Mining Single Nucleotide Polymorphism (SNP) Markers for Accurate Genotype Identification and Diversity Analysis of Chinese Jujube (Ziziphus jujuba Mill.) Germplasm
Abstract
1. Introduction
2. Materials and Methods
2.1. Discovering Jujube SNP Markers through Data Mining
2.2. Plant Materials and DNA Extraction
2.3. Validation of Putative SNPs
2.4. Data Analysis
3. Result
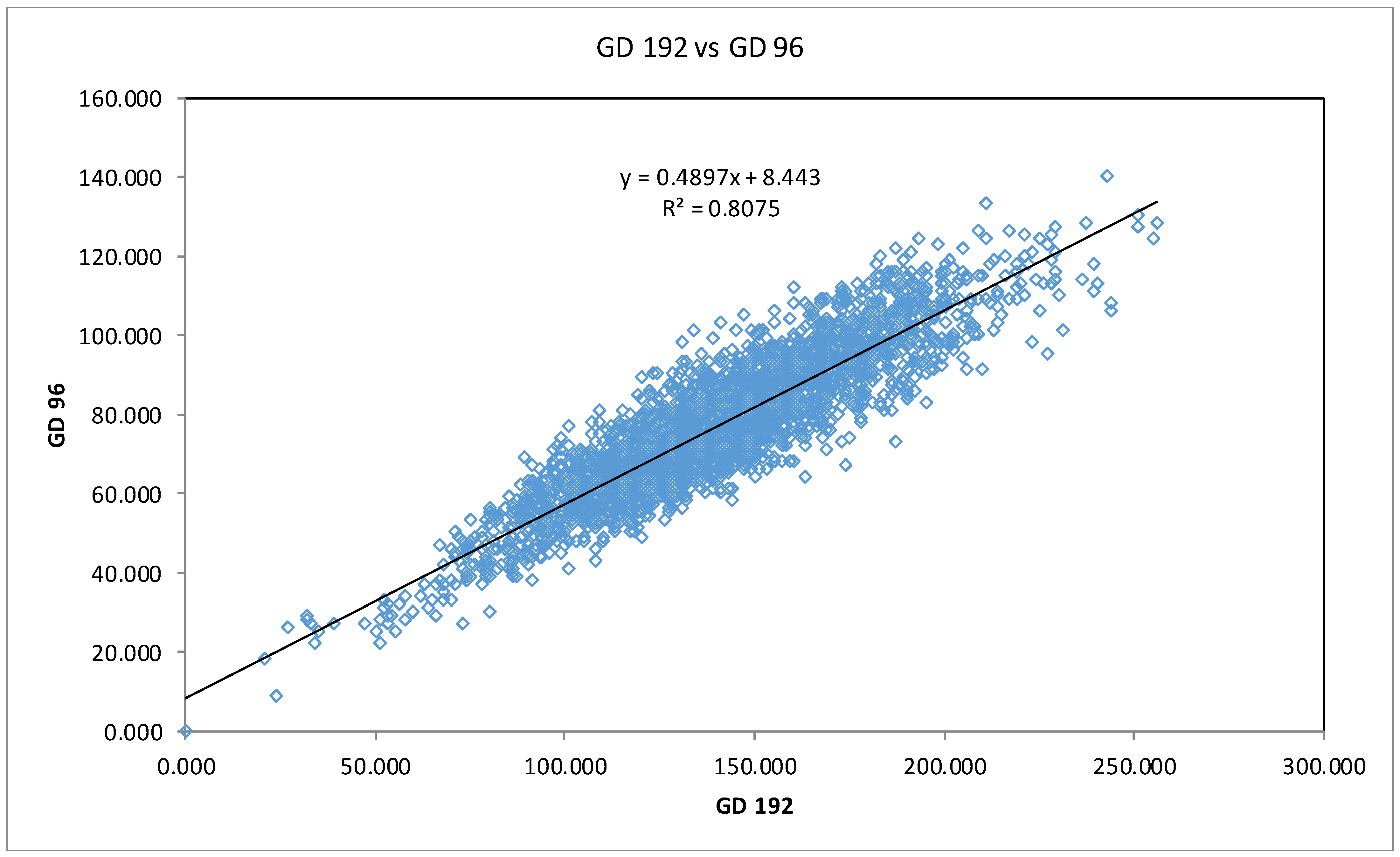
3.1. Data Mining and SNP Discovery
3.2. Cultivar Identification
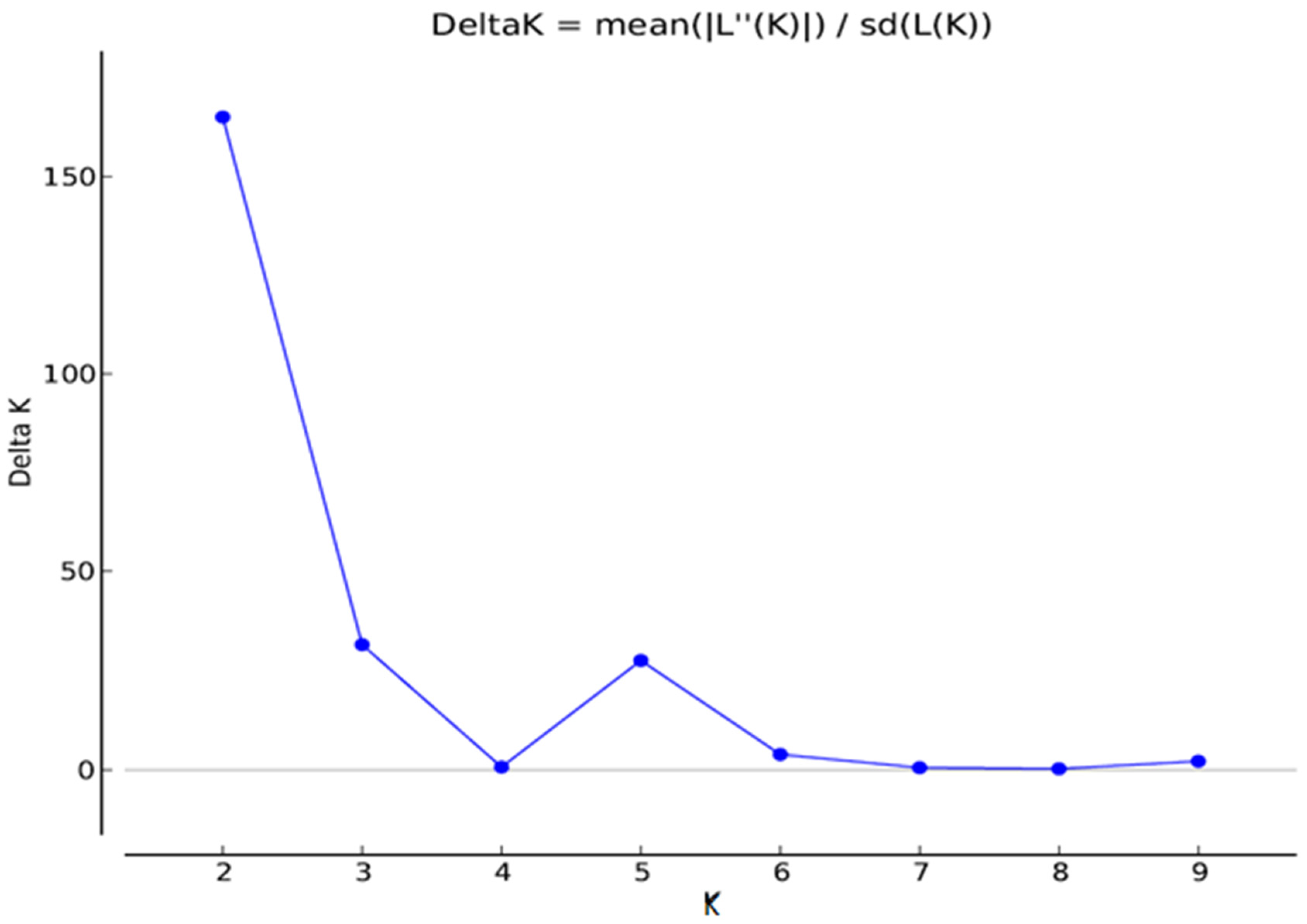
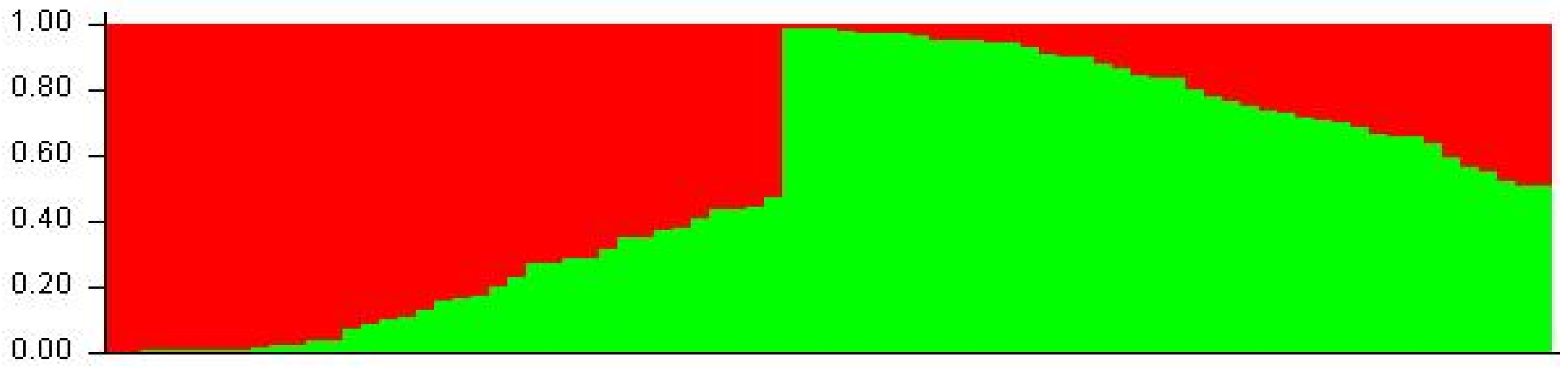
3.3. Population Stratification
3.4. PCoA and Clustering Analysis
3.5. Parentage Analysis
4. Discussion
4.1. Development of SNP Markers through Data Mining
4.2. Jujube Cultivar Identification Using SNP Markers
4.3. Parentage Verification for Improved Jujube Cultivars
4.4. The Core Set of SNP Markers for Universal Jujube Cultivar Identification
4.5. Genetic Relationships among the Different Germplasm Groups
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Qu, Z.Z.; Wang, Y.H.; Peng, S.Q.; Guo, Y.X. Monograph on Chinese jujube, In China Fruit-Plant; China Forestry Press: Beijing, China, 1993. [Google Scholar]
- Liu, M.J.; Wang, J.R.; Liu, P.; Zhao, J.; Zhao, Z.H.; Dai, L.; Li, X.S.; Liu, Z.G. Historical Achievements and Frontier Advances in the Production and Research of Chinese Jujube (Ziziphus jujube) in China. Acta Hortic. Sin. 2015, 42, 1683–1698. [Google Scholar] [CrossRef]
- Li, X.G. Chinese Jujube Industry; China Forestry Publishers: Beijing, China, 2015; p. 266. (In Chinese) [Google Scholar]
- Guo, Y.; Shan, G. The Chinese Jujube; Shanghai Scientific and Technical Publishers: Shanghai, China, 2010. (In Chinese) [Google Scholar]
- Nybom, H.; Weising, K.; Rotter, B. DNA Fingerprinting in Botany: Past, Present, Future. Investig. Genet. 2014, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Wang, G.; Liang, L. Development and Characterization of SSR Markers in Chinese Jujube (Ziziphus jujuba Mill.) and Its Related Species. Sci. Hortic. 2011, 129, 597–602. [Google Scholar] [CrossRef]
- Wang, S.; Liu, Y.; Ma, L.; Liu, H.; Tang, Y.; Wu, L.; Wang, Z.; Li, Y.; Wu, R.; Pang, X. Isolation and Characterization of Microsatellite Markers and Analysis of Genetic Diversity in Chinese jujube (Ziziphus Jujuba Mill.). PLoS ONE 2014, 9, e99842. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Zhao, J.; Liu, M.; Liu, P.; Dai, L.; Zhao, Z. Genome-Wide Characterization of Simple Sequence Repeat (SSR) Loci in Chinese Jujube and Jujube SSR Primer Transferability. PLoS ONE 2015, 10, e0127812. [Google Scholar] [CrossRef]
