Abstract
Plant growth-promoting rhizobacteria are often utilized to improve crop health and productivity. Nevertheless, their positive effects can be hindered if they fail to withstand the environmental and ecological conditions of the regions where they are applied. An alternative approach to circumvent this problem is a tailored selection of bacteria for specific agricultural systems. In this work, we evaluated the plant growth promoting and pathogen inhibition activity of rhizobacteria obtained from the rhizosphere of Mariola (Solanum hindsianum), an endemic shrub from Baja California. Eight strains were capable of inhibiting Fusarium oxysporum in vitro, and thirteen strains were found to possess three or more plant-growth-promotion traits. Molecular identification of these strains, using 16 s rRNA partial sequences, identified them as belonging to the genera Arthrobacter, Bacillus, Paenibacillus, Pseudomonas, and Streptomyces. Finally, the effect of selected plant growth-promoting rhizobacteria (PGPR) strains on the growth and suppression of Fusarium wilt in tomato was evaluated. Results showed that these strains improved tomato plants growth under greenhouse conditions and reduced Fusarium wilt effects, as reflected in several variables such as length and weight of roots and stem. This work highlights the potential of native plants related to regionally important crops as a valuable source of beneficial bacteria.
1. Introduction
Microbial communities from the soil are known to influence plant health and productivity and provide other critical ecosystemic services [1,2,3]. Plant growth-promoting rhizobacteria (PGPR) induce plant growth and protect plants against different pathogens through various mechanisms. Direct mechanisms include improving the availability of nutrients in the soil through the solubilization of minerals like phosphorus, potassium, and zinc or the production of phytohormones like indole-acetic acid, gibberellins, and ethylene. Indirect mechanisms involve disease reduction through antibiotic and siderophore production, competition for nutrients or space, and stimulation of resistance mechanisms [4,5,6]. Because of these properties, PGPR are an alternative for use in agriculture as biofertilizers and biological control agents, and their application has been shown to reduce the use of chemicals and minimize soil, air, and water pollution [7]. Soil and plant-associated bacteria belonging to the genera Bacillus, Burkholderia, Paenibacillus, Pseudomonas, and Streptomyces, have often been shown to act as PGPR and are commonly used in commercial formulations in agriculture [8]. However, the inoculation of crops with commercial PGPR often fails, as it does not produce the desired effects in growth promotion or disease control in plants [9]. This observation has been explained by the failure of microorganisms to adapt to the physical and chemical characteristics of the new location or to integrate into the local microbiome, all of which can be very different from the original conditions from which they were initially isolated [10]. Moreover, to establish a functional population on the roots of a new crop, bacteria must be compatible with the host root exudates [11]. These facts highlight the importance of inoculum selection, which ideally should be tailored for each specific soil and crop combination [12]. In this regard, the use of region-specific microbial strains has been shown to achieve higher yields, probably because of their adaptation to local ecological and environmental conditions [13,14].
The State of Baja California, Mexico, has a territorial extension of 71,445.9 km2, with an average temperature of 19 °C and a predominantly dry climate with 200 mm of total annual precipitation [15]. The ground cover consists of around 500 species of xerophilous shrubs, of which 20% are endemic. Until the 1950s, the extreme environmental conditions kept human presence at low levels, preserving the ecosystem. However, in recent decades there has been rapid population growth in the urban centers, and simultaneously some areas along the Pacific coast have been developed for intensive agricultural production. The state devotes 243,210.00 ha of land to agriculture, where up to 78 crops are grown. The main vegetable produced is red tomato (Solanum lycopersicum). Since 2009 this crop accounted for at least 22.4% of the value of total agricultural production in Baja California [15]. Tomato is a vulnerable crop; it requires constant care due to the attack of pests and pathogens, being tomato Fusarium wilt one of the most destructive diseases of this crop. It is caused by Fusarium oxysporum f. sp. lycopersici, a phytopathogenic fungus that penetrates the epidermis of the root, spreading through the vascular tissue, and colonizes the xylem of the plant obstructing it. The symptoms observed in infected individuals include growth reduction, wilting, yellowing of leaves and stems, defoliation, marginal leaf necrosis, and vascular necrosis [16,17]. The treatment of fusarium wilt, and other diseases and pests, requires the intensive application of agrochemicals like fungicides, insecticides, and foliar fertilizers [18]. Adverse effects from exposure to these compounds have already been demonstrated on the health of laborers [19], and in the quality of urban and agricultural water sources, through contamination [20,21]. These environmental and human costs, together with the shift of consumer preferences to sustainable agricultural practices [15], suggest the need for the development of more sustainable crop management alternatives.
Therefore, in recent years growers have adopted more environmentally friendly agricultural production methods such as organic agriculture that use low inputs, including beneficial microorganisms. These production methods require knowledge of the conservation and management of microbial biodiversity and the search for biological strategies for fertilization and crop protection tailored for the physical conditions of the environment where they are produced. In this sense, the feasibility of increasing agricultural ecosystems’ productivity through the use of native beneficial microorganisms has been proposed.
The potential of plants to adapt to different environmental conditions is highly dependent on their association with soil microorganisms [1,3]. This relationship is complementary because the structure of soil microbial communities is influenced by the exudates produced by roots and their composition, and the nutrients present in them [10,22,23]. The traditional approach for the use of PGPR in crop production is to apply one organism, or a mixture of organisms, to different crops growing under different climate and soil conditions. This approach produces mixed results because often microorganisms fail to establish a successful interaction with a new crop or to adapt to the physical and chemical characteristics of the soil, and even if a functional population establishes the growth-promoting ability may be highly specific to plant species, cultivars, and soil [24]. Previously, rhizobacteria associated with native plants from arid zones in Mexico have been isolated and characterized for plant growth-promoting traits. For example, strains identified as Rhizobium, Bacillus, and Streptomyces were isolated from the salinity resistant succulent Salicornia and Cactaceae like Opuntia, Mammillaria, and Stenocereus [25,26]. Moreover, in Tunisia, endophytic bacteria from the exotic Solanaceae S. elaeagnifolium were shown to reduce tomato fusarium wilt [27]. The Solanaceae Solanum hindsianum, commonly known as Mariola or Hinds Nightshade, is a perennial shrub native to the Sonoran Desert region of northern Mexico. It is widely distributed throughout the Baja California peninsula, where it can be found in coastal scrub and dunes near the shore, grows and blooms well in arid conditions, and has significant drought tolerance [28,29]. Given its environmental adaptation and that it belongs to the same taxonomic family of tomato, we considered that S. hindsianum could be a valuable source of PGPR biological control agents to be used in the sustainable production of this crop. We prospected soil associated with Solanum hindsianum plants growing at different locations of the Baja California Peninsula, obtaining a collection of more than 300 isolates. Next, isolates were screened for plant growth promotion-related traits and their ability to inhibit Fusarium oxysporum in vitro. Finally, greenhouse assays demonstrated that selected isolates had a significant positive effect on plant growth and reduced the effect of F. oxysporum on tomato plants.
