Long-Term Effects of Amendment with Olive Mill Wastewater on Soil Chemical Properties, Microbial Community, and Olive Tree Vegetative and Productive Activities
Abstract
1. Introduction
2. Materials and Methods
2.1. Olive Grove and Environmental Characteristics
2.2. Olive Mill Wastewater, Soil Amendment, and Sampling
2.3. OMWW Chemical Characterization
2.4. Soil Chemical Analysis
2.5. Leaf Net Photosynthesis (Pn)
2.6. Vegetative and Productive Activity, Characteristics of Fruit
2.7. Extraction of Total Bacterial DNA from Soil and OMWW
2.8. 16 S Library Preparation and Sequencing
2.9. Bioinformatics Analysis
2.10. Statistical Analysis
2.10.1. Microbial Diversity and Composition
2.10.2. Groves Characterization
2.10.3. Redundancy Analysis
3. Results
3.1. Effect of OMWs Spreading on Soil Chemical Properties
3.2. Leaf Net Photosynthesis (Pn)
3.3. Vegetative and Productive Activity, Characteristics of Fruit
3.4. High-Throughput Sequencing Results
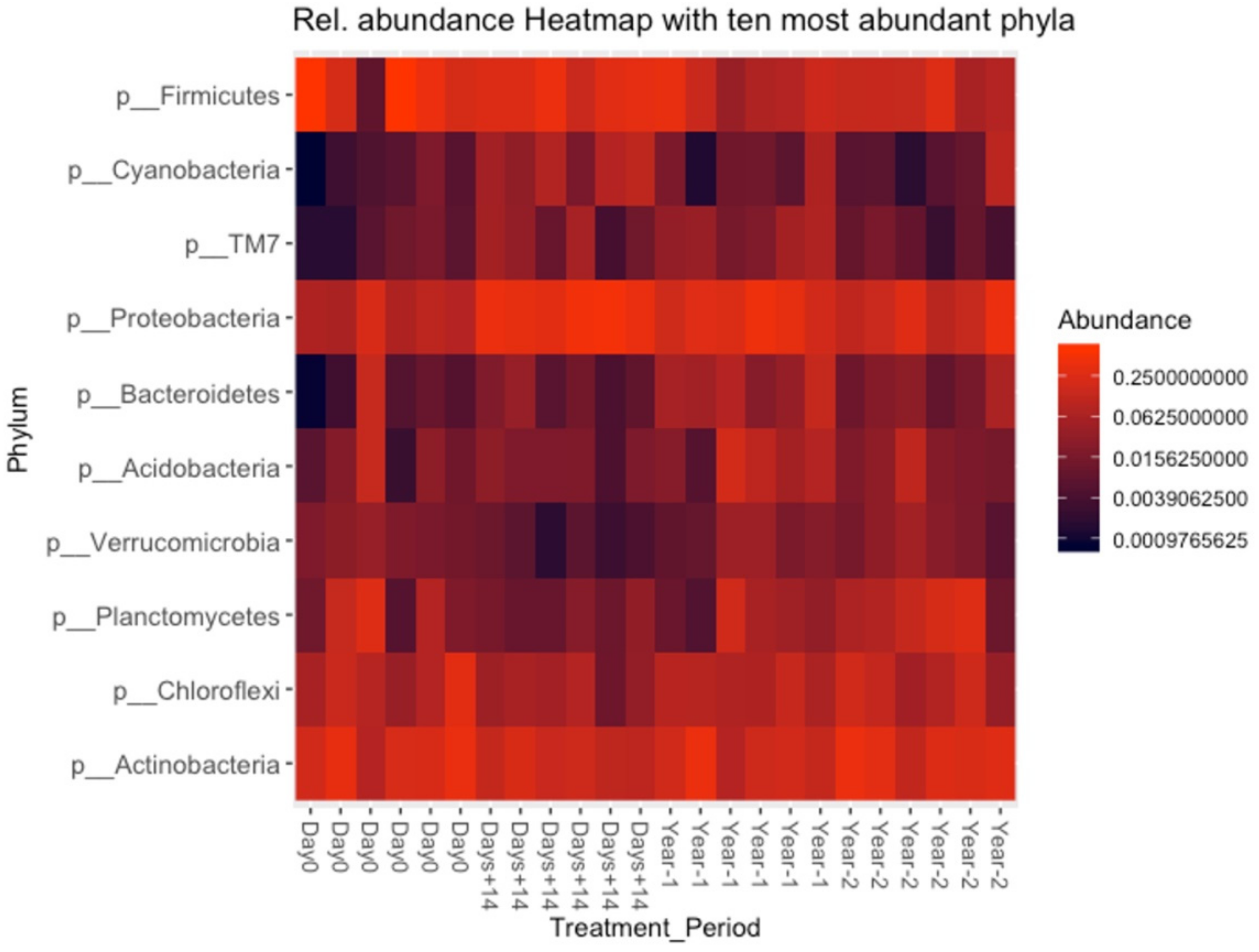
3.5. Bacterial Composition of Soil and OMWW
3.6. Microbial Richness
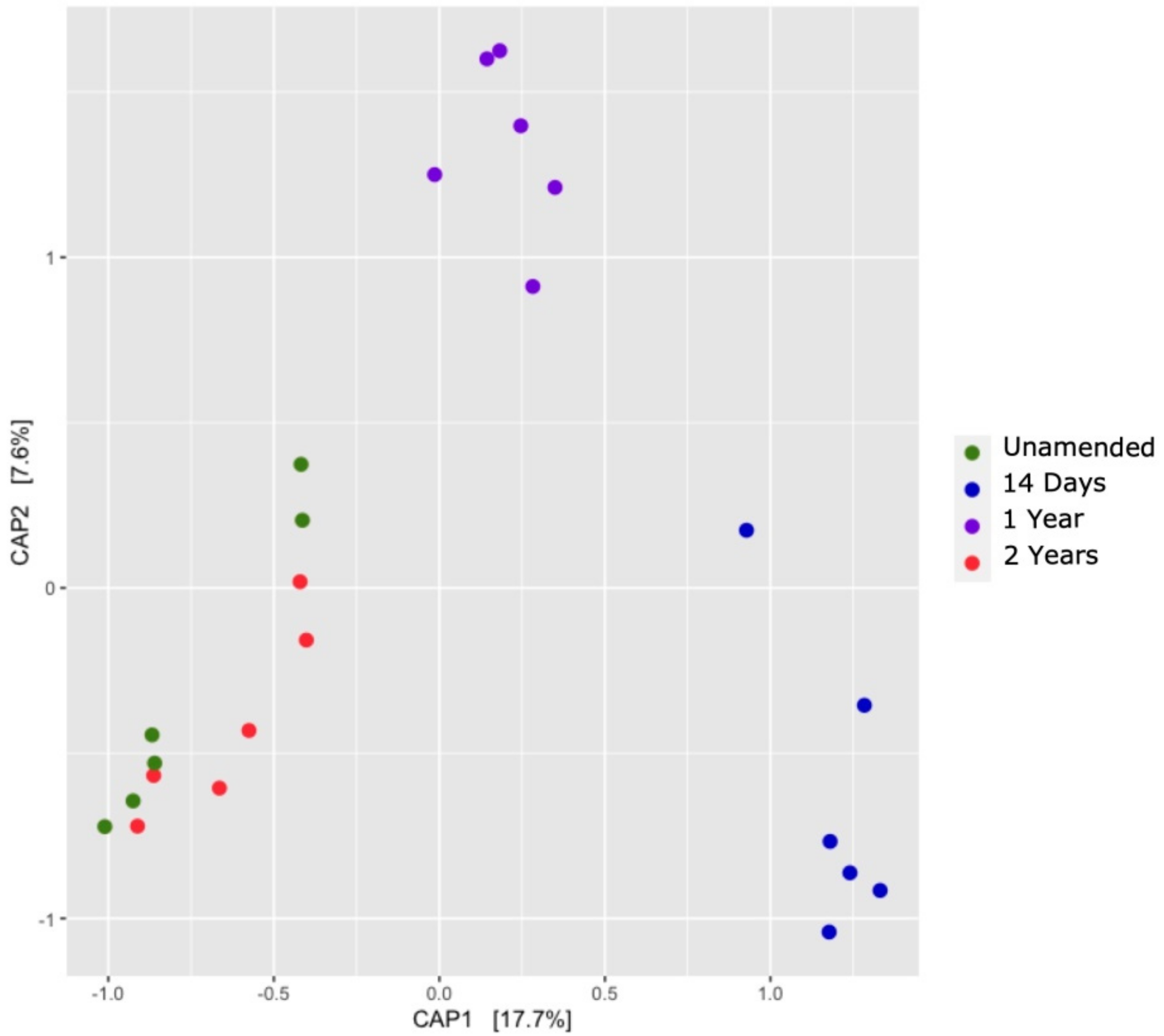
3.7. Comparison of Soil Bacterial Communities over Time (β-Diversity)
3.8. Differential Abundance Analysis (ANCOM)
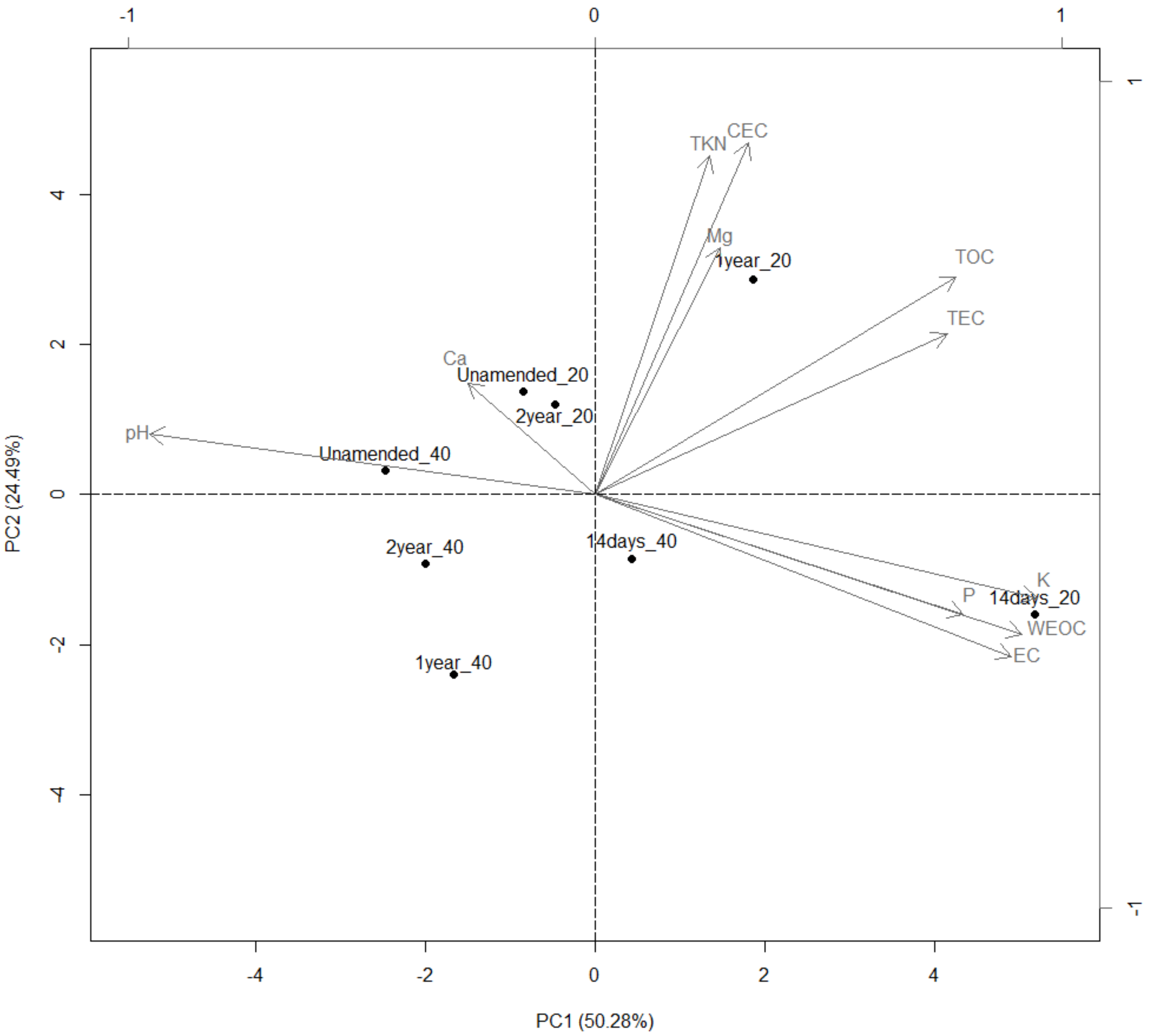
3.9. Groves Characterization
3.10. Redundancy Analysis
4. Discussion
4.1. Soil Chemical Changes after OMWW Application
4.2. Microbial Community Changes after OMWW Application
4.3. Olive Tree Production after OMWW Application
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nasini, L.; Gigliotti, G.; Balduccini, M.A.; Federici, E.; Cenci, G.; Proietti, P. Effect of solid olive-mill waste amendment on soil fertility and olive (Olea europaea L.) tree activity. Agric. Ecosyst. Environ. 2013, 164, 292–297. [Google Scholar] [CrossRef]
- Federici, E.; Massaccesi, L.B.; Pezzolla, D.; Fidati, L.; Montalbani, E.; Proietti, P.; Nasini, L.; Regni, L.; Scargetta, S.; Gigliotti, G. Short-term modifications of soil microbial community structure and soluble organic matter chemical composition following amendment with different solid olive mill waste and their derived composts. Appl. Soil Ecol. 2017, 119, 234–241. [Google Scholar] [CrossRef]
- Mechri, B.; Chehab, H.; Attia, F.; Mariem, F.; Braham, M.; Hammami, M. Olive mill wastewater effects on the microbial communities as studied in the field of olive trees by analysis of fatty acid signatures. Eur. J. Soil Biol. 2010, 46, 312–318. [Google Scholar] [CrossRef]
- Ouzounidou, G.; Zervakis, G.I.; Gaitis, F. Raw and microbiologically detoxified olive mill waste and their impact on plant growth. Terr. Aquat. Environ. Toxicol. 2010, 4, 21–38. [Google Scholar]
- Ntougias, S.; Bourtzis, K.; Tsiamis, G. The microbiology of olive mill wastes. BioMed Res. Int. 2013, 2013, 784591. [Google Scholar] [CrossRef] [PubMed]
- Regni, L.; Nasini, L.; Ilarioni, L.; Brunori, A.; Massaccesi, L.; Agnelli, A.; Proietti, P. Long Term Amendment with Fresh and Composted Solid Olive Mill Waste on Olive Grove Affects Carbon Sequestration by Prunings, Fruits, and Soil. Front. Plant Sci. 2017, 7, 2042. [Google Scholar] [CrossRef] [PubMed]
- Morillo, J.A.; Antizar-Ladislao, B.; Monteoliva-Sánchez, M.; Ramos-Cormenzana, A.; Russell, N.J. Bioremediation and biovalorisation of olive-mill wastes. Appl. Microbiol. Biotechnol. 2009, 82, 25–39. [Google Scholar] [CrossRef]
- Barbera, A.; Maucieri, C.; Cavallaro, V.; Ioppolo, A.; Spagna, G. Effects of spreading olive mill wastewater on soil properties and crops, a review. Agric. Water Manag. 2013, 119, 43–53. [Google Scholar] [CrossRef]
- Aviani, I.; Raviv, M.; Hadar, Y.; Saadi, I.; Dag, A.; Ben-Gal, A.; Yermiyahu, U.; Zipori, I.; Laor, Y. Effects of harvest date, irrigation level, cultivar type and fruit water content on olive mill wastewater generated by a laboratory scale ‘Abencor’ milling system. Bioresour. Technol. 2012, 107, 87–96. [Google Scholar] [CrossRef]
- Ammar, E.; Nasri, M.; Medhioub, K. Isolation of Enterobacteria able to degrade simple aromatic compounds from the wastewater from olive oil extraction. World J. Microbiol. Biotechnol. 2005, 21, 253–259. [Google Scholar] [CrossRef]
