DeepPaSTL: Spatio-Temporal Deep Learning Methods for Predicting Long-Term Pasture Terrains Using Synthetic Datasets
Abstract
:1. Introduction
2. Materials and Methods
2.1. Problem Formulation
2.2. Simulated Spatiotemporal Dataset
2.3. Pasture Construction for Evaluation
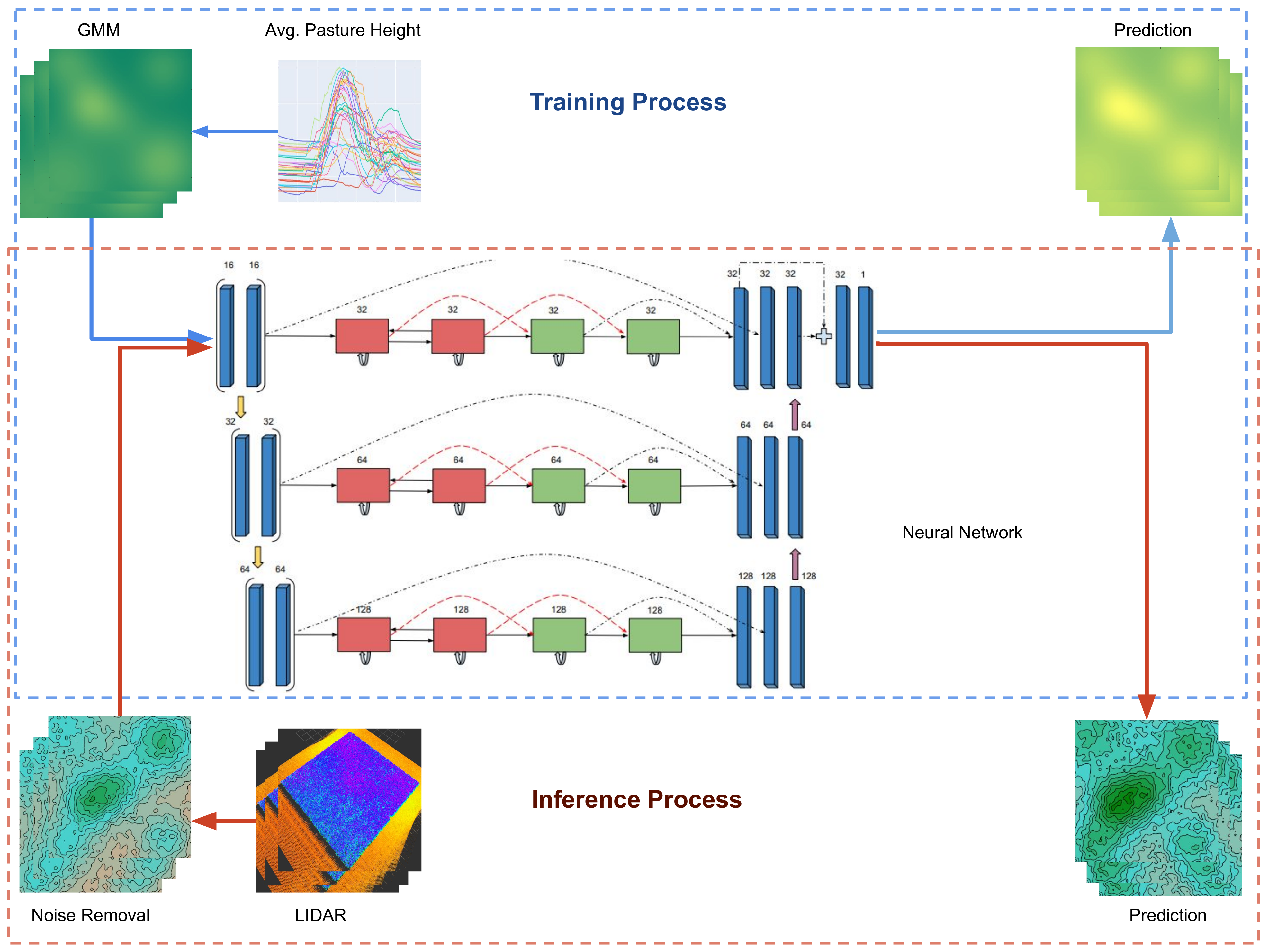
2.4. Data Processing for Training and Inference
- The use of convolution neural networks in deep learning introduces an unintended side effect popularly termed as boundary effects [47,48], where artifacts are introduced at the boundaries of the image due to no spatial information [49,50] available when CNN filters pass over boundaries of the image. We circumvent this issue by enlarging each image with size , pixels through mirror padding [51] to add spatial information on the boundaries of each pasture image in the dataset updating our new training dataset to .