2. Materials and Methods
2.1. Rhizosphere soil Sampling and Isolation of Solanum Hindsianum Rhizospheric Bacteria
Samples were collected from 12 sites along the Baja California peninsula, encompassing Baja California and Baja California Sur (Figure 1). Five specimens of S. hindsianum were located at each site, avoiding those with other adjacent plant species. One soil and roots sample was collected per plant at a depth of 20–30 cm and then pooled in a composite sample per site. Soil adhering to the roots was recovered. A working sample was prepared for each sampling site by resuspending 200 mg of bulked rhizosphere soil in 500 μL sterile distilled water using a vortex mixer. Subsequently, ten-folded serial dilutions in sterile water were prepared from each working sample, and 100 μL of dilutions 1:100, 1:1000, and 1:10,000 were streaked on culture plates containing LB Agar, YPD Agar, King medium B Agar (ATCC media No.1065, 1245, and 1213, respectively) and PY medium (tryptone 5.0 g, yeast extract. 3.0 g, CaCl2 0.9 g, pH 6.8 [30]. All the media were supplemented with cycloheximide (1 μg/mL) to prevent fungal growth. Streaked plates were incubated at 28 °C and monitored for colony formation for up to one week. The resulting colonies were examined morphologically for their shape, appearance, texture, color, and pigment secretion. Colonies with distinct morphological characteristics were selected and isolated by subculturing three times on PY and subsequently stored in a 35% glycerol solution at −20 °C and −80 °C for short- and long-term storage, respectively.
Figure 1.
Localization of sampling sites.
2.2. In Vitro Antifungal Activity against Fusarium Oxysporum f. sp. Lycopersici
All isolated bacterial strains were tested for antagonistic activity in triplicate. For the qualitative determination of antifungal activity, glycerol stocks were inoculated in PY liquid medium and incubated for 48 h at 30 °C and 110 rpm in a shaker incubator. F. oxysporum f. sp. lycopersici race 1 mycelium was obtained from the collection of Centro de Investigacion Cientifica y de Educacion Superior de Ensenada (CICESE) plant pathology laboratory. The fungus was grown for five days in PDA (ATCCTM medium No. 336), and one plug of 5 mm of diameter was taken from the border of placed on the center of a fresh PDA plate. When the fungus reached 1 cm in diameter, 5 µL of four bacterial cultures were inoculated at the edges, and incubated for seven days at 25 °C
The quantitative determination of antifungal activity was measured in triplicate for all strains that tested positive in the qualitative assay. Briefly, the fungus was placed at the Petri plate’s edge, and only one bacterial strain was inoculated directly opposed to it. The formula: %inhibition = [(R r)/R] × 100 where R is the radius of fungal growth of the control treatment, and r is the radius of fungal growth towards the bacterial treatment [3,31] was used to calculate the percentage of inhibition of mycelial growth. Two variations of inhibition were considered: those in which the strain was motile, covering the surface of the plate while limiting fungal growth, and those in which the strain was static and created an inhibition halo around itself.
2.3. In Vitro Screening of Plant Growth Promoting Traits
Glycerol stocks were inoculated in 96-tube T100 Biotube™ racks (Simport Scientific, Saint-Mathieu-de-Beloeil, QC, Canada) containing 1.5 mL PY liquid medium per tube. The cultures were grown for 48 h at 30 °C and 100 rpm in a shaker incubator. Unless otherwise stated, each colorimetric test was implemented in 96-well microplates. For solid test-media, 300 µL were added per well, and for liquid test-media, 200 µL were added. Inoculum of bacteria was adjusted to a concentration of ~1 × 104 CFU and inoculation was performed using a 48-tip metal replicator.
2.3.1. Phosphate Solubilization Assay
Inorganic phosphate solubilization was evaluated on modified Pikovskaya agar (0.5 g/L yeast extract, 10 g/L glucose, 5 g/L Ca3(PO4)2, 0.5 g/L (NH4)2SO4, 0.2 g/L KCl, 0.1 g/L MgSO4, 0.1 mg/L MnSO4, 0.1 mg/L FeSO4, 10 mg/L of bromocresol purple, 15 g/L agar, pH 7.2). Microplates were incubated in the dark for 72 h at 30 °C. The change in color from purple to yellow indicated a positive result [32,33]
2.3.2. Potassium Solubilization Assay
Inorganic potassium solubilization was determined on modified Pikovskaya agar prepared following the previous recipe [34], but using KNO3 instead of Ca3(PO4)2 and bromocresol green instead of bromocresol purple. Microplates were incubated in the dark for 72 h at 30 °C. The change in color from blue to yellow indicated a positive result.
2.3.3. Zinc Solubilization Assay
Zinc solubilization was assessed using Zinc-supplemented Pikovskaya medium prepared following the previous recipe complemented with 1.2 g/L ZnO instead of Ca3(PO4)2 substituting bromocresol purple with bromothymol blue. Microplates were incubated in the dark for 72 h at 30 °C. The change in color from blue to yellow indicated a positive result [35].
2.3.4. Indole Acetic Acid Production Assay
For the qualitative determination of indole-3-acetic acid (IAA) production, glycerol stocks were re-inoculated in PY liquid medium supplemented with 500 µg/mL tryptophan. Microplates were incubated for 48 h at 30 °C and 110 rpm in a shaker incubator. Afterward, 100 µL of Salkovsky reagent (50 mL, 35% of HClO4, 1 mL 0.5 M FeCl3) [36] were added per well, and cultures were incubated at room temperature for 30 min. The change of color to pink indicated a positive result.
For the quantitative determination of IAA production, three replicates of each strain were inoculated at a concentration of 1 × 106 CFU/mL in 3 mL of PY liquid medium supplemented with 500 μg/mL tryptophan. Cultures were incubated for 48 h at 30 °C and 110 rpm in a shaker incubator. Subsequently, the culture was centrifuged at 10,000 rpm for 3 min, and 200 µL of the supernatant were transferred to 96-well microplates. Next, 100 µL of Salkovsky reagent was added to each well, and the optical density was measured at 540 nm in a Multiskan Sky Microplate Spectrophotometer. Finally, IAA concentrations were calculated against a calibration curve (100, 50, 20, 10, and 5 mg/mL IAA) using the Thermo Scientific Skanlt PC software.
2.3.5. Hydrogen Cyanide Production Assay
After inoculating the bacteria in py, the microplate was covered with filter paper moistened with a solution of 0.5% sodium carbonate in 0.5% picric acid [36]. The plates were subsequently sealed with parafilm and incubated in the dark at 30 °C for 48 h; the development of orange-red color indicated a positive result.
2.3.6. ACC Deaminase Assay
The 1-Aminocyclopropane-1-carboxylate (ACC) deaminase activity was evaluated in DF minimal media (4.0 g/L KH2PO4, 6.0 g/L Na2HPO4, 0.2 g/L MgSO4·7H2O, 2.0 g/L glucose, 2 g/L gluconic acid, 2.0 g/L citric acid, pH 7.2) amended with 1 mL/L of a trace element solution (1 mg/L FeSO4·7H2O, 10 mg/L H3BO3, 11.19 mg/L MnSO4·H2O, 124.6 mg/L ZnSO4·7H2O, 78.22 mg/L CuSO4·5H2O, and 10 mg/L MoO3) and 3.0 mM ACC [37].