- Borja, R.; Sánchez, E.; Raposo, F.; Rincón, B.; Jiménez, A.; Martín, A. A study of the natural biodegradation of two-phase olive mill solid waste during its storage in an evaporation pond. Waste Manag. 2006, 26, 477–486. [Google Scholar] [CrossRef]
- Jarboui, R.; Sellami, F.; Kharroubi, A.; Gharsallah, N.; Ammar, E. Olive mill wastewater stabilization in open-air ponds: Impact on clay–sandy soil. Bioresour. Technol. 2008, 99, 7699–7708. [Google Scholar] [CrossRef]
- Mekki, A.; Dhouib, A.; Sayadi, S. Review: Effects of olive mill wastewater application on soil properties and plants growth. Int. J. Recycl. Org. Waste Agric. 2013, 2, 15–21. [Google Scholar] [CrossRef]
- Justino, C.I.L.; Pereira, R.; Freitas, A.C.; Rocha-Santos, T.A.P.; Panteleitchouk, T.S.L.; Duarte, A.C. Olive oil mill wastewaters before and after treatment: A critical review from the ecotoxicological point of view. Ecotoxicology 2011, 21, 615–629. [Google Scholar] [CrossRef]
- Belaqziz, M.; El-Abbassi, A.; Lakhal, E.K.; Agrafioti, E.; Galanakis, C.M. Agronomic application of olive mill wastewater: Effects on maize production and soil properties. J. Environ. Manag. 2016, 171, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Ayoub, S.; Al-Absi, K.; Al-Shdiefat, S.; Al-Majali, D.; Hijazean, D. Effect of olive mill wastewater land-spreading on soil properties, olive tree performance and oil quality. Sci. Hortic. Amst. 2014, 175, 160–166. [Google Scholar] [CrossRef]
- Di Serio, M.; Lanza, B.; Mucciarella, M.; Russi, F.; Iannucci, E.; Marfisi, P.; Madeo, A. Effects of olive mill wastewater spreading on the physico-chemical and microbiological characteristics of soil. Int. Biodeterior. Biodegrad. 2008, 62, 403–407. [Google Scholar] [CrossRef]
- Zbakh, H.; El Abbassi, A. Potential use of olive mill wastewater in the preparation of functional beverages: A review. J. Funct. Foods 2012, 4, 53–65. [Google Scholar] [CrossRef]
- Chaari, L.; Elloumi, N.; Mseddi, S.; Gargouri, K.; Ben Rouina, B.; Mechichi, T.; Kallel, M. Changes in Soil Macronutrients after a Long-Term Application of Olive Mill Wastewater. J. Agric. Chem. Environ. 2015, 4, 1–13. [Google Scholar] [CrossRef]
- Pezzolla, D.; Said-Pullicino, D.; Raggi, L.; Albertini, E.; Gigliotti, G. Short-term Variations in Labile Organic C and Microbial Biomass Activity and Structure After Organic Amendment of Arable Soils. Soil Sci. 2013, 178, 474–485. [Google Scholar] [CrossRef]
- Said-Pullicino, D.; Erriquens, F.G.; Gigliotti, G. Changes in the chemical characteristics of water-extractable organic matter during composting and their influence on compost stability and maturity. Bioresour. Technol. 2007, 98, 1822–1831. [Google Scholar] [CrossRef]
- Said-Pullicino, D.; Kaiser, K.; Guggenberger, G.; Gigliotti, G. Changes in the chemical composition of water-extractable organic matter during composting: Distribution between stable and labile organic matter pools. Chemosphere 2007, 66, 2166–2176. [Google Scholar] [CrossRef] [PubMed]
- Piotrowska, A.; Iamarino, G.; Rao, M.A.; Gianfreda, L. Short-term effects of olive mill waste water (OMW) on chemical and biochemical properties of a semiarid Mediterranean soil. Soil Biol. Biochem. 2006, 38, 600–610. [Google Scholar] [CrossRef]
- Di Bene, C.; Pellegrino, E.; Debolini, M.; Silvestri, N.; Bonari, E. Short- and long-term effects of olive mill wastewater land spreading on soil chemical and biological properties. Soil Biol. Biochem. 2013, 56, 21–30. [Google Scholar] [CrossRef]
- Mohawesh, O.; Al-Hamaiedeh, H.; Albalasmeh, A.; Qaraleh, S.; Haddadin, M. Effect of Olive Mill Wastewater (OMW) Application on Soil Properties and Wheat Growth Performance Under Rain-Fed Conditions. Water Air Soil Pollut. 2019, 230, 160. [Google Scholar] [CrossRef]
- Magdich, S.; Jarboui, R.; Ben Rouina, B.; Boukhris, M.; Ammar, E. A yearly spraying of olive mill wastewater on agricultural soil over six successive years: Impact of different application rates on olive production, phenolic compounds, phytotoxicity and microbial counts. Sci. Total Environ. 2012, 430, 209–216. [Google Scholar] [CrossRef]