- Training and inference of the neural network on original dimensions of the training dataset may potentially increase accuracy. However, it severely limits the capability of the neural network to adapt to variable input dimensions while also increasing computational requirements as GPU memory is a limited resource, specifically when training inputs with large dimensions. To this end, we quantize the training data into smaller sized patches of with an overlap of between them. The overlapping of the images and subsequent reconstruction of the image post inference through a weighted average allows us to mitigate boundary effects between each cropped frame, an undesirable artifact of CNN output that would occur if they were to be naively cropped without any overlaps. This methodology requires the neural network to only learn over small patches of the field and can be practically used to predict field sizes of any size , as long as the original image is appropriately processed to meet the input size of , where .
- We fix the sequence length of the training inputs and output prediction to trajectories of time . The final input training set is then defined as input sequences of , where is the number of data points in the quantized dataset . Each individual sequence for the backward propagation is , where . Similarly, the target values dataset is created for training. Each input sequence has a corresponding target value , where .
2.5. Deep Learning Model for Long-Term Prediction
2.6. Uncertainty Estimation of the Model
2.7. Experiment Details
2.8. Model Training and Evaluation
3. Results
- DeepPaSTL predictions perform within a error rate for long horizon predictions up to 60 days in the future, and approximately with a error rate for predictions closer to its historical data.
- Allowing the model to have regular observations, i.e., with smaller intervals, is essential for capturing large dynamic changes in the pasture growth.
- DeepPaSTL prediction uncertainty increases as the volatility in pasture growth increases.
- We show that DeepPaSTL has the capacity to predict and generate future pasture terrains that replicate the growth and surface characteristics of ground truth data.
3.1. Effect of Input Quantization
3.2. Effect of Intervals between Observations
3.3. Uncertainty over Pasture Dynamics
3.4. Imputation of Missing Data
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Computer Code and Software
Conflicts of Interest
Abbreviations
| APSIM | Agricultural Production Systems sIMulator |
| BiConvLSTM | Bidirectional Convolutional Long Short Term Memory |
| BPTT | Back Propagation Through Time |
| CNN | Convolution Neural Network |
| ConvLSTM | Convolutional Long Short Term Memory |
| DeepPaSTL | Deep Pasture SpatioTemporal Learning |
| DOAJ | Directory of Open Access Journals |
| GMM | Gaussian Mixture Model |
| LIDAR | Light Detection and Ranging |
| LSTM | Long Short Term Memory |
| MAE | Mean Absolute Error |
| MAPE | Mean Absolute Percentage Error |
| MaxPool | Maximum Pooling |
| MCMC | Markov Chain Monte Carlo |
| MDPI | Multidisciplinary Digital Publishing Institute |
| MSE | Mean Squared Error |
| UAV | Unmanned Aerial Vehicle |
| VRAM | Video Random Access Memory |
References
- Garcia, S.C.; Clark, C.E.; Kerrisk, K.L.; Islam, M.R.; Fariña, S.; Evans, J. Gaps and variability in pasture utilisation in Australian pasture-based dairy systems. In Proceedings of the XXII International Grassland Congress (Revitalising Grasslands to Sustain Our Communities), Sydney, Australia, 15–19 September 2013; pp. 1709–1716. [Google Scholar]