2.3.7. Microplate Biofilm Formation Assay
Three replicates of each strain were inoculated at a concentration of 1 × 106 CFU/mL in 300 µL of PY. Then, the microplates were incubated at 30 °C and 50 rpm for 48 h in a shaker incubator. Afterward, 75 μL of 10% crystal violet were added to each well, and plates were left to stand for 15 min at room temperature, followed by three washes with distilled water. The formation of a violet ring indicated the formation of biofilm [38,39].
2.3.8. Siderophores Production Assay
Chrome azurol S agar (CAS) solid medium [40] was prepared as described previously [41]. After inoculation, the plates were incubated in the dark for 96 h at 30 °C. The change in color from blue to yellow indicated a positive result.
2.3.9. Chitinase Production Assay
Basal detection medium (0.3 g/L MgSO4·H2O, 3 g/L NH4 (SO4), 2 g/L KH2PO4), 1 g/L citric acid, 15 g/L agar, 0.2 g/L Tween-80, pH 4.7) was supplemented with 4.5 g/L colloidal chitin and 0.15 g/L bromocresol purple [42]. After inoculation, the plates were incubated in the dark for 48 h at 30 °C. The change in color from yellow to purple indicated a positive result.
2.4. Effect of Rhizobacteria Inoculation on Tomato Growth
Seeds of the Bonny Best cultivar [43] were surface-sterilized by sequential immersions in 70% (v/v) ethanol for 3 min and 2.0% (v/v) NaOCl for 2 min, followed by three rinses with sterile water. Sterile seeds were grown, in a greenhouse, for 35 days on Cosmopeat substrate (Cosmocel SA, San Nicolás de los Garza N.L., Mexico) in 72-cavity germination trays. After the initial growth period, seedlings with an average height of 5.8 cm with the appearance of a true leaf were selected. To prepare the bacterial inoculums, 25 mL of PY were inoculated from overnight pre-cultures of the appropriate strains adjusted to A600 of 0.05 and incubated at 30 °C and 110 rpm in a shaker. When cultures reached mid-log phase (~A600 of 0.02) they were used to inoculate sterile water to a concentration of ~2 × 104 CFU/mL in a final volume of 50 mL.
The seedlings were inoculated through a 10-s immersion of the root ball in 50 mL of inoculum solution (2 × 104 CFU/mL) and transplanted to 6 inches plastic pots filled with Cosmopeat substrate. Each treatment consisted of the individual inoculation of each of the selected strains. Two negative controls were included in the assay; in the first one, the root ball was submerged in sterile water, and in the second in an inoculum solution (2 × 104 CFU/mL) of the commercial product Bacillioss (Bioamin SA, Coah, Mexico), a commercial Bacillus subtillis-based biological product. Eight replicates were prepared for each treatment, and pots were arranged in the greenhouse in a completely randomized design and kept under semi controlled conditions at an average temperature of 24 °C in the day and 20 °C at night. Eight days after the first inoculation, a booster inoculation was administered through 20 mL of inoculum solution (2 × 104 CFU/mL) applied directly to the substrate near the plant stem. Fifty days after sowing, plants were harvested, and stem and root length, diameter, fresh and dry weight, and thickness at the plant stem base were assessed. Comparisons between treatments were performed through a one-way ANOVA and Tukey’s post-hoc test using SPSS version 25 (IBM). Data was tested for compliance with ANOVA assumptions. All results were considered significant at p ≤ 0.05.
Effect of Rhizobacteria Inoculation on Tomato Fusarium Wilt
Experimental design, seed germination, and rhizobacteria inoculation were performed as described for the in-vivo plant growth promotion assays. The evaluated treatments consisted of a single bacterial strain applied in plants inoculated, or not, with F. oxysporum f. sp. lycopersici race 1, as control the commercial Bacillus subtillis-based biological product Bacillioss (Bioamin SA, Mexico) was used, for the negative control sterile distilled water was used. The first application of bacteria was done 35 days after sowing with a second inoculation after eight days. F. oxysporum f. sp. lycopersici race 1 was inoculated a concentration of ~1 × 106 CFU when plants were 43 days. The fungal inoculum was obtained by adding five mycelium discs to a moistened substrate composed of sorghum seeds (20.0 g) with vermiculite (5.0 g) and incubated for 10 days at 30 °C. Inoculum was placed through three holes in the substrate near the plant stem. After inoculation plants were kept under greenhouse with semi controlled conditions at an average temperature of 24 °C in the day and 20 °C at night.
Sixty days after sowing, plants were harvested, and stem and root length, diameter, fresh and dry weight, and thickness at the plant stem base as well as the presence or absence of symptoms such as root necrosis, leaf chlorosis, and crown necrosis were assessed. For the re-isolation of F. oxysporum from tomato plants, fragments of roots and the crown were flame-sterilized and inoculated in PDA. Comparisons between treatments were performed through a one-way ANOVA using SPSS version 25 (IBM). Data was tested for compliance with ANOVA assumptions. All results were considered significant at p ≤ 0.05.
2.5. Molecular Identification of Bacterial Strains
Strains with a minimum Fusarium inhibition in vitro percentage of 50% for motile and 15% for static, or that showed a positive result for at least four of the evaluated Plant growth promoting (PGP) traits, were identified through the sequencing of the 16S rRNA gene as follows. Genomic DNA was purified using the Gentra Puregen kit (Qiagen, Cary, NC, USA), following the manufacturer’s instructions. The 16S rRNA gene was PCR-amplified using the 27F (5-AGAGTTTGA TCMTGGCTCAG-3′) and 1492R (5′TACGGYTACCTTGTTACGACTT-3′) primers [44]. Each PCR reaction was carried out in a total volume of 25 µL, containing 1 µL of genomic DNA (50 µg/µL), 2.5 µL of Taq Buffer10×, 0.5 µL of dNTP mix (10 mM), 0.5 µL of each primer (10 µM ), 0.2 µL of Taq DNA polymerase (1U/μL) (Thermo Fisher, Waltham, MA, USA) and 19.8 μL of distilled water. Amplification was carried out in a Bio-Rad T-100 thermocycler, under the following conditions: an initial denaturation step of 95 °C for 3 min, followed by 30 cycles of 95 °C 30 s, 48 °C 30 s, 72 °C 1 min, and a final step of 72 °C 10 min. PCR products were verified by electrophoresis on a 1% agarose gel. Those that showed the expected size and quality were purified using the GenElute™ PCR Clean-Up Kit (Sigma-Aldrich, St. Louis, MI, USA) and sent to Eton Biocience Inc. for sequencing with primers 27F and 1492R. The resulting sequences were manually checked in GeneStudio™ Pro v2.2 (GeneStudio Inc., Suwanee, GA, USA). Preliminary taxonomic assignations were obtained using the Ribosomal Database Project Classifier [45]. To increase quality and coverage, the 16s rRNA gene of strains selected for in planta assays was sequenced twice. Next, the edited sequences were blasted against the GenBank 16S Ribosomal RNA sequences database, and the closest matches were used to construct the alignment using MEGA X [46] and the multiple alignment program MUSCLE. A Maximum-Likelihood tree was constructed using Kimura’s two-parameter model with Gamma distribution and Bootstrap values based on 1000 replicates. Sequences were deposited in GenBank (accession numbers MW391472 to MW391481).
