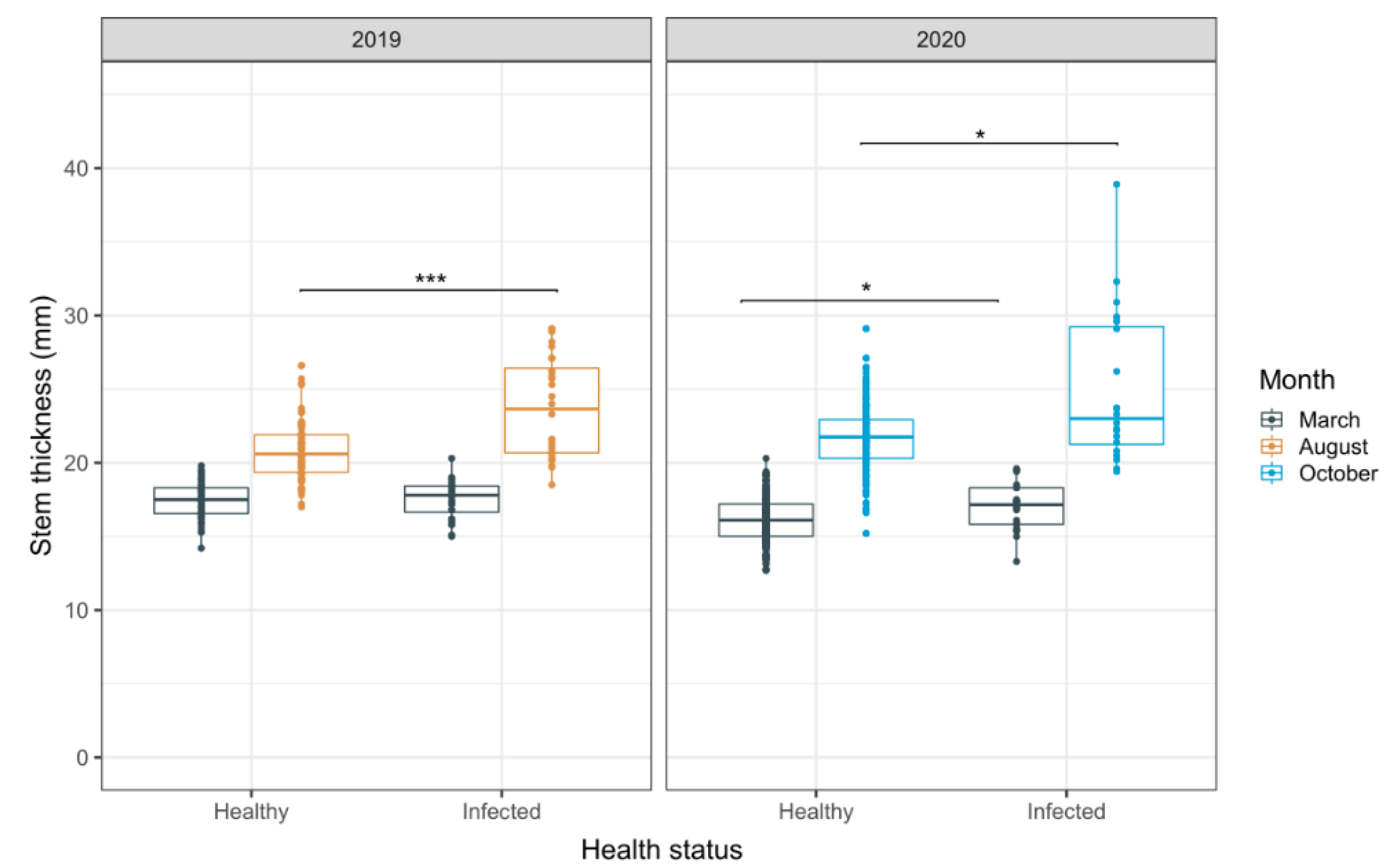
3.3. Evaluation of Stem Thickness as an Alternative Indicator for HRD
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bosmans, L.; Moerkens, R.; Wittemans, L.; De Mot, R.; Rediers, H.; Lievens, B. Rhizogenic agrobacteria in hydroponic crops: Epidemics, diagnostics and control. Plant Pathol. 2017, 66, 1043–1053. [Google Scholar] [CrossRef]
- Vargas, P.; Van Kerckhove, S.; Van Calenberge, B.; Bosmans, L.; Lievens, B.; Rediers, H. First Report of Hairy Root Disease, Caused by Rhizogenic Agrobacterium Biovar 1, in Hydroponic Bell Pepper Crop in South Korea. Plant Dis. 2020, 104, 968. [Google Scholar] [CrossRef]
- Weller, S.A.; Stead, D.E.; Young, J.P.W. Recurrent outbreaks of root mat in cucumber and tomato are associated with a monomorphic, cucumopine, Ri-plasmid harboured by various Alphaproteobacteria. FEMS Microbiol. Lett. 2006, 258, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Bosmans, L.; Álvarez-Pérez, S.; Moerkens, R.; Wittemans, L.; Van Calenberge, B.; Kerckhove, S.V.; Paeleman, A.; De Mot, R.; Rediers, H.; Lievens, B. Assessment of the genetic and phenotypic diversity among rhizogenic Agrobacterium biovar 1 strains infecting solanaceous and cucurbit crops. FEMS Microbiol. Ecol. 2015, 91, fiv081. [Google Scholar] [CrossRef] [PubMed]
- Hyakumachi, M.; Takahashi, H.; Matsubara, Y.; Someya, N.; Shimizu, M.; Kobayashi, K.; Nishiguchi, M. Recent studies on biological control of plant diseases in Japan. J. Gen. Plant Pathol. 2014, 80, 287–302. [Google Scholar] [CrossRef]
- Grady, E.N.; MacDonald, J.; Liu, L.; Richman, A.; Yuan, Z.-C. Current knowledge and perspectives of Paenibacillus: A review. Microb. Cell Factories 2016, 15, 203. [Google Scholar] [CrossRef]
- Lal, S.; Chiarini, L.; Tabacchioni, S. New insights in plant-associated Paenibacillus species: Biocontrol and plant growth-promoting activity. In Bacilli and Agrobiotechnology; Islam, T.M., Rahman, M., Pandey, P., Jha, C.K., Aeron, A., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 237–279. [Google Scholar]
- Padda, K.P.; Puri, A.; Chanway, C.P. Paenibacillus polymyxa: A prominent biofertilizer and biocontrol agent for sustainable agriculture. In Agriculturally Important Microbes for Sustainable Agriculture Volume 2 Applications in Crop Production and Protection; Meena, V.S., Mishra, P.K., Bisht, J.K., Eds.; Springer: Singapore, 2017; pp. 165–191. [Google Scholar]
- Sato, I.; Yoshida, S.; Iwamoto, Y.; Aino, M.; Hyakumachi, M.; Shimizu, M.; Takahashi, H.; Ando, S.; Tsushima, S. Suppressive Potential of Paenibacillus Strains Isolated from the Tomato Phyllosphere against Fusarium Crown and Root Rot of Tomato. Microbes Environ. 2014, 29, 168–177. [Google Scholar] [CrossRef]
- Zhang, F.; Li, X.-L.; Zhu, S.-J.; Ojaghian, M.R.; Zhang, J.-Z. Biocontrol potential of Paenibacillus polymyxa against Verticillium dahliae infecting cotton plants. Biol. Control 2018, 127, 70–77. [Google Scholar] [CrossRef]