- Xu, C.; Gao, J.; Du, Z.; Li, D.; Wang, Z.; Li, Y.; Pang, X. Identifying the Genetic Diversity, Genetic Structure and a Core Col lection of Ziziphus Jujuba Mill. Var. Jujuba Accessions Using Microsatellite Markers. Sci. Rep. 2016, 6, e31503. [Google Scholar] [CrossRef]
- Liu, M.J. The Challenges and Countermeasures of Jujube Industry during Transition Period. China Fruits 2018, 1, 1–4. [Google Scholar] [CrossRef]
- Zhou, L.; Matsumoto, B.T.K.; Tan, H.; Meinhardt, L.W.; Mischke, B.S.; Wang, B.; Zhang, D. Developing Single Nucleotide Polymorphism (SNP) Markers for the Identification of Pineapple (Ananas Comosus) Germplasm. Hortic. Res. 2015, 2, 15056. [Google Scholar] [CrossRef]
- Fang, W.; Meinhardt, L.W.; Mischke, B.S.; Bellato, C.; Motilal, L.; Zhang, D. Accurate Determination of Genetic Identity for a Single Cacao Bean, Using Molecular Markers with a Nanofluidic System, Ensures Cocoa Authenticity and Traceability. J. Agric. Food Chem. 2013, 62, 481–487. [Google Scholar] [CrossRef]
- Fang, W.; Meinhardt, L.W.; Tan, H.; Zhou, L.; Wang, X.; Mischke, B.S.; Zhang, D. Identification of the Varietal Origin of Loose Leaf Tea Based on Analysis of a Single Leaf by SNP Nanofluidic Array. Crop. J. 2016, 4, 304–312. [Google Scholar] [CrossRef][Green Version]
- Zhang, Z.; Wei, T.; Zhong, Y.; Li, X.; Huang, J. Construction of a High-Density Genetic Map of Ziziphus Jujuba Mill. Using Genotyping by Sequencing Technology. Tree Genet. Genomes 2016, 4, 1–10. [Google Scholar] [CrossRef]
- Zhao, J.; Jian, J.; Liu, G.; Wang, J.; Lin, M.; Ming, Y.; Liu, Z.; Chen, Y.; Liu, X.; Liu, M. Rapid SNP Discovery and a RAD-Based High-Density Linkage Map in Jujube (Ziziphus Mill.). PLoS ONE 2014, 10, e109850. [Google Scholar] [CrossRef][Green Version]
- Chen, W.; Hou, L.; Zhang, Z.; Pang, X.; Li, Y. Genetic Diversity, Population Structure, and Linkage Disequilibrium of A Core Collection of Ziziphus Jujuba Assessed with Genome-Wide SNPS Developed by Genotyping-by-Sequencing and SSR Markers. Front. Plant Sci. 2017, 18, 575. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhao, J.; Cai, Q.; Liu, G.; Wang, J.R.; Zhao, Z.H.; Liu, P.; Dai, L.; Yan, G.; Wang, W.J.; et al. The Complex Jujube Genome Provides Insights into Fruit Tree Biology. Nat. Commun. 2014, 5, 5315. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce Framework for Analyzing Next-Generation DNA Sequencing Data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software Structure: A Simulation Study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; Vonholdt, B.M. Structure Harvester: A Website And Program for Visualizing Structure Output and Implementing the Evanno Method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A Cluster Matching and Permutation Program for Dealing with Label Switching and Multimodality in Analysis of Population Structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, N.A. DISTRUCT: A Program for the Graphical Display of Population Structure. Mol. Ecol. 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Nei, M. Genetic Distance between Populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Dieringer, D.; Schlötterer, C. Microsatellite Analyser (MSA): A platform Independent Analysis Tool for Large Microsatellite Datasets. Mol. Ecol. Notes 2003, 3, 167–169. [Google Scholar] [CrossRef]
- Felsenstein, J. PHYLIP-Phylogeny Inference Package (Version 3.2). Cladistics 1989, 5, 164–166. [Google Scholar]
- Rambaut, A. Molecular Evolution, Phylogenetics and Epidemiology: FigTree v1.3.1 2006–2009; China Forestry Press: Beijing, China, 2009; Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 9 May 2018).
- Marshall, T.C.; Slate, J.; Kruuk, L.E.B.; Pemberton, J.M. Statistical Confidence for Likelihood-Based Paternity Inference in Natural Populations. Mol. Ecol. 1998, 7, 639–655. [Google Scholar] [CrossRef]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising How the Computer Program CERVUS Accommodates Genotyping Error Increases Success in Paternity Assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef]
- Patel, R.K.; Jain, M. NGS QC Toolkit: A Toolkit for Quality Control of Next Generation Sequencing Data. PLoS ONE 2012, 7, e30619. [Google Scholar]
- Wang, B.; Tan, H.; Fang, W.; Zhang, D. Developing Single Nucleotide Polymorphism (SNP) Markers from Transcriptome Sequences for Identification of Longan (Dimocarpus Longan) Germplasm. Hortic. Res. 2015, 2, 14065. [Google Scholar] [CrossRef]
- Irish, B.M.; Cuevas, H.E.; Simpson, S.A.; Scheffler, B.E.; Sardos, J.; Ploetz, R.; Goenaga, R.J. Musa spp. Germplasm Management: Microsatellite Fingerprinting of USDA-ARS National Plant Germplasm System (NPGS) Collection. Crop Sci. 2014, 54, 2140–2151. [Google Scholar] [CrossRef]



| Name of Cultivars | Source (Province) | Name of Cultivars | Source (Province) |
|---|---|---|---|
| Fuyangmutouzao | Anhui | Zaoqiuhong | Ningxia |
| Beijingbenzao | Beijing | Changtanzao | Ningxia |
| Beijingpaoapaozao | Beijing | Zhongningyuanzao | Ningxia |