3. Results
3.1. Antagonistic Effect of Mariola Rhizobacteria against F. oxysporum In Vitro
In total, 343 bacterial strains with distinct characteristics were obtained from the soil samples. The most significant proportion came from the M-10 sample (20.40%), which was collected in El Vizcaíno Biosphere Reserve in Baja California Sur. In comparison, the lowest number of strains were obtained from samples M-2 (3.79%) located near the Picacho Prieto hill in Baja California (Figure 1 and Table S1). All strains were able to grow in solid and liquid PY media regardless of the solid media they were initially isolated in.
First, the 343 strains were screened in vitro for antagonistic activity against F. oxysporum. We evaluated inhibition considering two variations: those in which the strain was motile, covering the surface of the plate while limiting fungal growth, and those in which the strain was static and created an inhibition halo around itself, probably by the action of a diffusible antifungal compound (Figure S1). A total of 44 strains were found to inhibit F. oxysporum in the qualitative tests (Table S1) and were selected for quantitative evaluation. We established an arbitrary minimum cut-off for the inhibition percentage of 50% for motile strains and 15% for static strains. Eight strains met the minimum values and were selected for molecular identification, six were motile, and two were static (Table 1). The initial taxonomic assignment indicated that the first belonged to the genera Bacillus, while the latter were identified as Glutamicibacter and Streptomyces (Table 1 and Figure 2).

Table 1.
Plant growth promotion associated traits of selected strains.

Figure 2.
Maximum-Likelihood tree of partially sequenced 16S rRNA gene. Arrows indicate strains described in this work. Values above nodes are bootstrap values obtained from 1000 replicates.
3.2. In Vitro Identification of Mariola Associated Rhizobacteria with Plant Growth-Promoting Traits
The 96-well microplate format provided a fast and reliable way to screen the 343 strains simultaneously for plant growth promotion traits (Figure S2). First, we evaluated potassium and phosphate solubilization and indoleacetic acid, chitinases, siderophores, and hydrogen cyanide production. Although several strains showed a positive result in at least one test (Table S1), only 19 showed a positive result for at least four of the evaluated traits and were selected for molecular identification (Table 1).
The taxonomic assignment indicated that these belong to seven genera of bacteria: Chryseobacterium, Bacillus, Lysinibacillus, Microbacterium, Paenibacillus, Pseudomonas, and Staphylococcus (Table 1 and Figure 2), that are commonly associated with plant growth promotion. Remarkably, strain rbES182, belonging to the genera Bacillus, was selected in both the PGPR and the antifungal activity screenings. This motile strain, which tested positive for IAA and siderophore production and potassium and phosphate solubilization, showed an inhibition percentage of 50% against F. oxysporum. The strain was isolated from the Town of Santa Agueda in Baja California Sur.
To further characterize the fungal antagonistic and the putative plant growth-promoting strains, IAA production was quantified, and additional plant growth promotion traits like zinc solubilization, biofilm formation, and ACC deaminase activity were evaluated. Only strain rbES182 tested positive for all the assays, although it produced the third-lowest level of IAA under our experimental conditions. Strains Pseudomonas sp. rbES090 and Bacillus sp. rbES015 produced the highest and lowest levels of IAA, respectively (Table 1).
3.3. Inoculation of Tomato with Mariola Associated Rhizobacteria Increases Plant Root and Aerial Biomass
Altogether, the results from the in vitro evaluations suggested that our strains could act as PGPR but did not allow us to pinpoint those strains with higher potential to promote plant growth. Therefore, we performed a greenhouse in vivo assay using Bacillus sp. strains rbES015 and rbES182, Pseudomonas sp. strains rbES061, rbES087and rbES090, and Paenibacillus sp. rbES075. Our results showed that all seven strains elicited a strong positive effect on S. lycopersicum growth (Figure S3). For the aerial section, inoculated plants showed an increase, when compared with the control, of up to160%, 290%, 380%, and 420% in fresh weight, dry weight, stem length, and thickness (Figure 3A–D). Interestingly, although root length was unaffected (Figure 3E), we observed up to a 620% and 450% increase in root fresh and dry weight for all treatments when compared to the negative controls (Figure 3F,G). These results are consistent with an increase in root biomass, which can be explained by an increase in secondary root growth. For all the treatments, the bacterial strains were successfully re-isolated (data not shown).
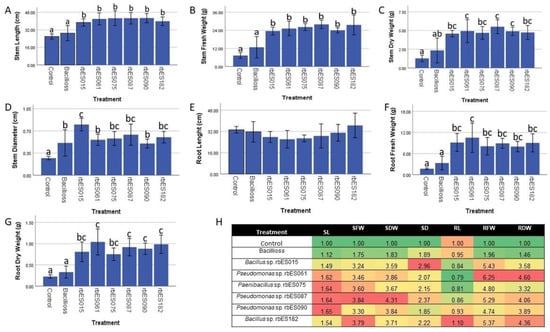
Figure 3.
Effect of rhizobacteria inoculation on S. lycopersicum growth. (A–G) Effect of treatment of tomato plants with rhizobacteria strains on different plant growth indicators. Bars represent the mean of 8 plants ± SD per treatment. Different letters over bars indicate significant differences (p < 0.05). (H) Fold change of evaluated variables of plants inoculated with different strains when compared to the negative control. Stem length (SL), stem fresh weight (SFW), stem dry weight (SDW), stem diameter (SD), root length (RL), root fresh weight (RFW), root dry weight (RDW).
3.4. Mariola Associated Rhizobacteria Decrease the Effects of Fusarium on Tomato Growth
Finally, we tested in vivo the effect of antagonist strains on Fusarium of the two mobile strains with the higher Fusarium inhibition in vitro percentage (rbES245 and rbES262), the only two static strains with a Fusarium inhibition in vitro percentage higher than 15% (rbES158 and rbES331), and the only strain that was selected on both, the plant growth promotion and the pathogen inhibition screenings (rbES182). Our results revealed significant variation in plant growth, depending on the tested strains. Untreated plants and plants treated with Bacillus sp. rbES182 showed a consistent and significant decrease in all evaluated plant traits when inoculated with F. oxysporum f. sp. lycopersici (Figure 4F). Plants treated with Bacillus sp. strains rbES182 and rbES245, and the commercial product Bacillioss showed inconsistent results regarding root traits (Figure 4E–G) but exhibited a significant decrease in stem fresh and dry weight when inoculated with F. oxysporum (Figure 4B,C), previously it has been shown that these two parameters correlate negatively to Fusarium wilt severity [27]. On the other hand, plants treated with Bacillus sp. rbES262 were not significantly different for all the measured traits, whether or not inoculated with the pathogen (Figs. 4 and S4). Moreover, when the plants inoculated with F. oxysporum were assessed for Fusarium wilt symptoms, only those treated with isolate rbES262 were asymptomatic (Table 2). Altogether, these results suggest that the treatment with strain rbES262 effectively decreased the effects of Fusarium wilt on plant growth in inoculated plants.

Figure 4.