- Rybakova, D.; Cernava, T.; Köberl, M.; Liebminger, S.; Etemadi, M.; Berg, G. Endophytes-assisted biocontrol: Novel insights in ecology and the mode of action of Paenibacillus. Plant Soil 2015, 405, 125–140. [Google Scholar] [CrossRef]
- Song, J.; Seo, H.-J. Antifungal Activity of Agro-Materials against Pear Scab (Venturia nashicola) and Pear Rust (Gymnosporangium asiaticum) Fungi. Res. Plant Dis. 2018, 24, 33–40. [Google Scholar] [CrossRef]
- Hong, C.E.; Kwon, S.Y.; Park, J.M. Biocontrol activity of Paenibacillus polymyxa AC-1 against Pseudomonas syringae and its interaction with Arabidopsis thaliana. Microbiol. Res. 2016, 185, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Bosmans, L.; De Bruijn, I.; Gerards, S.; Moerkens, R.; Van Looveren, L.; Wittemans, L.; Van Calenberge, B.; Paeleman, A.; Van Kerckhove, S.; De Mot, R.; et al. Potential for Biocontrol of Hairy Root Disease by a Paenibacillus Clade. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Bosmans, L.; Paeleman, A.; Moerkens, R.; Wittemans, L.; Van Calenberge, B.; Van Kerckhove, S.; De Mot, R.; Rediers, H.; Lievens, B. Development of a qPCR assay for detection and quantification of rhizogenic Agrobacterium biovar 1 strains. Eur. J. Plant Pathol. 2016, 145, 719–730. [Google Scholar] [CrossRef]
- Bosmans, L.; Lievens, B.; Rediers, H.; Raaijmakers, J.; De Bruijn, I. Biocontrol Organism. US Patent 2019/0357541 A1, 28 November 2019. [Google Scholar]
- Bosmans, L. Epidemiology, Ecology and Management of Rhizogenic Agrobacteria, Causative Agents of Hairy Root Disease in Vegetable Crops. Ph.D. Thesis, KU Leuven, Leuven, Belgium, September 2017. [Google Scholar]
- RSTudio Team. RStudio: Integrated Development for R; RStudio PBC: Boston, MA, USA, 2020. [Google Scholar]
- Patil, I. Ggstatsplot: ’ggplot2′ based plots with statistical details. CRAN 2018. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Montealegre, J.R.; Herrera, R.; Velasquez, J.C.; Silva, P.; Besoain, X.; Perez, L.M. Biocontrol of root and crown rot in tomatoes under greenhouse conditions using Trichoderma harzianum and Paenibacillus lentimorbus. Additional effect of solarization. Electron. J. Biotechnol. 2005, 8, 249–257. [Google Scholar] [CrossRef]
- Kim, Y.C.; Hur, J.Y.; Park, S.K. Biocontrol of Botrytis cinerea by chitin-based cultures of Paenibacillus elgii HOA73. Eur. J. Plant Pathol. 2019, 155, 253–263. [Google Scholar] [CrossRef]
- Hong, S.H.; Anees, M.; Kim, K.Y. Biocontrol of Meloidogyne incognita inciting disease in tomato by using a mixed compost inoculated with Paenibacillus ehimensis RS820. Biocontrol Sci. Technol. 2013, 23, 1024–1039. [Google Scholar] [CrossRef]
- Roselló, G.; Francés, J.; Daranas, N.; Montesinos, E.; Bonaterra, A. Control of fire blight of pear trees with mixed inocula of two Lactobacillus plantarum strains and lactic acid. J. Plant Pathol. 2017, 99, 111–120. [Google Scholar]