| Jingzao60 | Beijing | Dalixiaodundunzao | Shaanxi |
| Shaizao | Beijing | Dieyazao | Shaanxi |
| Yingluozao | BeiJing | Dongzao | Shaanxi |
| Beibeixiaozao | Chongqing | Ganweibazao | Shaanxi |
| Gansudongzao | Gansu | Goutouzao | Shaanxi |
| Minqinxiaozao | Gansu | Jiaxianyazao | Shaanxi |
| Zunyitianzao | Guizhou | Muzao | Shaanxi |
| Cangxiantunzizao | Hebei | Pingshunbenzao | Shaanxi |
| Chuanlingzao | Hebei | Pingshunjunzao | Shaanxi |
| Fengmizao | Hebei | Qiyuexian | Shaanxi |
| Hebei 13# | Hebei | Zhongcaobenzao | Shaanxi |
| Jinsixiaozao | Hebei | Dalingzao | Shandong |
| Longzao | Hebei | Fucuimizao | Shandong |
| Malianxiaozao | Hebei | Laolingxiaozao | Shandong |
| Pozaozhibian 1# | Hebei | Liaochengyuanlingzao | Shandong |
| Shulutangzao | Hebei | Linqinzao | Shandong |
| Xianxianmatouzao | Hebei | Luzao 2# | Shandong |
| Zanhuangchangzao | Hebei | Luzao 5# | Shandong |
| Zaocuiwang | Hebei | Suyuanlingzao | Shandong |
| Bianhesuanzao | Henan | Tengxiantangzao | Shandong |
| Dongzao 2# | Henan | Baodeyouzao | Shanxi |
| Henanlongzao | Henan | Mamazao | Shanxi |
| Jinmangguozao | Henan | Jianjianzao | Shanxi |
| Lingbaoyuanzao | Henan | Jiaochengduanzao | Shanxi |
| Fuyangtangzao | Henan | Jingudazao | Shanxi |
| Fuyangxiaozao | Henan | Jinzao | Shanxi |
| Xinzhengjixinzao | Henan | Jiuyuehan | Shanxi |
| Hunanchangzao | Hunan | Lizao | Shanxi |
| Mifengzao | Hunan | Linfenmizao | Shanxi |
| Ruchengzao | Hunan | Linfenmugedazao | Shanxi |
| Tangtouzao | Hunan | Linyibenzao | Shanxi |
| Xupuchengtuozao | Hunan | Linyitiansuanzao | Shanxi |
| Xupumizao | Hunan | Linglingzao | Shanxi |
| Xupushatangzao | Hunan | Mopanzao | Shanxi |
| Xupushibingzao | Hunan | Puchengjinzao | Shanxi |
| Xupusuanyuanzao | Hunan | Sumuzao | Shanxi |
| Lengsizao | Jiangsu | Taigulinglingzao | Shanxi |
| Penzao 2# | Jiangsu | Taiyuanyuanzao | Shanxi |
| Dasuanzao | Ningxia | Tuanzao | Shanxi |
| Dayuanzao | Ningxia | Wanrongcuizao | Shanxi |
| Jinchang 1# | Ningxia | Xiangfenyuanzao | Shanxi |
| Lingwusuanzao | Ningxia | Yongjijidanzao | Shanxi |
| Lingwuyuanzao | Ningxia | Yucijiuyueqing | Shanxi |
| Lingwuchangzao | Ningxia | Yucituanzao | Shanxi |
| Longzhu 1# | Ningxia | Yuanquzao | Shanxi |
| Longzhu 2# | Ningxia | Yunchengjinzao | Shanxi |
| Longzhu 3# | Ningxia | Yunchengpopozao | Shanxi |
| Suanzao | Ningxia | Alaayuancuizao | Xinjiang |
| Tongxinyuanzao | Ningxia | Hamidazao | Xinjiang |
| Tongxinyuanzao | Ningxia | Huizao | Xinjiang |
| Yuanzao | Ningxia | Kunmingzao | Yunnan |