Effect of rhizobacteria inoculation on Fusarium wilt in tomato. Effect on stem length (A), stem fresh weight (B), stem dry weight (C), stem diameter (D), root length (E), root fres weight (F), and root dry weight (G) in plants treated with different rhizobacterial strains and inoculated (red bars) or not (blue bars) with F.oxysporum.Bars represent the mean of 8 plants ± SD per treatment. An asterisk over a pair of bars indicates significant differences (p < 0.05).

Table 2.
Fusarium wilt symptoms observed in plants from the in vivo pathogen suppression assays.
Interestingly, plants treated with Streptomyces sp. rbES158 and Glutamicibacter sp. rbES331 showed a high mortality rate as 33% and 66%, respectively, died during the experiment (Table 2). The mortality rate for plants treated with other strains was 0%; this suggested that these strains affected the plant’s health directly and were dropped from the assay. For all the treatments, the bacterial strains and F. oxysporum were successfully re-isolated.
4. Discussion
In this work, we have shown that S. hindsianum is a potential source of bacteria with plant-growth promoting and pathogen inhibition features that could be used as a tailored alternative for the production of this legume in semi-arid regions.
The molecular identification indicated a close relationship to previously identified PGPR in different crops, mainly Pseudomonas and Bacillus [47,48,49,50,51]. Overall, these are congruent with a recent work that characterized the bacterial community’s structure from the rhizosphere of tomato plants growing in arid regions at the south of the Baja California peninsula. It was found that the most abundant families were the Pseudomonadaceae, Enterobacteriaceae, and Bacillaceae and that the most represented genera were Pseudomonas and Bacillus [52]. Historically, these genera were among the first bacterial isolates to show promising plant growth promotion and biocontrol characteristics. Both genera promote plant growth through the synthesis and release of phytostimulants such as IAA, ABA, and cytokinins [24]. Pseudomonads produce diverse compounds with antimicrobial activity, such as phenazine-1-carboxylic acid, 2,4-diacetylphloroglucinol (DAPG), pyrrolnitrin, and pyoluteorin [53]. Moreover, they often play critical roles in several characterized suppressive soils. For example, DAPG producing fluorescent Pseudomonas sp. play a prominent role in the development of Take-all decline [54]. Also, several strains of Bacillus synthesize lipopeptide-type compounds, like fengycin, surfactin, and iturin, that not only effectively suppress the growth of soil-borne pathogens but also stimulate induced systemic resistance [55]. For example, B. subtilis RB14 controls damping-off of tomato caused by Rhizoctonia solani through the secretion of iturin A [56].
We successfully identified several strains that possess traits associated with plant growth promotion. Moreover, the six strains tested in the greenhouse assays increased plant stem length, weight, and diameter significantly. The observed increase in secondary root growth is one of the known effects that indole-3-acetic acid (IAA) producing plant growth-promoting rhizobacteria elicit on plants [57,58]. Interestingly, strains rbES087 and rbES090, identified as Pseudomonas, showed 100% identity in their 16s rRNA gene, but they tested positive for different plant growth-promoting traits (Table 1). Both strains came from El Vizcaíno Biosphere Reserve, close to the city of Guerrero Negro. Although each strain was isolated from different media (PY and King B, respectively), they shared similar colony morphology when grown in the same media.
Although the screening process successfully identified PGPR, there was no clear correlation between plant growth promotion in vivo and the results of either the colorimetric or the germination assays. For example, Bacillus sp. rbES015 had the lowest quantified IAA levels, but there were no significant differences with other strains in most of the measured plant variables. Moreover, this strain induced a significant 30% increase in stem diameter when compared with other strains and 300% when compared with the inoculated control. Previous works have shown that growth promotion-associated traits assessed in vitro are not necessarily a robust indicator of the potential effects of bacteria on plant growth [58]. For example, several environmental factors like pH, osmotic stress, and carbon limitation may influence IAA biosynthesis in bacteria. Additionally, the optimal concentration of this auxin required for plant growth promotion may be precise for different plant phenotypes [57]. The same observation was true for the antagonistic activity against F. oxysporum. Although the three tested Bacillus sp. strains suppressed the pathogen in vitro, only strain rbES262 effectively reduced the effects of Fusarium on plant growth in vivo while the rest of the treatments produced inconsistent results, even the commercial product. This strain tested negative in the in vitro tests for the production of siderophores and chitinases, traits commonly associated with antagonistic activity. Interestingly we re-isolated F. oxysporum from inoculated plants treated with strain Bacillus sp. rbES262 even though no adverse effects on plant growth were observed, indicating that the pathogen was not completely suppressed but reduced.
There are several examples of the successful utilization of native Bacillus and Pseudomonas strains associated with wild plants of non-temperate regions for plant growth promotion and pathogen suppression. For example, cold-adapted Bacillus and Pseudomonas strains isolated from the rhizosphere of native potatoes from the Andean Highlands of Peru and Bolivia have been shown to promote plant growth and protect a commercial potato cultivar against Rhizoctonia solani [59]. Bacillus strains isolated from three halophyte species native to desetic regions in Utah (Salicornia rubra, Sarcocornia utahensis, and Allenrolfea occidentalis) were used to stimulate growth of alfalfa in salty soils [60] and strains of Bacillus, Pseudomonas, and Cellulomonas, isolated from the rhizosphere of different wild plants of arid regions in Turkey, were found to elicit a positive effect on plant height, dry matter, and protein content on wheat [61]. Moreover, a recent screening of endophytic bacteria from the wild Solanaceae Solanum sodomaeum and Solanum lead to the isolation of a Bacillus, a Serratia, and several Stenotrophomonas strains capable of stimulating growth and controlling Fusarium wilt in tomato [62]. Stenotrophomonas and Serratia are known opportunistic human pathogens, so their use as PGPR is questionable. In this work, we isolated six strains (rbES001, rbES005, rbES050, rbES053, and rbES057) that were identified as Staphylococcus sciuri. Although this species has been characterized previously as a PGPR, it has also been reported as an opportunistic human pathogen, and several environmental and clinical strains have been shown to carry antibiotic resistance determinants. Therefore, they were dropped from this study [63]. Nevertheless, our extensive geographical sampling, and the fact that we screened rhizospheric communities, produced a more diverse collection of PGPR isolates.
Finally, of particular interest was the lethal effect on plants elicited by Streptomyces sp. rbES158 and Glutamicibacter sp. rbES331 (Table 2) that also showed antifungal activity through diffusible compounds. Actinobacteria produce diverse secondary metabolites that inhibit different biological pathways in microorganisms and plants [64]. For example, the non-proteinogenic amino acid 4-Chlorothreonine, isolated from Streptomyces sp. OH-5093 inhibits the growth of radish and sorghum by inhibiting amino acid metabolism [65]; Bialaphos is a peptide produced by Streptomyces hygroscopicus and Streptomyces viridochromogenes that releases the glutamic acid analog glufosinate when metabolized by the plant resulting in the accumulation of ammonium and the consequent disruption of primary metabolism [66]. Moreover, often the same molecule with antimicrobial activity can act as an herbicide, for example, the macrolide Phthoramycin produced by Streptomyces sp. WK-1875 and identified by its activity against the Phytophthora parasitica through cellulose biosynthesis inhibition, was also found to inhibit radish and rice [67]. More work is needed to identify the secondary metabolites produced by these bacteria, their mechanism of action, and determine if the same compound is responsible for the antifungal and herbicidal activities. Even though these strains may not be useful as inoculants, they can be valuable sources of metabolites with a potential application in agriculture or other fields. On the other hand, the observed inconsistencies between the results of in vitro and in vivo assays raise the question of the missed potential of strains discarded by arbitrary cut-off values during our screening process and stress the importance of performing in vivo assays to exploit the potential of our extensive strain collection completely.