- Proietti, P.; Federici, E.; Fidati, L.; Scargetta, S.; Massaccesi, L.; Nasini, L.; Regni, L.; Ricci, A.; Cenci, G.; Gigliotti, G. Effects of amendment with oil mill waste and its derived-compost on soil chemical and microbiological characteristics and olive (Olea europaea L.) productivity. Agric. Ecosyst. Environ. 2015, 207, 51–60. [Google Scholar] [CrossRef]
- Altieri, R.; Esposto, A. Olive orchard amended with two experimental olive mill wastes mixtures: Effects on soil organic carbon, plant growth and yield. Bioresour. Technol. 2008, 99, 8390–8393. [Google Scholar] [CrossRef]
- Roig, A.; Cayuela, M.L.; Sánchez-Monedero, M.A. An overview on olive mill wastes and their valorisation methods. Waste Manag. 2006, 26, 960–969. [Google Scholar] [CrossRef] [PubMed]
- Amaral, C.; Lucas, M.S.; Coutinho, J.; Crespí, A.L.; do Rosário Anjos, M.; Pais, C. Microbiological and physicochemical characterization of olive mill wastewaters from a continuous olive mill in Northeastern Portugal. Bioresour. Technol. 2008, 99, 7215–7223. [Google Scholar] [CrossRef]
- El Hadrami, A. Physico-chemical Characterization and Effects of Olive Oil Mill Wastewaters Fertirrigation on the Growth of Some Mediterranean Crops. J. Agron. 2004, 3, 247–254. [Google Scholar] [CrossRef]
- Ayed, L.; Assas, N.; Sayadi, S.; Hamdi, M. Involvement of lignin peroxidase in the decolourization of black olive mill wastewaters by Geotrichum candidum. Lett. Appl. Microbiol. 2005, 40, 7–11. [Google Scholar] [CrossRef]
- Saadi, I.; Laor, Y.; Raviv, M.; Medina, S. Land spreading of olive mill wastewater: Effects on soil microbial activity and potential phytotoxicity. Chemosphere 2007, 66, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Karpouzas, D.G.; Rousidou, C.; Papadopoulou, K.K.; Bekris, F.; Zervakis, G.I.; Singh, B.K.; Ehaliotis, C. Effect of continuous olive mill wastewater applications, in the presence and absence of nitrogen fertilization, on the structure of rhizosphere-soil fungal communities. FEMS Microbiol. Ecol. 2009, 70, 388–401. [Google Scholar] [CrossRef] [PubMed]
- Kavroulakis, N.; Ntougias, S. Bacterial and β-proteobacterial diversity in Olea europaea var. mastoidis- and Olea europaea var. koroneiki-generated olive mill wastewaters: Influence of cultivation and harvesting practice on bacterial community structure. World J. Microbiol. Biotechnol. 2010, 27, 57–66. [Google Scholar] [CrossRef]
- Rincón, B.; Raposo, F.; Borja, R.; Gonzalez, J.; Portillo, M.; Saiz-Jimenez, C. Performance and microbial communities of a continuous stirred tank anaerobic reactor treating two-phases olive mill solid wastes at low organic loading rates. J. Biotechnol. 2006, 121, 534–543. [Google Scholar] [CrossRef]
- Morillo, J.A.; Aguilera, M.; Antízar-Ladislao, B.; Fuentes, S.; Ramos-Cormenzana, A.; Russell, N.J.; Monteoliva-Sánchez, M. Molecular microbial and chemical investigation of the bioremediation of two-phase olive mill waste using laboratory-scale bioreactors. Appl. Microbiol. Biotechnol. 2008, 79, 309–317. [Google Scholar] [CrossRef]
- Tsiamis, G.; Tzagkaraki, G.; Chamalaki, A.; Xypteras, N.; Andersen, G.; Vayenas, D.; Bourtzis, K. Olive-Mill Wastewater Bacterial Communities Display a Cultivar Specific Profile. Curr. Microbiol. 2011, 64, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Lakshmanan, V.; Selvaraj, G.; Bais, H.P. Functional Soil Microbiome: Belowground Solutions to an Aboveground Problem. Plant Physiol. 2014, 166, 689–700. [Google Scholar] [CrossRef]
- Jansson, J.; Hofmockel, K.S. The soil microbiome—From metagenomics to metaphenomics. Curr. Opin. Microbiol. 2018, 43, 162–168. [Google Scholar] [CrossRef]
- Fausto, C.; Mininni, A.N.; Sofo, A.; Crecchio, C.; Scagliola, M.; Dichio, B.; Xiloyannis, C. Olive orchard microbiome: Characterisation of bacterial communities in soil-plant compartments and their comparison between sustainable and conventional soil management systems. Plant Ecol. Divers. 2018, 11, 597–610. [Google Scholar] [CrossRef]