| Yuanzao 1# | Ningxia | Yiwudazao | Zhejiang |
| Yuanzao 2# | Ningxia | Yiwuezizao | Zhejiang |
| Yuanzao 3# | Ningxia | Yiwumianxuzao | Zhejiang |
| Cultivars | Zj002 | Zj018 | Zj019 | Zj022 | Zj025 | Zj026 | Zj028 | Zj030 | Zj031 | Zj032 | Zj033 | Zj034 | Zj035 | Zj036 | Zj037 | Zj038 | Zj039 | Zj040 | Zj041 | Zj043 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alaayuancuizao | C T | T T | A A | G G | G T | G G | C T | C C | T T | C C | T T | A A | G G | G T | C T | G G | C C | T T | C C | A A |
| Baodeyouzao | C T | C T | A A | A G | G T | G G | C T | C C | T T | C C | T T | A A | G G | G T | C T | G G | C C | T T | A C | A A |
| Beibeixiaozao | C C | T T | T T | A A | G G | G G | C C | C C | T T | C C | T T | A G | G G | G G | C T | G G | C C | T T | C C | A A |
| Beijingpaoapaozao | C C | T T | A A | A A | G G | G G | C C | T T | C T | C C | T T | A G | G G | G G | C T | C G | C G | C T | A C | A T |
| Chuanlingzao | C C | T T | T T | A A | G G | G G | C C | T T | C T | C C | T T | A A | G G | G G | C C | C G | C G | C T | A C | A T |
| Dalingzao | T T | C T | A T | A G | G G | G G | T T | C C | C T | C C | T T | A A | G G | G G | C C | C G | C G | C T | A A | A T |
| Dalixiaodundunzao | T T | C T | T T | A A | G G | G T | C C | C C | T T | C C | T T | A A | G G | G G | C C | C G | C G | C T | A C | A T |
| Dasuanzao | C C | T T | T T | A A | G G | G T | C C | C C | T T | C C | C T | A A | A G | G G | C C | G G | C C | T T | A C | A A |
| Dieyazao | T T | C T | A A | G G | G G | G G | T T | T T | C T | C C | T T | A A | G G | G T | C C | C G | C G | C T | A A | A T |
| Dongzao | C C | T T | T T | A A | G T | G G | C T | C C | T T | C C | T T | A A | G G | G T | C T | G G | C C | T T | A C | A A |
| Fengmizao | T T | C T | A T | A G | G G | G G | C C | C T | T T | C C | T T | A G | G G | G G | C T | C G | C G | C T | C C | A T |
| Fuyangmutouzao | C C | T T | T T | A A | G G | G G | C T | C C | T T | C C | T T | A G | G G | G T | C T | G G | C C | T T | A C | A A |
| Gansudongzao | T T | C T | T T | A A | G G | G G | C C | C C | T T | C C | C T | A A | A G | G T | C C | C G | C G | C C | C C | A T |
| Hamidazao | C C | T T | T T | A A | G G | G T | C C | C C | T T | C C | T T | A A | G G | G G | C C | G G | C C | T T | A C | A A |
| Hebei 13# | C T | T T | A A | A A | G G | G G | C T | C C | T T | C C | T T | A A | G G | G G | C C | G G | C C | T T | A C | A A |
| Huizao | C T | C T | A A | A G | G T | G G | T T | C C | T T | C T | T T | A A | G G | G T | C T | G G | C C | T T | A A | A A |