In conclusion, by prospecting soil associated with Solanum hindsianum plants growing at different locations of the Baja California Peninsula, we obtained a collection of more than 300 bacterial strains, some with growth promotion-related traits and others with the ability to inhibit Fusarium oxysporum in vitro. Moreover, we demonstrated that tomato plants’ inoculation with selected strains has a significant positive effect on plant growth and ameliorated tomato Fusarium wilt, confirming that plants of the arid region of Baja California are an important source of microbes for agriculture.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4395/11/3/579/s1, Figure S1: Fusarium oxysporum (FO) inhibition by isolates rbES158 and rbES245, Figure S2: Microplate assays for the determination of plant growth promotion-associated traits, Figure S3: Effect of the inoculation of different bacterial isolates on the growth of S. lycopersicum, Figure S4: Effect of Fusarium oxysporum on the growth of S. lycopersicum inoculated with different bacterial isolates. Table S1: Plant Growth Promotion Associated Traits of Isolated Strains.
Author Contributions
C.S.D.-R.: Investigation, Methodology, Formal analysis, Writing—Review & Editing. R.H.-M.: Conceptualization, Supervision, Methodology, Writing—Review & Editing. E.S.: Conceptualization, Supervision, Methodology, Formal analysis, Writing—Original Draft. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in Table S1.
Acknowledgments
We are thankful to Edith González, Jessica Osorio, Andrea Guzmán, and Rodrigo Torres for their assistance in soil sampling and greenhouse experiments. CSDR was supported during the master program (Programa de Posgrado en Ciencias de la Vida, CICESE) by scholarship 724689 from Consejo Nacional de Ciencia y Tecnología (México).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Hassani, M.A.; Durán, P.; Hacquard, S. Microbial interactions within the plant holobiont. Microbiome 2018, 6, 58. [Google Scholar] [CrossRef]
- Méndez-Bravo, A.; Cortazar-Murillo, E.M.; Guevara-Avendaño, E.; Ceballos-Luna, O.; Rodríguez-Haas, B.; Kiel-Martínez, A.L.; Hernández-Cristóbal, O.; Guerrero-Analco, J.A.; Reverchon, F. Plant growth-promoting rhizobacteria associated with avocado display antagonistic activity against Phytophthora cinnamomi through volatile emissions. PLoS ONE 2018, 13, e0194665. [Google Scholar] [CrossRef] [PubMed]
- Avis, T.J.; Gravel, V.; Antoun, H.; Tweddell, R.J. Multifaceted beneficial effects of rhizosphere microorganisms on plant health and productivity. Soil Biol. Biochem. 2008, 40, 1733–1740. [Google Scholar] [CrossRef]
- Hayat, R.; Ali, S.; Amara, U.; Khalid, R.; Ahmed, I. Soil beneficial bacteria and their role in plant growth promotion: A review. Ann. Microbiol. 2010, 60, 579–598. [Google Scholar] [CrossRef]
- Khatoon, Z.; Huang, S.; Rafique, M.; Fakhar, A.; Kamran, M.A.; Santoyo, G. Unlocking the potential of plant growth-promoting rhizobacteria on soil health and the sustainability of agricultural systems. J. Environ. Manag. 2020, 273, 111118. [Google Scholar] [CrossRef]
- Berg, G. Plant-microbe interactions promoting plant growth and health: Perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 2009, 84, 11–18. [Google Scholar] [CrossRef]
- Glick, B.R. Plant Growth-Promoting Bacteria: Mechanisms and Applications. Scientifica 2012, 2012, 963401. [Google Scholar] [CrossRef]
- Collinge, D.B.; Jørgensen, H.J.; Latz, M.A.; Manzotti, A.; Ntana, F.; Rojas Tayo, E.C.; Jensen, B. Searching for novel fungal biological control agents for plant disease control among endophytes. In Endophytes for a Growing World, 1st ed.; Hodkinson, T., Doohan, F., Saunders, M., Murphy, B., Eds.; Cambridge University Press: Cambridge, UK, 2019; pp. 25–51. [Google Scholar]
- Raaijmakers, J.M.; Paulitz, T.C.; Steinberg, C.; Alabouvette, C.; Moënne-Loccoz, Y. The Rhizosphere: A Playground and Battlefield for Soilborne Pathogens and Beneficial Microorganisms. Plant Soil 2009, 321, 341–361. [Google Scholar] [CrossRef]
- Dutta, S.; Rani, T.S.; Podile, A.R. Root exudate-induced alterations in Bacillus cereus cell wall contribute to root colonization and plant growth promotion. PLoS ONE 2013, 8, e78369. [Google Scholar] [CrossRef]
- Martínez-Viveros, O.; Jorquera, M.A.; Crowley, D.E.; Gajardo, G.; Mora, M.L. Mechanisms and practical considerations involved in plant growth promotion by Rhizobacteria. J. Plant Nutr. 2010, 10, 293–319. [Google Scholar] [CrossRef]
- Requena, N.; Jimenez, I.; Toro, M.; Barea, J.M. Interactions between plant-growth-promoting rhizobacteria (PGPR), arbuscular mycorrhizal fungi and Rhizobium spp. in the rhizosphere of Anthyllis cytisoides, a model legume for revegetation in mediterranean semi-arid ecosystems. New Phytol. 1997, 136, 667–677. [Google Scholar] [CrossRef]
- Zahid, M.; Kaleem Abbasi, M.; Hameed, S.; Rahim, N. Isolation and identification of indigenous plant growth promoting rhizobacteria from Himalayan region of Kashmir and their effect on improving growth and nutrient contents of maize (Zea mays L.). Front. Microbiol. 2015, 6, 207. [Google Scholar] [CrossRef] [PubMed]
- Borja-Bravo, M.; García-Salazar, J.A.; Skaggs, R.K. Mexican fresh tomato exports in the North American market: A case study of the effects of productivity on competitiveness. Can. J. Plant Sci. 2013, 93, 839–850. [Google Scholar] [CrossRef][Green Version]
- Singh, V.K.; Singh, H.B.; Upadhyay, R.S. Role of fusaric acid in the development of “Fusarium wilt” symptoms in tomato: Physiological, biochemical and proteomic perspectives. Plant Physiol. Bioch. 2017, 118, 320–332. [Google Scholar] [CrossRef]
- Blancard, D. Tomato Diseases, 2nd ed.; CRC Press: London, UK, 2013; p. 668. [Google Scholar] [CrossRef]