- Sofo, A.; Ricciuti, P.; Fausto, C.; Mininni, A.N.; Crecchio, C.; Scagliola, M.; Malerba, A.D.; Xiloyannis, C.; Dichio, B. The metabolic and genetic diversity of soil bacterial communities depends on the soil management system and C/N dynamics: The case of sustainable and conventional olive groves. Appl. Soil Ecol. 2019, 137, 21–28. [Google Scholar] [CrossRef]
- Mekki, A.; Dhouib, A.; Sayadi, S. Changes in microbial and soil properties following amendment with treated and untreated olive mill wastewater. Microbiol. Res. 2006, 161, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Mechri, B.; Echbili, A.; Issaoui, M.; Braham, M.; Ben Elhadj, S.; Hammami, M. Short-term effects in soil microbial community following agronomic application of olive mill wastewaters in a field of olive trees. Appl. Soil Ecol. 2007, 36, 216–223. [Google Scholar] [CrossRef]
- Jarboui, R.; Chtourou, M.; Azri, C.; Gharsallah, N.; Ammar, E. Time-dependent evolution of olive mill wastewater sludge organic and inorganic components and resident microbiota in multi-pond evaporation system. Bioresour. Technol. 2010, 101, 5749–5758. [Google Scholar] [CrossRef]
- Hentati, O.; Oliveira, V.; Sena, C.; Bouji, M.S.M.; Wali, A.; Ksibi, M. Soil contamination with olive mill wastes negatively affects microbial communities, invertebrates and plants. Ecotoxicology 2016, 25, 1500–1513. [Google Scholar] [CrossRef]
- Peikert, B.; Schaumann, G.E.; Bibus, D.; Fischer, J.; Braun, U.; Brunkhardt, J. Effects of olive oil mill wastewater on chemical, microbiological, and physical properties of soil incubated under four different climatic conditions. Biol. Fertil. Soils 2016, 53, 89–102. [Google Scholar] [CrossRef]
- Rossi, L.; Regni, L.; Rinaldi, S.; Sdringola, P.; Calisti, R.; Brunori, A.; Dini, F.; Proietti, P. Long-Term Water Footprint Assessment in a Rainfed Olive Tree Grove in the Umbria Region, Italy. Agric. Lond. 2019, 10, 8. [Google Scholar] [CrossRef]
- Bacon, J.R.; Hudson, G. A flexible methodology for the characterization of soils: A case study of the heavy metal status of a site at Dornach. Sci. Total Environ. 2001, 264, 153–162. [Google Scholar] [CrossRef]
- Tian, G.; Vose, J.M.; Coleman, D.; Geron, C.D.; Walker, J.T. Evaluation of the effectiveness of riparian zone restoration in the southern Appalachians by assessing soil microbial populations. Appl. Soil Ecol. 2004, 26, 63–68. [Google Scholar] [CrossRef][Green Version]
- Nelson, D.W.; Sommers, L.E. Total Carbon, Organic Carbon, and Organic Matter. In Methods of Soil Analysis: Part 3 Chemical Methods; Bigham, J.M., Ed.; SSSA: Madison, WI, USA, 1996; Volume 5, pp. 961–1010. [Google Scholar]
- ANPA (Agenzia Nazionale per la Protezione Dell’Ambiente). Metodi di Analisi del Compost. Manuali e Linee Guida 3; ANPA: Roma, Italy, 2001. [Google Scholar]
- Carter, M.R.; Gregorich, E.G. (Eds.) Soil Sampling and Methods of Analysis; CRC Press: Boca Raton, FL, USA, 2008. [Google Scholar]
- Ciavatta, C.; Govi, M.; Antisari, L.V.; Sequi, P. An enzymatic approach to the determination of the degree of stabilization of organic carbon in fertilizers. Nutr. Cycl. Agroecosyst. 1990, 25, 167–174. [Google Scholar] [CrossRef]
- Pent, M.; Põldmaa, K.; Bahram, M. Bacterial Communities in Boreal Forest Mushrooms Are Shaped Both by Soil Parameters and Host Identity. Front. Microbiol. 2017, 8, 836. [Google Scholar] [CrossRef] [PubMed]
- Bolano, A.; Stinchi, S.; Preziosi, R.; Bistoni, F.; Allegrucci, M.; Baldelli, F.; Martini, A.; Cardinali, G. Rapid methods to extract DNA and RNA fromCryptococcus neoformans. FEMS Yeast Res. 2001, 1, 221–224. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Amir, A.; McDonald, D.; Navas-Molina, J.A.; Kopylova, E.; Morton, J.T.; Xu, Z.Z.; Kightley, E.P.; Thompson, L.R.; Hyde, E.R.; Gonzalez, A.; et al. Deblur Rapidly Resolves Single-Nucleotide Community Sequence Patterns. mSystems 2017, 2, e00191-16. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl. Environ. Microbiol. 2016, 71, 672–685. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Vanderplas, J. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria; Available online: http://www.R-project.org (accessed on 8 December 2021).