| Synon. Group | Cultivar Name | Origin | Synon. Group | Cultivar Name | Origin |
|---|---|---|---|---|---|
| 1 | Cangxiantunzizao | Hebei | 8 | Pozaozhibian1# | Hebei |
| 1 | Jinsixiaozao | Hebei | 8 | Dalingzao | Shandong |
| 1 | Laolingxiaozao | Shandong | |||
| 9 | Jingudazao | Shanxi | |||
| 2 | Shulutangzao | Hebei | 9 | Jinchang1# | Ningxia |
| 2 | Chuanlingzao | Hebei | |||
| 2 | Bianhesuanzao | Henan | 10 | Yingluozao | Beijing |
| 10 | Baodeyouzao | Shanxi | |||
| 3 | Zanhuangchangzao | Hebei | 10 | Xiangfenyuanzao | Shanxi |
| 3 | Tongxinyuanzao | Ningxia | |||
| 3 | Dayuanzao | Ningxia | 11 | Goutouzao | Shaanxi |
| 3 | Zhongcaobenzao | Shaanxi | 11 | Jianjianzao | Shanxi |
| 3 | Wanrongcuizao | Shanxi | |||
| 12 | Xinzhengjixinzao | Henan | |||
| 4 | Shaizao | Beijing | 12 | Huizao | Xinjiang |
| 4 | Yiwudazao | Zhejiang | |||
| 13 | Sumuzao | Shanxi | |||
| 5 | Minqinxiaozao | Gansu | 13 | Beijingbenzao | Beijing |
| 5 | Zhongningyuanzao | Ningxia | |||
| 5 | Yuanzao | Ningxia | 14 | Yunchengpopozao | Shanxi |
| 5 | Yuanzao1# | Ningxia | 14 | Mopanzao | Shanxi |
| 5 | Yuanzao2# | Ningxia | |||
| 5 | Yuanzao3# | Ningxia | 15 | Zaocuiwang | Hebei |
| 5 | Lingwuyuanzao | Ningxia | 15 | Luzao5# | Shandong |
| 5 | Changtanzao | Ningxia | |||
| 16 | Linfenmugeda | Shanxi | |||
| 6 | Malianxiaozao | Hebei | 16 | Jiaxianyazao | Shaanxi |
| 6 | Ganweibazao | Shaanxi | 16 | Muzao | Shaanxi |
| 6 | Jinzao | Shanxi | |||
| 6 | Linyibenzao | Shanxi | 17 | Longzhu2# | Ningxia |
| 17 | Longzhu3# | Ningxia | |||
| 7 | Dongzao | Shaanxi | |||
| 7 | Dongzao2# | Henan |
| Cultivar | Origin | Q-Value Group 1 | Q-Value Group 2 | Group |
|---|---|---|---|---|
| Beijingpaoapaozao | Beijing | 0.909 | 0.091 | Group 1 |
| Chuanlingzao | Hebei | 0.898 | 0.102 | Group 1 |
| Lingbaoyuanzao | Henan | 0.868 | 0.132 | Group 1 |
| Muzao | Shaanxi | 0.995 | 0.005 | Group 1 |
| Pingshunbenzao | Shaanxi | 0.994 | 0.006 | Group 1 |
| Qiyuexian | Shaanxi | 0.836 | 0.164 | Group 1 |
| Dalingzao | Shandong | 0.991 | 0.009 | Group 1 |
| Linqinzao | Shandong | 0.987 | 0.013 | Group 1 |
| Liaochengyuanlingzao | Shandong | 0.832 | 0.168 | Group 1 |
| Yucituanzao | Shanxi | 0.992 | 0.008 | Group 1 |
| Yuanquzao | Shanxi | 0.989 | 0.011 | Group 1 |
| Jiaochengduanzao | Shanxi | 0.989 | 0.011 | Group 1 |
| Jingudazao | Shanxi | 0.988 | 0.012 | Group 1 |
| Mamazao | Shanxi | 0.983 | 0.017 | Group 1 |
| Linglingzao | Shanxi | 0.978 | 0.022 | Group 1 |
| Yucijiuyueqing | Shanxi | 0.977 | 0.023 | Group 1 |