- Sala, O.E.; Paruelo, J.M.; Sala, O.E.; Paruelo, J.M. Ecosystem services in grasslands. In Nature’s Services: Societal Dependence on Natural Ecosystems; Island Press: Washington, DC, USA, 1997; pp. 237–251. [Google Scholar]
- Insua, J.R.; Utsumi, S.A.; Basso, B. Estimation of spatial and temporal variability of pasture growth and digestibility in grazing rotations coupling unmanned aerial vehicle (UAV) with crop simulation models. PLoS ONE 2019, 14, e0212773. [Google Scholar] [CrossRef] [Green Version]
- Fulkerson, W.; McKean, K.; Nandra, K.; Barchia, I. Benefits of accurately allocating feed on a daily basis to dairy cows grazing pasture. Aust. J. Exp. Agric. 2005, 45, 331–336. [Google Scholar] [CrossRef]
- De Rosa, D.; Basso, B.; Fasiolo, M.; Friedl, J.; Fulkerson, B.; Grace, P.R.; Rowlings, D.W. Predicting pasture biomass using a statistical model and machine learning algorithm implemented with remotely sensed imagery. Comput. Electron. Agric. 2021, 180, 105880. [Google Scholar] [CrossRef]
- Liu, J.; Williams, R.K. Submodular optimization for coupled task allocation and intermittent deployment problems. IEEE Robot. Autom. Lett. 2019, 4, 3169–3176. [Google Scholar]
- Liu, J.; Williams, R.K. Monitoring over the long term: Intermittent deployment and sensing strategies for multi-robot teams. In Proceedings of the IEEE International Conference on Robotics and Automation, Paris, France, 31 May 2020; pp. 7733–7739. [Google Scholar]
- Sung, Y.; Budhiraja, A.K.; Williams, R.K.; Tokekar, P. Distributed assignment with limited communication for multi-robot multi-target tracking. Auton. Robots 2010, 44, 57–73. [Google Scholar] [CrossRef] [Green Version]
- Heintzman, L.; Williams, R.K. Multi-agent intermittent interaction planning via sequential greedy selections over position samples. IEEE Robot. Autom. Lett. 2021, 6, 534–541. [Google Scholar] [CrossRef]
- Heintzman, L.; Hashimoto, A.; Abaid, N.; Williams, R.K. Anticipatory Planning and Dynamic Lost Person Models for Human-Robot Search and Rescue. In Proceedings of the IEEE International Conference on Robotics and Automation, Xi’an, China, 30 May 2021; pp. 8252–8258. [Google Scholar]
- Liu, J.; Williams, R.K. Coupled temporal and spatial environment monitoring for multi-agent teams in precision farming. In Proceedings of the IEEE Conference on Control Technology and Applications, Montreal, QC, Canada, 24–26 August 2020; pp. 273–278. [Google Scholar]
- Liu, J.; Williams, R.K. Optimal intermittent deployment and sensor selection for environmental sensing with multi-robot teams. In Proceedings of the IEEE International Conference on Robotics and Automation, Brisbane, Australia, 21–25 May 2018; pp. 1078–1083. [Google Scholar]
- Williams, R.K.; Gasparri, A.; Ulivi, G. Decentralized matroid optimization for topology constraints in multi-robot allocation problems. In Proceedings of the IEEE International Conference on Robotics and Automation, Marina Bay Sands, Singapore, 29 May 2017; pp. 293–300. [Google Scholar]
- Tao, F.; Yokozawa, M.; Zhang, Z. Modelling the impacts of weather and climate variability on crop productivity over a large area: A new process-based model development, optimization, and uncertainties analysis. Agric. For. Meteorol. 2009, 149, 831–850. [Google Scholar] [CrossRef]
- Iizumi, T.; Yokozawa, M.; Nishimori, M. Parameter estimation and uncertainty analysis of a large-scale crop model for paddy rice: Application of a Bayesian approach. Agric. For. Meteorol. 2009, 149, 333–348. [Google Scholar] [CrossRef]