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- De Mendiburu, F. Una Herramienta de Analisis Estadistico para la Investigacion Agricola. Master’s Thesis, Lima-PERU, Facultad de Economia y Planificacion Departamento Academico de Estadistica e Informatica (UNI-PERU), Universidad Nacional Agraria La Molina, La Molina, Peru, 2009. [Google Scholar]
- Oksanen, J. Vegan: An Introduction to Ordination. 2017. Available online: https://cran.r-project.org/web/packages/vegan/vignettes/intro-vegan.pdf (accessed on 8 December 2021).
- Weiss, S.; Xu, Z.Z.; Peddada, S.; Amir, A.; Bittinger, K.; Gonzalez, A.; Lozupone, C.; Zaneveld, J.R.; Vázquez-Baeza, Y.; Birmingham, A.; et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 2017, 5, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.C.; Ma, K.H.; Chao, A. iNEXT: An R package for rarefaction and extrapolation of species diversity (H ill numbers). Methods Ecol. Evol. 2016, 7, 1451–1456. [Google Scholar] [CrossRef]
- Anderson, M.J.; Willis, T.J. Canonical analysis of principal coordinates: A useful method of constrained ordination for ecology. Ecology 2003, 84, 511–525. [Google Scholar] [CrossRef]
- Mandal, S.; Van Treuren, W.; White, R.A.; Eggesbø, M.; Knight, R.T.; Peddada, S.D. Analysis of composition of microbiomes: A novel method for studying microbial composition. Microb. Ecol. Health Dis. 2015, 26, 27663. [Google Scholar] [CrossRef] [PubMed]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: AnRPackage for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Zar, J.H. Multiple repression and correlation. In Biostatistical Analysis; Zar, J.H., Ed.; Prentice-Hall International: Upper Saddle River, NJ, USA, 1996; pp. 407–445. [Google Scholar]
- Akaike, H. A New Look at the Statistical Model Identification. In Selected Papers of Hirotugu Akaike; Springer: New York, NY, USA, 1974; pp. 215–222. [Google Scholar]
- Venables, W.N.; Ripley, B.D. Modern Applied Statistics with S, 4th ed.; Springer: New York, NY, USA, 2002; ISBN 0-387-95457-0. [Google Scholar]
- Legendre, P.; Legendre, L. Numerical Ecology; Elsevier: Amsterdam, The Netherlands, 1998; p. 853. [Google Scholar]
- Acland, A.; Agarwala, R.; Barrett, T.; Beck, J.; Benson, D.A.; Bollin, C.; Bolton, E.; Bryant, S.H.; Canese, K.; Church, D.M.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2014, 42, D7–D17. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Sierra, J.; Martí, E.; Montserrat, G.; Cruañas, R.; Garau, M. Characterisation and evolution of a soil affected by olive oil mill wastewater disposal. Sci. Total Environ. 2001, 279, 207–214. [Google Scholar] [CrossRef]
- Meftah, O.; Guergueb, Z.; Braham, M.; Sayadi, S.; Mekki, A. Long term effects of olive mill wastewaters application on soil properties and phenolic compounds migration under arid climate. Agric. Water Manag. 2019, 212, 119–125. [Google Scholar] [CrossRef]
- Mahmoud, M.; Janssen, M.; Haboub, N.; Nassour, A.; Lennartz, B. The impact of olive mill wastewater application on flow and transport properties in soils. Soil Tillage Res. 2010, 107, 36–41. [Google Scholar] [CrossRef]
- Rousk, J.; Brookes, P.C.; Bååth, E. Contrasting Soil pH Effects on Fungal and Bacterial Growth Suggest Functional Redundancy in Carbon Mineralization. Appl. Environ. Microbiol. 2009, 75, 1589–1596. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.; Fierer, N.; Lauber, C.L.; Caporaso, J.G.; Knight, R.; Grogan, P. Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ. Microbiol. 2010, 12, 2998–3006. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Hu, X.; Li, X.; Zhang, Y.; Jiang, L.; Li, J.; Guan, Z.; Cai, Y.; Liao, X. The rhizospheric microbial community structure and diversity of deciduous and evergreen forests in Taihu Lake area, China. PLoS ONE 2017, 12, e0174411. [Google Scholar] [CrossRef]
- Pezzolla, D.; Marconi, G.; Turchetti, B.; Zadra, C.; Agnelli, A.; Veronesi, F.; Onofri, A.; Benucci, G.M.N.; Buzzini, P.; Albertini, E.; et al. Influence of exogenous organic matter on prokaryotic and eukaryotic microbiota in an agricultural soil. A multidisciplinary approach. Soil Biol. Biochem. 2015, 82, 9–20. [Google Scholar] [CrossRef]
- Marschner, B. Structure and function of the soil microbial community in a long-term fertilizer experiment. Soil Biol. Biochem. 2003, 35, 453–461. [Google Scholar] [CrossRef]
- Francioli, D.; Schulz, E.; Lentendu, G.; Wubet, T.; Buscot, F.; Reitz, T. Mineral vs. Organic Amendments: Microbial Community Structure, Activity and Abundance of Agriculturally Relevant Microbes Are Driven by Long-Term Fertilization Strategies. Front. Microbiol. 2016, 7, 1446. [Google Scholar] [CrossRef]
- de Gannes, V.; Eudoxie, G.; Hickey, W.J. Prokaryotic successions and diversity in composts as revealed by 454-pyrosequencing. Bioresour. Technol. 2013, 133, 573–580. [Google Scholar] [CrossRef]
- Neher, D.A.; Weicht, T.R.; Bates, S.T.; Leff, J.W.; Fierer, N. Changes in Bacterial and Fungal Communities across Compost Recipes, Preparation Methods, and Composting Times. PLoS ONE 2013, 8, e79512. [Google Scholar] [CrossRef] [PubMed]
- Storey, S.; Chualain, D.N.; Doyle, O.; Clipson, N.; Doyle, E. Comparison of bacterial succession in green waste composts amended with inorganic fertiliser and wastewater treatment plant sludge. Bioresour. Technol. 2015, 179, 71–77. [Google Scholar] [CrossRef]
- Bertin, L.; Colao, M.C.; Ruzzi, M.; Fava, F. Performances and microbial features of a granular activated carbon packed-bed biofilm reactor capable of an efficient anaerobic digestion of olive mill wastewaters. FEMS Microbiol. Ecol. 2004, 48, 413–423. [Google Scholar] [CrossRef]