- Padilla-Bernal, L.E.; Lara-Herrera, A.; Reyes-Rivas, E.; González-Hernández, J. Roberto Assessing Environmental Management of Tomato Production under Protected Agriculture. Int. Food Agribus. Manag. Rev. 2015, 18, 193–210. [Google Scholar] [CrossRef]
- Arellano, E.; Montaño-Soto, T.; Arellano-García, E.; Ojinaga, L.C.; Vonglascoe, C.; Ruiz-Ruiz, B. Genotoxic Biomonitoring and Exposure to Pesticides in Women Laborers at Maneadero Valley in Baja California, Mexico. Int. J. Appl. Nat. Sci. 2014, 3, 89–96. [Google Scholar]
- Espejel, I.; Fischer, D.W.; Hinojosa, A.; García, C.; Leyva, C. Land-use planning for the Guadalupe Valley, Baja California, Mexico. Landsc. Urban Plan. 1999, 45, 219–232. [Google Scholar] [CrossRef]
- Gilabert-Alarcón, C.; Daesslé, L.W.; Salgado-Méndez, S.O.; Pérez-Flores, M.A.; Knöller, K.; Kretzschmar, T.G.; Stumpp, C. Effects of reclaimed water discharge in the Maneadero coastal aquifer, Baja California, Mexico. J. Appl. Geochem. 2018, 92, 121–139. [Google Scholar] [CrossRef]
- Hartmann, A.; Schmid, M.; Van Tuinen, D.; Berg, G. Plant-driven selection of microbes. Plant Soil 2009, 321, 235–257. [Google Scholar] [CrossRef]
- Zhang, N.; Wang, D.; Liu, Y.; Li, S.; Shen, Q.; Zhang, R. Effects of different plant root exudates and their organic acid components on chemotaxis, biofilm formation and colonization by beneficial rhizosphere-associated bacterial strains. Plant Soil 2014, 374, 689–700. [Google Scholar] [CrossRef]
- Santoyo, G.; Hernández-Pacheco, C.; Hernández-Salmerón, J.; Hernández-León, R. The role of abiotic factors modulating the plant-microbe-soil interactions: Toward sustainable agriculture. A review. Span. J. Agric. Res. 2017, 15, 13. [Google Scholar] [CrossRef]
- Rueda-Puente, E.O.; Castellanos-Cervantes, T.; Díaz de León-Álvarez, J.L.; Preciado-Rangel, P.; Almaguer-Vargas, G. Bacterial community of rhizosphere associated to the annual halophite Salicornia bigelovii (Torr). Terra Latinoam. 2010, 28, 345–353. [Google Scholar]
- Aguirre-Garrido, J.F.; Montiel-Lug, D.; Hernández-Rodríguez, C.; Torres-Cortés, G.; Millán, V.; Toro, N.; Martínez-Abarca, F.; Ramírez-Saad, H. Bacterial community structure in the rhizosphere of three cactus species from semi-arid highlands in central Mexico. Antonie Leeuwenhoek 2012, 101, 891–904. [Google Scholar] [CrossRef] [PubMed]
- Aydi Ben Abdallah, R.; Jabnoun-Khiareddine, H.; Nefzi, A.; Mokni-Tlili, S.; Daami-Remadi, M. Biocontrol of Fusarium Wilt and Growth Promotion of Tomato Plants Using Endophytic Bacteria Isolated from Solanum elaeagnifolium Stems. J. Phytopathol. 2016, 164, 811–824. [Google Scholar] [CrossRef]
- Knapp, S.; Sagona, E.; Carbonell, A.K.Z.; Chiarini, F. A revision of the Solanum elaeagnifolium clade (Elaeagnifolium clade; subgenus Leptostemonum, Solanaceae). PhytoKeys 2017, 2017, 1–104. [Google Scholar] [CrossRef] [PubMed]
- Rebman, J.; ROberts, N.C. Baja California Plant Field Guide, 3rd ed.; San Diego Natural History Museum: San Diego, CA, USA, 2012; p. 480. ISBN 9780916251185. [Google Scholar]
- Noel, K.D.; Sanchez, A.; Fernandez, L.; Leemans, J.; Cevallos, M.A. Rhizobium phaseoli symbiotic mutants with transposon Tn5 insertions. J. Bacteriol. 1984, 158, 148–155. [Google Scholar] [CrossRef]
- Guevara-Avendaño, E.; Carrillo, J.D.; Ndinga-Muniania, C.; Moreno, K.; Méndez-Bravo, A.; Guerrero-Analco, J.A.; Eskalen, A.; Reverchon, F. Antifungal activity of avocado rhizobacteria against Fusarium euwallaceae and Graphium spp., associated with Euwallacea spp. nr. fornicatus, and Phytophthora cinnamomi. Antonie Leeuwenhoek 2018, 111, 563–572. [Google Scholar] [CrossRef]
- Gupta, R.; Singal, R.; Shankar, A.; Kuhad, R.C.; Saxena, R.K. A modified plate assay for screening phosphate solubilizing microorganisms. J. Gen. Appl. Microbiol. 1994, 40, 255–260. [Google Scholar] [CrossRef]
- Zheng, B.X.; Ibrahim, M.; Zhang, D.P.; Bi, Q.F.; Li, H.Z.; Zhou, G.W.; Ding, K.; Peñuelas, J.; Zhu, Y.G.; Yang, X.R. Identification and characterization of inorganic-phosphate-solubilizing bacteria from agricultural fields with a rapid isolation method. AMB Express 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Pikovskaya, R.I. Mobilization of Phosphorus in Soil Connection with the Vital Activity of Some Microbial Species. Microbiologiya 1948, 17, 362–370. [Google Scholar]
- Bapiri, A.; Asgharzadeh, A.; Mujallali, H.; Khavazi, K.; Pazira, E. Evaluation of Zinc solubilization potential by different strains of Fluorescent Pseudomonads. J. Appl. Sci. Environ. Manag. 2012, 16, 295–298. [Google Scholar]
- Ahmad, F.; Ahmad, I.; Khan, M.S. Screening of free-living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiol. Res. 2008, 163, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Penrose, D.M.; Glick, B.R. Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol. Plant 2003, 118, 10–15. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, G.A.; Pratt, L.A.; Watnick, P.I.; Newman, D.K.; Weaver, V.B.; Kolter, R. Genetic approaches to study of biofilms. Meth. Enzymol. 1999, 310, 91–109. [Google Scholar] [CrossRef]
- Hamon, M.A.; Lazazzera, B.A. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol. Microbiol. 2001, 42, 1199–1209. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Lynne, A.M.; Haarmann, D.; Louden, B.C. Use of Blue Agar CAS Assay for Siderophore Detection. J. Microbiol. Biol. Edu. 2011, 12, 51–53. [Google Scholar] [CrossRef]
- Agrawal, T.; Kotasthane, A.S. Chitinolytic assay of indigenous Trichoderma isolates collected from different geographical locations of Chhattisgarh in central India. SpringerPlus 2012, 1, 1–10. [Google Scholar] [CrossRef]
- Hanson, P.; Lu, S.F.; Wang, J.F.; Chen, W.; Kenyon, L.; Tan, C.W. Conventional and molecular marker-assisted selection and pyramiding of genes for multiple disease resistance in tomato. Sci. Hortic. 2016, 201, 346–354. [Google Scholar] [CrossRef]