- De Vrieze, M.; Germanier, F.; Vuille, N.; Weisskopf, L. Combining Different Potato-Associated Pseudomonas Strains for Improved Biocontrol of Phytophthora infestans. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Eke, P.; Wakam, L.N.; Fokou, P.V.T.; Ekounda, T.V.; Sahu, K.P.; Kamdem Wankeu, T.H.; Boyom, F.F. Improved nutrient status and Fusarium root rot mitigation with an inoculant of two biocontrol fungi in the common bean (Phaseolus vulgaris L.). Rhizosphere 2019, 12, 100172. [Google Scholar] [CrossRef]
- Matos, A.; Garland, J.L. Effects of Community Versus Single Strain Inoculants on the Biocontrol of Salmonella and Microbial Community Dynamics in Alfalfa Sprouts. J. Food Prot. 2005, 68, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Vanlommel, W.; Moerkens, R.; Bosmans, L.; Lievens, B.; Rediers, H.; Wittemans, L.; Van Calenberge, B.; Paeleman, A.; Van Kerckhove, S. Management of hairy root disease in protected tomato crops: A biological and chemical approach. Acta Hortic. 2020, 41–50. [Google Scholar] [CrossRef]
- Langendries, S.; Goormachtig, S. Paenibacillus polymyxa, a jack of all trades. Environ. Microbiol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Vargas, P.; Bosmans, L.; Van Calenberge, B.; Van Kerckhove, S.; Lievens, B.; Rediers, H. Bacterial community dynamics of tomato hydroponic greenhouses infested with hairy root disease (HRD). FEMS Microb. Ecol. Under review.

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vargas, P.; Bosmans, L.; Van Kerckhove, S.; Van Calenberge, B.; Raaijmakers, J.M.; Lievens, B.; Rediers, H. Optimizing Biocontrol Activity of Paenibacillus xylanexedens for Management of Hairy Root Disease in Tomato Grown in Hydroponic Greenhouses. Agronomy 2021, 11, 817. https://doi.org/10.3390/agronomy11050817
Vargas P, Bosmans L, Van Kerckhove S, Van Calenberge B, Raaijmakers JM, Lievens B, Rediers H. Optimizing Biocontrol Activity of Paenibacillus xylanexedens for Management of Hairy Root Disease in Tomato Grown in Hydroponic Greenhouses. Agronomy. 2021; 11(5):817. https://doi.org/10.3390/agronomy11050817
Chicago/Turabian StyleVargas, Pablo, Lien Bosmans, Stefan Van Kerckhove, Bart Van Calenberge, Jos M. Raaijmakers, Bart Lievens, and Hans Rediers. 2021. "Optimizing Biocontrol Activity of Paenibacillus xylanexedens for Management of Hairy Root Disease in Tomato Grown in Hydroponic Greenhouses" Agronomy 11, no. 5: 817. https://doi.org/10.3390/agronomy11050817
APA StyleVargas, P., Bosmans, L., Van Kerckhove, S., Van Calenberge, B., Raaijmakers, J. M., Lievens, B., & Rediers, H. (2021). Optimizing Biocontrol Activity of Paenibacillus xylanexedens for Management of Hairy Root Disease in Tomato Grown in Hydroponic Greenhouses. Agronomy, 11(5), 817. https://doi.org/10.3390/agronomy11050817