- Bertin, L.; Colao, M.C.; Ruzzi, M.; Marchetti, L.; Fava, F. Performances and microbial features of an aerobic packed-bed biofilm reactor developed to post-treat an olive mill effluent from an anaerobic GAC reactor. Microb. Cell Factories 2006, 5, 16. [Google Scholar] [CrossRef]
- Ntougias, S.; Zervakis, G.I.; Ehaliotis, C.; Kavroulakis, N.; Papadopoulou, K.K. Ecophysiology and molecular phylogeny of bacteria isolated from alkaline two-phase olive mill wastes. Res. Microbiol. 2006, 157, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Pozo, C.; Rodelas, B.; Martínez-Toledo, M.V.; Vílchez, R.; González-López, J. Removal of organic load from olive washing water by an aerated submerged biofilter and profiling of the bacterial community involved in the process. J. Microbiol. Biotechnol. 2007, 17, 18051300. [Google Scholar]
- Vivas, A.; Moreno, B.; Garcia-Rodriguez, S.; Benitez, E. Assessing the impact of composting and vermicomposting on bacterial community size and structure, and microbial functional diversity of an olive-mill waste. Bioresour. Technol. 2009, 100, 1319–1326. [Google Scholar] [CrossRef] [PubMed]
- Rousk, J.; Bååth, E.; Brookes, P.C.; Lauber, C.L.; Lozupone, C.; Caporaso, J.G.; Knight, R.; Fierer, N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef]
- Blagodatskaya, E.; Blagodatsky, S.; Anderson, T.-H.; Kuzyakov, Y. Priming effects in Chernozem induced by glucose and N in relation to microbial growth strategies. Appl. Soil Ecol. 2007, 37, 95–105. [Google Scholar] [CrossRef]
- Fierer, N.; Breitbart, M.; Nulton, J.; Salamon, P.; Lozupone, C.; Jones, R.; Robeson, M.; Edwards, R.; Felts, B.; Rayhawk, S.; et al. Metagenomic and Small-Subunit rRNA Analyses Reveal the Genetic Diversity of Bacteria, Archaea, Fungi, and Viruses in Soil. Appl. Environ. Microbiol. 2007, 73, 7059–7066. [Google Scholar] [CrossRef] [PubMed]
- Ullah, A.; Mushtaq, H.; Fahad, S.; Shah, A.; Chaudhary, H.J. Plant growth promoting potential of bacterial endophytes in novel association with Olea ferruginea and Withania coagulans. Microbiology 2017, 86, 119–127. [Google Scholar] [CrossRef]
- Kumar, A.; Maurya, B.R.; Raghuwanshi, R.; Meena, V.S.; Islam, M.T. Co-inoculation with Enterobacter and Rhizobacteria on Yield and Nutrient Uptake by Wheat (Triticum aestivum L.) in the Alluvial Soil Under Indo-Gangetic Plain of India. J. Plant Growth Regul. 2017, 36, 608–617. [Google Scholar] [CrossRef]
- Bhise, K.K.; Bhagwat, P.K.; Dandge, P.B. Plant Growth-Promoting Characteristics of Salt Tolerant Enterobacter cloacae Strain KBPD and Its Efficacy in Amelioration of Salt Stress in Vigna radiata L. J. Plant Growth Regul. 2016, 36, 215–226. [Google Scholar] [CrossRef]
- Zhong, W.; Gu, T.; Wang, W.; Zhang, B.; Lin, X.; Huang, Q.; Shen, W. The effects of mineral fertilizer and organic manure on soil microbial community and diversity. Plant Soil 2010, 326, 511–522. [Google Scholar] [CrossRef]
- Lazcano, C.; Gómez-Brandón, M.; Revilla, P.; Domínguez, J. Short-term effects of organic and inorganic fertilizers on soil microbial community structure and function. Biol. Fertil. Soils 2013, 49, 723–733. [Google Scholar] [CrossRef]
- Siles, J.; Pascual, J.; González-Menéndez, V.; Sampedro, I.; García-Romera, I.; Bills, G. Short-term dynamics of culturable bacteria in a soil amended with biotransformed dry olive residue. Syst. Appl. Microbiol. 2013, 37, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Nemergut, D.R.; Cleveland, C.C.; Wieder, W.; Washenberger, C.L.; Townsend, A.R. Plot-scale manipulations of organic matter inputs to soils correlate with shifts in microbial community composition in a lowland tropical rain forest. Soil Biol. Biochem. 2010, 42, 2153–2160. [Google Scholar] [CrossRef]
- Pascault, N.; Ranjard, L.; Kaisermann, A.; Bachar, D.; Christen, R.; Terrat, S.; Mathieu, O.; Lévêque, J.; Mougel, C.; Henault, C.; et al. Stimulation of Different Functional Groups of Bacteria by Various Plant Residues as a Driver of Soil Priming Effect. Ecosystems 2013, 16, 810–822. [Google Scholar] [CrossRef]
- Liu, J.J.; Sui, Y.Y.; Yu, Z.H.; Shi, Y.; Chu, H.Y.; Jin, J.; Linu, X.; Wang, G. High throughput sequencing analysis of biogeographical distribution of bacterial communities in the black soils of northeast China. Soil Biol. Biochem. 2014, 70, 113–122. [Google Scholar] [CrossRef]
- Li, C.H.; Yan, K.; Tang, L.S.; Jia, Z.J.; Li, Y. Change in deep soil microbial communities due to long-term fertilization. Soil Biol. Biochem. 2014, 75, 264–272. [Google Scholar] [CrossRef]
- Karpouzas, D.G.; Ntougias, S.; Iskidou, E.; Rousidou, C.; Papadopoulou, K.K.; Zervakis, G.I.; Ehaliotis, C. Olive mill wastewater affects the structure of soil bacterial communities. Appl. Soil Ecol. 2010, 45, 101–111. [Google Scholar] [CrossRef]
- Siles, J.A.; Rachid, C.T.C.C.; Sampedro, I.; García-Romera, I.; Tiedje, J.M. Microbial Diversity of a Mediterranean Soil and Its Changes after Biotransformed Dry Olive Residue Amendment. PLoS ONE 2014, 9, e103035. [Google Scholar] [CrossRef]
- Sampedro, I.; Giubilei, M.; Cajthaml, T.; Federici, E.; Federici, F.; Petruccioli, M.; D’Annibale, A. Short-term impact of dry olive mill residue addition to soil on the resident microbiota. Bioresour. Technol. 2009, 100, 6098–6106. [Google Scholar] [CrossRef]
- Rousidou, C.; Papadopoulou, K.; Zervakis, G.; Singh, B.K.; Ehaliotis, C.; Karpouzas, D.G. Repeated application of diluted olive mill wastewater induces changes in the structure of the soil microbial community. Eur. J. Soil Biol. 2010, 46, 34–40. [Google Scholar] [CrossRef]