- Frank, J.A.; Reich, C.I.; Sharma, S.; Weisbaum, J.S.; Wilson, B.A.; Olsen, G.J. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 2008, 74, 2461–2470. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. R Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Habibi, S.; Djedidi, S.; Prongjunthuek, K.; Mortuza, M.F.; Ohkama-Ohtsu, N.; Sekimoto, H.; Yokoyoma, T. Physiological and genetic characterization of rice nitrogen fixer PGPR isolated from rhizosphere soils of different crops. Plant Soil 2014, 379, 51–66. [Google Scholar] [CrossRef]
- Liu, D.; Yang, Q.; Ge, K.; Hu, X.; Qi, G.; Du, B.; Liu, K.; Ding, Y. Promotion of iron nutrition and growth on peanut by Paenibacillus illinoisensis and Bacillus sp. strains in calcareous soil. Braz. J. Microbiol. 2017, 48, 656–670. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.-G.; Mun, B.-G.; Kang, S.-M.; Hussain, A.; Shahzad, R.; Seo, C.-W.; Kim, A.-Y.; Lee, S.-U.; Oh, K.Y.; Lee, D.Y.; et al. Bacillus aryabhattai SRB02 tolerates oxidative and nitrosative stress and promotes the growth of soybean by modulating the production of phytohormones. PLoS ONE 2017, 12, e0173203. [Google Scholar] [CrossRef]
- Kang, S.M.; Khan, A.L.; Waqas, M.; Asaf, S.; Lee, K.E.; Park, Y.G.; Kim, A.Y.; Khan, M.A.; You, Y.H.; Lee, I.J. Integrated phytohormone production by the plant growth-promoting rhizobacterium Bacillus tequilensis SSB07 induced thermotolerance in soybean. J. Plant Interact. 2019, 14, 416–423. [Google Scholar] [CrossRef]
- Barraza, A.; Caamal-Chan, M.G.; Castellanos, T.; Loera-Muro, A. Bacterial community characterization of the rhizobiome of plants belonging to Solanaceae family cultivated in desert soils. Ann. Microbiol. 2020, 70, 34. [Google Scholar] [CrossRef]
- Hernández-León, R.; Rojas-Solís, D.; Contreras-Pérez, M.; Orozco-Mosqueda, M.C.; Macías-Rodríguez, L.I.; Cruz, H.R.-D.L.; Valencia-Cantero, E.; Santoyo, G. Characterization of the antifungal and plant growth-promoting effects of diffusible and volatile organic compounds produced by Pseudomonas fluorescens strains. Biol. Contr. 2015, 81, 83–92. [Google Scholar] [CrossRef]
- Schlatter, D.; Kinkel, L.; Thomashow, L.; Weller, D.; Paulitz, T. Disease Suppressive Soils: New Insights from the Soil Microbiome. Phytopathology 2017, 107, 1284–1297. [Google Scholar] [CrossRef]
- Ongena, M.; Jacques, P. Bacillus lipopeptides: Versatile weapons for plant disease biocontrol. Trends Microbiol. 2008, 16, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Asaka, O.; Shoda, M. Biocontrol of Rhizoctonia solani Damping-Off of Tomato with Bacillus subtilis RB14. Appl. Environ. Microbiol. 1996, 62, 4081–4085. [Google Scholar] [CrossRef] [PubMed]
- Spaepen, S.; Vanderleyden, J.; Remans, R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 2007, 31, 425–448. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.F.; Wei, J.Y.; Chen, H.W.; Liu, Y.Y.; Lu, H.Y.; Chou, J.Y. Indole-3-acetic acid: A widespread physiological code in interactions of fungi with other organisms. Plant. Signal. Behav. 2015, 10, e1048052. [Google Scholar] [CrossRef] [PubMed]
- Smyth, E.M.; McCarthy, J.; Nevin, R.; Khan, M.R.; Dow, J.M.; O’Gara, F.; Doohan, F.M. In vitro analyses are not reliable predictors of the plant growth promotion capability of bacteria; a Pseudomonas fluorescens strain that promotes the growth and yield of wheat. J. Appl. Microbiol. 2011, 111, 683–692. [Google Scholar] [CrossRef]
- Ghyselinck, J.; Velivelli, S.L.; Heylen, K.; O’Herlihy, E.; Franco, J.; Rojas, M.; Prestwich, B.D. Bioprospecting in potato fields in the Central Andean Highlands: Screening of rhizobacteria for plant growth-promoting properties. Syst. Appl. Microbiol. 2013, 36, 116–127. [Google Scholar] [CrossRef]
- Kearl, J.; McNary, C.; Lowman, J.S.; Mei, C.; Aanderud, Z.T.; Smith, S.T.; West, J.; Colton, E.; Hamson, M.; Nielsen, B.L. Salt-Tolerant Halophyte Rhizosphere Bacteria Stimulate Growth of Alfalfa in Salty Soil. Front. Microbiol. 2019, 10, 1849. [Google Scholar] [CrossRef]
- Sezen, A.; Ozdal, M.; Koc, K.; Algur, O.F. Isolation and Characterization of Plant Growth Promoting Rhizobacteria (PGPR) and Their Effects on Improving Growth of Wheat. J. Appl. Biol. Sci. 2019, 10, 41–46. Available online: http://jabsonline.org/index.php/jabs/article/view/485 (accessed on 17 March 2021).
- Aydi-Ben-Abdallah, R.; Jabnoun-Khiareddine, H.; Daami-Remadi, M. Fusarium wilt biocontrol and tomato growth stimulation, using endophytic bacteria naturally associated with Solanum sodomaeum and S. bonariense plants. Egypt. J. Biol. Pest Cont. 2020, 30, 113. [Google Scholar] [CrossRef]
- Akram, M.S.; Shahid, M.; Tariq, M.; Azeem, M.; Javed, M.T.; Saleem, S.; Riaz, S. Deciphering Staphylococcus sciuri SAT-17 Mediated Anti-oxidative Defense Mechanisms and Growth Modulations in Salt Stressed Maize (Zea mays L.). Front. Microbiol. 2016, 7, 867. [Google Scholar] [CrossRef]
- Liqiao, S.; Zhaoyuan, W.; Yani, Z.; Zhigang, Z.; Wei, F.; Yueying, W.; Zhongyi, W.; Kaimei, W.; Shaoyong, K. Herbicidal Secondary Metabolites from Actinomycetes: Structure Diversity, Modes of Action, and Their Roles in the Development of Herbicides. J. Agric. Food Chem. 2020, 68, 17–32. [Google Scholar] [CrossRef]
- Yoshida, H.; Arai, N.; Sugoh, M.; Iwabuchi, J.; Shiomi, K.; Shinose, M.; Tanaka, Y.; Omura, S. 4-chlorothreonine, a herbicidal antimetabolite produced by Streptomyces sp. OH-5093. J. Antibiot. 1994, 47, 1165–1166. [Google Scholar] [CrossRef][Green Version]
- Duke, S.O.; Dayan, F.E. Modes of Action of Microbially-Produced Phytotoxins. Toxins 2011, 3, 1038–1064. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Sugoh, M.; Ji, W.; Iwabuchi, J.; Yoshida, H.; Omura, S. Screening method for cellulose biosynthesis inhibitors with herbicidal activity. J. Antibiot. 1995, 48, 720–724. [Google Scholar] [CrossRef] [PubMed][Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).