| Taiyuanyuanzao | Shanxi | 0.961 | 0.039 | Group 1 |
| Mopanzao | Shanxi | 0.960 | 0.040 | Group 1 |
| Jianjianzao | Shanxi | 0.924 | 0.076 | Group 1 |
| Tuanzao | Shanxi | 0.888 | 0.112 | Group 1 |
| Kunmingzao | Yunnan | 0.822 | 0.178 | Group 1 |
| Gansudongzao | Gansu | 0.011 | 0.989 | Group 2 |
| Longzao | Hebei | 0.049 | 0.951 | Group 2 |
| Fengmizao | Hebei | 0.051 | 0.949 | Group 2 |
| Zanhuangchangzao | Hebei | 0.087 | 0.913 | Group 2 |
| Xupusuanyuanzao | Hunan | 0.016 | 0.984 | Group 2 |
| Xupuchengtuozao | Hunan | 0.051 | 0.949 | Group 2 |
| Mifengzao | Hunan | 0.116 | 0.884 | Group 2 |
| Xupushatangzao | Hunan | 0.152 | 0.848 | Group 2 |
| Xupumizao | Hunan | 0.160 | 0.840 | Group 2 |
| Penzao | Jiangsu | 0.011 | 0.989 | Group 2 |
| Zhongningyuanzao | Ningxia | 0.014 | 0.986 | Group 2 |
| Lingwuchangzao | Ningxia | 0.022 | 0.978 | Group 2 |
| Longzhu | Ningxia | 0.022 | 0.978 | Group 2 |
| Longzhu | Ningxia | 0.023 | 0.977 | Group 2 |
| Dongzao | Shaanxi | 0.049 | 0.951 | Group 2 |
| Luzao | Shandong | 0.050 | 0.950 | Group 2 |
| Tengxiantangzao | Shandong | 0.093 | 0.907 | Group 2 |
| Fucuimizao | Shandong | 0.100 | 0.900 | Group 2 |
| Suyuanlingzao | Shandong | 0.163 | 0.837 | Group 2 |
| Jinzao | Shanxi | 0.035 | 0.965 | Group 2 |
| Puchengjinzao | Shanxi | 0.067 | 0.933 | Group 2 |
| Lizao | Shanxi | 0.131 | 0.869 | Group 2 |
| Yiwuezizao | Zhejiang | 0.199 | 0.801 | Group 2 |
| Alaayuancuizao | Xinjiang | 0.217 | 0.783 | Admixture |
| Jingzao60 | Beijing | 0.233 | 0.767 | Admixture |
| Jinsixiaozao | Hebei | 0.243 | 0.757 | Admixture |
| Linfenmizao | Shanxi | 0.262 | 0.738 | Admixture |
| Pingshunjunzao | Shaanxi | 0.266 | 0.734 | Admixture |
| Zunyitianzao | Guizhou | 0.281 | 0.719 | Admixture |
| Baodeyouzao | Shanxi | 0.288 | 0.712 | Admixture |
| Huizao | Xinjiang | 0.300 | 0.700 | Admixture |
| Taigulinglingzao | Shanxi | 0.308 | 0.692 | Admixture |
| Hebei | Hebei | 0.335 | 0.665 | Admixture |
| Dieyazao | Shaanxi | 0.340 | 0.660 | Admixture |
| Hamidazao | Xinjiang | 0.341 | 0.659 | Admixture |
| Suanzao | Ningxia | 0.363 | 0.637 | Admixture |
| Henanlongzao | Henan | 0.406 | 0.594 | Admixture |
| Zaoqiuhong | Ningxia | 0.429 | 0.571 | Admixture |
| Lingwusuanzao | Ningxia | 0.444 | 0.556 | Admixture |
| Xianxianmatouzao | Hebei | 0.473 | 0.527 | Admixture |
| Zaocuiwang | Hebei | 0.490 | 0.510 | Admixture |
| Luzao | Shandong | 0.492 | 0.508 | Admixture |
| Dalixiaodundunzao | Shaanxi | 0.526 | 0.474 | Admixture |
| Yiwumianxuzao | Zhejiang | 0.550 | 0.450 | Admixture |