- Proietti, P.; Cartechini, A.; Tombesi, A. Influenza delle acque reflue di frantoi oleari su olivi in vaso ed in campo. L’informatore Agrario 1988, 45, 87–91. [Google Scholar]
- Chartzoulakis, K.; Psarras, G.; Moutsopoulou, M.; Stefanoudaki, E. Application of olive mill wastewater to a Cretan olive orchard: Effects on soil properties, plant performance and the environment. Agric. Ecosyst. Environ. 2010, 138, 293–298. [Google Scholar] [CrossRef]
- Ferri, D.; Convertini, G.; Montemurro, F.; Rinaldi, M.; Rana, G. Olive Wastes Spreading in Southern Italy: Effects on Crops and Soil. In Proceedings of the 12th ISCO Conference, Beijing, China, 26–31 May 2002; pp. 593–600. [Google Scholar]
- Gioffré, D.; Cannavò, S.; Smorto, D. Risultati sugli effetti delle acque reflue olearie somministrate con diverse modalità su terreno ulivetato in pieno campo e su piante di olivo allevate in mastello. In Valorizzazione di Acque Reflue e Sottoprodotti Dell’industria Agrumaria e Olearia; Laruffa: Reggio Calabria, Italy, 2004; pp. 81–98. [Google Scholar]
- Magdich, S.; Abid, W.; Boukhris, M.; Ben Rouina, B.; Ammar, E. Effects of long-term olive mill wastewater spreading on the physiological and biochemical responses of adult Chemlali olive trees (Olea europaea L.). Ecol. Eng. 2016, 97, 122–129. [Google Scholar] [CrossRef]

| Parameters | OMWW |
|---|---|
| Moisture (%) | 87.1 ± 0.1 |
| Volatile solids (%) | 94.6 ± 0.0 |
| pH | 5.4 ± 0.0 |
| EC (mS/cm) | 3.7 ± 0.0 |
| TOC (%) | 48.7 ± 3.3 |
| TKN (%) | 0.39 ± 0.05 |
| Ammonium N (%) | 0.12 ± 0.01 |
| Organic N (%) | 0.27 |
| Organic N (% on total N) | 70 |
| WEOC (%) | 11.2 ± 0.0 |
| Total P (%) | 0.11 ± 0.00 |
| Total K (%) | 0.01 ± 0.00 |
| Parameters | Time | |||
|---|---|---|---|---|
| Unamended_20 | 14 Days_20 | 1 Year_20 | 2 Year_20 | |
| pH | 8.2 ± 0.1 | 7.5 ± 0.1 | 8.0 ± 0.0 | 8.1 ± 0.0 |
| EC (µS/cm) | 342 ± 21 | 2660 ± 30 | 630 ± 125 | 333 ± 5 |
| TOC (%) | 2.00 ± 0.23 | 2.22 ± 0.02 | 2.27 ± 0.01 | 1.35 ± 0.06 |
| TEC (%) | 1.27 ± 0.38 | 2.04 ± 0.13 | 2.70 ± 0.03 | 1.27 ± 0.03 |
| WEOC (mg/kg) | 316.8 ± 0.2 | 3574.0 ± 6.0 | 815.4 ± 65.4 | 455.5 ± 4.9 |
| TKN (%) | 0.17 ± 0.04 | 0.18 ± 0.00 | 0.39 ± 0.02 | 0.29 ± 0.04 |
| CEC (meq/100 g) | 16.1 ± 0.5 | 14.1 ± 0.3 | 17.1 ± 0.1 | 13.8 ± 0.0 |
| Available P (mg/kg) | 10.5 ± 0.7 | 86.7 ± 1.5 | 44.7 ± 12.7 | 48.5 ± 3.3 |
| Exchangeable K (mg/kg) | 725 ± 38 | 2389 ± 109 | 1090 ± 169 | 767 ± 13 |
| Exchangeable Mg (mg/kg) | 168 ± 7 | 162 ± 20 | 169 ± 15 | 180 ± 6 |
| Exchangeable Ca (mg/kg) | 8698 ± 990 | 7526 ± 586 | 6296 ± 280 | 10394 ± 954 |
| Parameters | Time | |||
|---|---|---|---|---|
| Unamended_40 | 14 Days_40 | 1 Year_40 | 2 Year_40 | |
| pH | 8.4 ± 0.0 | 7.9 ± 0.0 | 8.2 ± 0.0 | 8.4 ± 0.0 |
| EC (µS/cm) | 352 ± 4 | 1171 ± 47 | 516 ± 50 | 257 ± 10 |
| TOC (%) | 0.66 ± 0.04 | 1.02 ± 0.01 | 0.58 ± 0.21 | 0.60 ± 0.09 |
| TEC (%) | 0.63 ± 0.01 | 0.98 ± 0.03 | 1.30 ± 0.17 | 0.67 ± 0.03 |
| WEOC (mg/kg) | 284.3 ± 15.5 | 1134 ± 9.2 | 408.7 ± 32.5 | 414.0 ± 14.4 |
| TKN (%) | 0.19 ± 0.08 | 0.15 ± 0.01 | 0.13 ± 0.02 | 0.19 ± 0.01 |
| CEC (meq/100 g) | 13.7 ± 0.7 | 11.9 ± 0.1 | 11.5 ± 2.0 | 12.6 ± 0.3 |
| Available P (mg/kg) | 6.3 ± 0.8 | 65.7 ± 2.4 | 26.1 ± 8.4 | 46.4 ± 0.9 |
| Exchangeable K (mg/kg) | 603 ± 8 | 987 ± 80 | 715 ± 232 | 717 ± 8 |
| Exchangeable Mg (mg/kg) | 169 ± 16 | 183 ± 32 | 121 ± 20 | 144 ± 16 |
| Exchangeable Ca (mg/kg) | 9852 ± 80 | 8688 ± 96 | 5296 ± 60 | 8566 ± 1014 |
| ANCOM Results (ASVs) | Reject Null Hypothesis | W | Taxonomy |
|---|---|---|---|
| 54006fd3be9a8609d4a2d04f12573a0c | True | 7524 | k_Bacteria;p_Proteobacteria;c_Gammaproteobacteria;o_Enterobacteriales;f_Enterobacteriaceae; |
| 040ff57af2e17018f490b96f4451a3e4 | True | 7458 | k_Bacteria;p_Proteobacteria;c_Gammaproteobacteria;o_Enterobacteriales;f_Enterobacteriaceae; |
| 1ae7ead5138fa5145ad71abc1bb2e776 | True | 7424 | k_Bacteria;p_Firmicutes;c_Bacilli;o_Bacillales;f_Sporolactobacillaceae;g_Sporolactobacillus; |
| 1157032931d4b8ce4a851e95cfcc3c33 | True | 7232 | k_Bacteria;p_Firmicutes;c_Clostridia;o_Clostridiales;f_Clostridiaceae;g_Clostridium;s_butyricum |
| 8afe5e8dbfe1fbab3b787103dba1ce81 | True | 7514 | k_Bacteria;p_Proteobacteria;c_Alphaproteobacteria;o_Sphingomonadales;f_Sphingomonadaceae;g_Novosphingobium |
| 1653005b37d2ba3490c18db3359794ff | True | 7116 | k_Bacteria;p_Firmicutes;c_Clostridia;o_Clostridiales;f_Lachnospiraceae;g_Coprococcus |
| 7da1250b471f87c40c9fc162c3ead212 | True | 7256 | k_Bacteria;p_Firmicutes;c_Clostridia;o_Clostridiales;f_Clostridiaceae;g_Clostridium;s_butyricum |
| 67465099c7e7995ae38f287944b6094f | True | 7399 | k_Bacteria;p_Proteobacteria;c_Gammaproteobacteria;o_Enterobacteriales;f_Enterobacteriaceae;g_Trabulsiella |
| b95025e3d0cb9ce8c4807ce2737bdf1a | True | 7499 | k_Bacteria;p_Firmicutes;c_Bacilli;o_Bacillales;f_Sporolactobacillaceae;g_Sporolactobacillus |
| c98061c433607fe4b2f06fceb61c7458 | True | 7442 | k_Bacteria;p_Proteobacteria;c_Gammaproteobacteria;o_Enterobacteriales;f_Enterobacteriaceae;g_Enterobacter |
| e4a1ebdc1ea26629eca90e0fb4c34245 | True | 7266 | p_Proteobacteria;c_Gammaproteobacteria;o_Enterobacteriales;f_Enterobacteriaceae |
| 311a58a1ae5008a2ccde8f7a41710be7 | True | 7160 | k_Bacteria;p_Proteobacteria;c_Alphaproteobacteria;o_Rhodospirillales;f_Acetobacteraceae;g_Gluconobacter |
| Percentile | 0 | 25 | 50 | 75 | 100 | 0 | 25 | 50 | 75 | 100 | 0 | 25 | 50 | 75 | 100 | 0 | 25 | 50 | 75 | 100 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group | Unam. | Unam. | Unam. | Unam. | Unam. | 14D | 14D | 14D | 14D | 14D | 1Y | 1Y | 1Y | 1Y | 1Y | 2Y | 2Y | 2Y | 2Y | 2Y |
| 54006fd3be9a8609d4a2d04f12573a0c | 1 | 1 | 1 | 1 | 1 | 160 | 495.5 | 725 | 901.25 | 1867 | 1 | 1 | 1 | 1 | 52 | 1 | 1 | 1 | 1 | 1 |
| 040ff57af2e17018f490b96f4451a3e4 | 1 | 1 | 1 | 1 | 1 | 8 | 113.75 | 165.5 | 176.75 | 539 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 1ae7ead5138fa5145ad71abc1bb2e776 | 1 | 1 | 1 | 1 | 1 | 48 | 80.25 | 179.5 | 216.5 | 296 | 1 | 1 | 1 | 8.5 | 14 | 1 | 1 | 1 | 1 | 2 |
| 1157032931d4b8ce4a851e95cfcc3c33 | 1 | 1 | 1 | 1 | 2 | 109 | 178.75 | 219.5 | 417 | 676 | 1 | 1 | 7 | 15.25 | 23 | 1 | 1 | 1 | 9.25 | 25 |
| 8afe5e8dbfe1fbab3b787103dba1ce81 | 1 | 1 | 1 | 1 | 1 | 166 | 397.5 | 583.5 | 899.25 | 1014 | 1 | 1 | 1 | 1 | 64 | 1 | 1 | 1 | 1 | 3 |
| 1653005b37d2ba3490c18db3359794ff | 1 | 1 | 1 | 1 | 1 | 21 | 22.5 | 40 | 75.5 | 150 | 1 | 1 | 1 | 2.5 | 4 | 1 | 1 | 1 | 1 | 5 |
| 7da1250b471f87c40c9fc162c3ead212 | 1 | 1 | 1 | 1 | 9 | 113 | 129.75 | 162 | 258 | 342 | 1 | 2.25 | 6.5 | 15.25 | 19 | 1 | 1 | 1.5 | 2 | 13 |
| 67465099c7e7995ae38f287944b6094f | 1 | 1 | 1 | 1 | 1 | 19 | 33.5 | 67 | 90 | 151 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| b95025e3d0cb9ce8c4807ce2737bdf1a | 1 | 1 | 1 | 1 | 1 | 194 | 353.75 | 550.5 | 975.25 | 1523 | 1 | 1 | 5 | 24.75 | 39 | 1 | 1 | 1 | 1 | 20 |
| c98061c433607fe4b2f06fceb61c7458 | 1 | 1 | 1 | 1 | 1 | 24 | 42.75 | 92 | 108.25 | 226 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| e4a1ebdc1ea26629eca90e0fb4c34245 | 1 | 1 | 1 | 1 | 1 | 13 | 26.25 | 45.5 | 76 | 158 | 1 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 1 |
| 311a58a1ae5008a2ccde8f7a41710be7 | 1 | 1 | 1 | 1 | 1 | 1 | 433.75 | 649.5 | 900.5 | 1378 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Pairwise Comparison | Group | Reject Null Hypothesis | W | Taxonomy |
|---|---|---|---|---|
| Unamended vs. 14 days | 1157032931d4b8ce4a851e95cfcc3c33 | True | 5770 | k_Bacteria;p_Firmicutes;c_Clostridia;o_Clostridiales;f_Clostridiaceae;g_Clostridium;s_butyricum |
| Unamended vs. 14 days | 7da1250b471f87c40c9fc162c3ead212 | True | 5346 | k_Bacteria;p_Firmicutes;c_Clostridia;o_Clostridiales;f_Clostridiaceae;g_Clostridium;s_butyricum |
| Unamended vs. 14 days | b95025e3d0cb9ce8c4807ce2737bdf1a | True | 5844 | k_Bacteria;p_Firmicutes;c_Bacilli;o_Bacillales;f_Sporolactobacillaceae;g_Sporolactobacillus |
| Unamended vs. 14 days | 54006fd3be9a8609d4a2d04f12573a0c | True | 5852 | k_Bacteria;p_Proteobacteria;c_Gammaproteobacteria;o_Enterobacteriales;f_Enterobacteriaceae |
| Unamended vs. 14 days | 8afe5e8dbfe1fbab3b787103dba1ce81 | True | 5847 | k_Bacteria;p_Proteobacteria;c_Alphaproteobacteria;o_Sphingomonadales;f_Sphingomonadaceae;g_Novosphingobium |
| Unamended vs. 14 days | 1ae7ead5138fa5145ad71abc1bb2e776 | True | 5466 | k_Bacteria;p_Firmicutes;c_Bacilli;o_Bacillales;f_Sporolactobacillaceae;g_Sporolactobacillus; |
| 14 days vs. 1 year | 67465099c7e7995ae38f287944b6094f | True | 3729 | k_Bacteria;p_Proteobacteria;c_Gammaproteobacteria;o_Enterobacteriales;f_Enterobacteriaceae;g_Trabulsiella |
| 14 days vs. 1 year | c98061c433607fe4b2f06fceb61c7458 | True | 3931 | k_Bacteria;p_Proteobacteria;c_Gammaproteobacteria;o_Enterobacteriales;f_Enterobacteriaceae;g_Enterobacter |
| 14 days vs. 2 years | b95025e3d0cb9ce8c4807ce2737bdf1a | True | 5497 | k_Bacteria;p_Firmicutes;c_Bacilli;o_Bacillales;f_Sporolactobacillaceae;g_Sporolactobacillus;s_ |
| 14 days vs. 2 years | 54006fd3be9a8609d4a2d04f12573a0c | True | 5922 | k_Bacteria;p_Proteobacteria;c_Gammaproteobacteria;o_Enterobacteriales;f_Enterobacteriaceae;; |
| 14 days vs. 2 years | 8afe5e8dbfe1fbab3b787103dba1ce81 | True | 5910 | k_Bacteria;p_Proteobacteria;c_Alphaproteobacteria;o_Sphingomonadales;f_Sphingomonadaceae;g_Novosphingobium;s_ |
| 14 days vs. 2 years | e3d383866de994753086e21bf41b6947 | True | 5368 | k_Bacteria;p_Cyanobacteria;c_Chloroplast;o_Chlorophyta;f_;g_;s_ |
| 14 days vs. 2 years | 1ae7ead5138fa5145ad71abc1bb2e776 | True | 5409 | k_Bacteria;p_Firmicutes;c_Bacilli;o_Bacillales;f_Sporolactobacillaceae;g_Sporolactobacillus;s_ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Regni, L.; Pezzolla, D.; Ciancaleoni, S.; Marozzi, G.; Albertini, E.; Gigliotti, G.; Proietti, P. Long-Term Effects of Amendment with Olive Mill Wastewater on Soil Chemical Properties, Microbial Community, and Olive Tree Vegetative and Productive Activities. Agronomy 2021, 11, 2562. https://doi.org/10.3390/agronomy11122562
Regni L, Pezzolla D, Ciancaleoni S, Marozzi G, Albertini E, Gigliotti G, Proietti P. Long-Term Effects of Amendment with Olive Mill Wastewater on Soil Chemical Properties, Microbial Community, and Olive Tree Vegetative and Productive Activities. Agronomy. 2021; 11(12):2562. https://doi.org/10.3390/agronomy11122562
Chicago/Turabian StyleRegni, Luca, Daniela Pezzolla, Simona Ciancaleoni, Giorgio Marozzi, Emidio Albertini, Giovanni Gigliotti, and Primo Proietti. 2021. "Long-Term Effects of Amendment with Olive Mill Wastewater on Soil Chemical Properties, Microbial Community, and Olive Tree Vegetative and Productive Activities" Agronomy 11, no. 12: 2562. https://doi.org/10.3390/agronomy11122562
APA StyleRegni, L., Pezzolla, D., Ciancaleoni, S., Marozzi, G., Albertini, E., Gigliotti, G., & Proietti, P. (2021). Long-Term Effects of Amendment with Olive Mill Wastewater on Soil Chemical Properties, Microbial Community, and Olive Tree Vegetative and Productive Activities. Agronomy, 11(12), 2562. https://doi.org/10.3390/agronomy11122562