| Sumuzao | Shanxi | 0.558 | 0.442 | Admixture |
| Ruchengzao | Hunan | 0.562 | 0.438 | Admixture |
| Dasuanzao | Ningxia | 0.589 | 0.411 | Admixture |
| Shaizao | Beijing | 0.619 | 0.381 | Admixture |
| Linyitiansuanzao | Shanxi | 0.625 | 0.375 | Admixture |
| Yunchengjinzao | Shanxi | 0.644 | 0.356 | Admixture |
| Lengsizao | Jiangsu | 0.649 | 0.351 | Admixture |
| Beibeixiaozao | Chongqing | 0.680 | 0.320 | Admixture |
| Jinmangguozao | Henan | 0.708 | 0.292 | Admixture |
| Yongjijidanzao | Shanxi | 0.714 | 0.286 | Admixture |
| Jiuyuehan | Shanxi | 0.723 | 0.277 | Admixture |
| Fuyangmutouzao | Anhui | 0.728 | 0.272 | Admixture |
| Tangtouzao | Hunan | 0.768 | 0.232 | Admixture |
| Xipushibingzao | Hunan | 0.793 | 0.207 | Admixture |
| Cultivar | Recorded Parent | Identified Parent | Pair Top LOD |
|---|---|---|---|
| Jing 60 | Uknown landraces from Beijing | Yingluozao (Syn. 10) | 1.91 |
| Zaocuiwang | Jinsixiaozao | Jinsixiaozao (Syn. 1) | 4.91 |
| Qiyuexian | Known landraces | Yongjijidanzao | 3.66 |
| Longzhu 1 | Lizao and/or Dongzao | Lizao | 1.41 |
| Longzhu 2 | Lizao and/or Dongzao | Dongzao (Syn. 7) | 3.01 |
| Zaoqiuhong | Dalingzao | Dalixiaodundunzao | 7.18 |
| Luzao 2 | Liuyuexian | Zanhuangchangzao (Syn. 3) | 2.52 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, L.; Cao, B.; Zhang, Y.; Meinhardt, L.W.; Zhang, D. Mining Single Nucleotide Polymorphism (SNP) Markers for Accurate Genotype Identification and Diversity Analysis of Chinese Jujube (Ziziphus jujuba Mill.) Germplasm. Agronomy 2021, 11, 2303. https://doi.org/10.3390/agronomy11112303
Song L, Cao B, Zhang Y, Meinhardt LW, Zhang D. Mining Single Nucleotide Polymorphism (SNP) Markers for Accurate Genotype Identification and Diversity Analysis of Chinese Jujube (Ziziphus jujuba Mill.) Germplasm. Agronomy. 2021; 11(11):2303. https://doi.org/10.3390/agronomy11112303
Chicago/Turabian StyleSong, Lihua, Bing Cao, Yue Zhang, Lyndel W. Meinhardt, and Dapeng Zhang. 2021. "Mining Single Nucleotide Polymorphism (SNP) Markers for Accurate Genotype Identification and Diversity Analysis of Chinese Jujube (Ziziphus jujuba Mill.) Germplasm" Agronomy 11, no. 11: 2303. https://doi.org/10.3390/agronomy11112303
APA StyleSong, L., Cao, B., Zhang, Y., Meinhardt, L. W., & Zhang, D. (2021). Mining Single Nucleotide Polymorphism (SNP) Markers for Accurate Genotype Identification and Diversity Analysis of Chinese Jujube (Ziziphus jujuba Mill.) Germplasm. Agronomy, 11(11), 2303. https://doi.org/10.3390/agronomy11112303