- Lobell, D.B.; Burke, M.B. On the use of statistical models to predict crop yield responses to climate change. Agric. For. Meteorol. 2010, 150, 1443–1452. [Google Scholar] [CrossRef]
- Chen, Y.; Guerschman, J.; Shendryk, Y.; Henry, D.; Harrison, M.T. Estimating Pasture Biomass Using Sentinel-2 Imagery and Machine Learning. Remote Sens. 2021, 13, 603. [Google Scholar] [CrossRef]
- Gargiulo, J.; Clark, C.; Lyons, N.; de Veyrac, G.; Beale, P.; Garcia, S. Spatial and temporal pasture biomass estimation integrating electronic plate meter, planet cubesats and sentinel-2 satellite data. Remote Sens. 2020, 12, 3222. [Google Scholar] [CrossRef]
- Ali, I.; Greifeneder, F.; Stamenkovic, J.; Neumann, M.; Notarnicola, C. Review of machine learning approaches for biomass and soil moisture retrievals from remote sensing data. Remote Sens. 2015, 7, 16398–16421. [Google Scholar]
- Dang, A.T.N.; Nandy, S.; Srinet, R.; Luong, N.V.; Ghosh, S.; Kumar, A.S. Forest aboveground biomass estimation using machine learning regression algorithm in Yok Don National Park, Vietnam. Ecol. Inform. 2019, 50, 24–32. [Google Scholar] [CrossRef]
- Ghosh, S.M.; Behera, M.D. Aboveground biomass estimation using multi-sensor data synergy and machine learning algorithms in a dense tropical forest. Appl. Geogr. 2018, 96, 29–40. [Google Scholar] [CrossRef]
- Rangwala, M.; Williams, R. Learning Multi-Agent Communication through Structured Attentive Reasoning. Adv. Neural Inf. Process. Syst. 2020, 33, 10088–10098. [Google Scholar]
- Wehbe, R.; Williams, R.K. A Deep Learning Approach for Probabilistic Security in Multi-Robot Teams. IEEE Robot. Autom. Lett. 2019, 4, 4262–4269. [Google Scholar] [CrossRef]
- Neal, R.M. Bayesian Learning for Neural Networks; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012; Volume 118. [Google Scholar]
- Fukushima, K.; Miyake, S. Neocognitron: A self-organizing neural network model for a mechanism of visual pattern recognition. In Competition and Cooperation in Neural Nets; Springer: Berlin/Heidelberg, Germany, 1982; pp. 267–285. [Google Scholar]
- LeCun, Y.; Haffner, P.; Bottou, L.; Bengio, Y. Object recognition with gradient-based learning. In Shape, Contour and Grouping in Computer Vision; Springer: Berlin/Heidelberg, Germany, 1999; pp. 319–345. [Google Scholar]
- Lin, Z.; Li, M.; Zheng, Z.; Cheng, Y.; Yuan, C. Self-attention convlstm for spatiotemporal prediction. In Proceedings of the AAAI Conference on Artificial Intelligence, New York, NY, USA, 7 February 2020; Volume 34, pp. 11531–11538. [Google Scholar]
- Xu, Y.; Gao, L.; Tian, K.; Zhou, S.; Sun, H. Non-local convlstm for video compression artifact reduction. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Korea, 27 October 2019; pp. 7043–7052. [Google Scholar]
- Azad, R.; Asadi-Aghbolaghi, M.; Fathy, M.; Escalera, S. Bi-directional ConvLSTM U-Net with densley connected convolutions. In Proceedings of the IEEE/CVF International Conference on Computer Vision Workshops, Seoul, Korea, 27 October 2019; pp. 406–415. [Google Scholar]
- Xu, N.; Yang, L.; Fan, Y.; Yang, J.; Yue, D.; Liang, Y.; Price, B.; Cohen, S.; Huang, T. Youtube-vos: Sequence-to-sequence video object segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 585–601. [Google Scholar]
- Oliu, M.; Selva, J.; Escalera, S. Folded recurrent neural networks for future video prediction. In Proceedings of the European Conference on Computer Vision, Munich, Germany, 8–14 September 2018; pp. 716–731. [Google Scholar]
- Michalski, V.; Memisevic, R.; Konda, K. Modeling deep temporal dependencies with recurrent grammar cells. Adv. Neural Inf. Process. Syst. 2014, 27, 1925–1933. [Google Scholar]
- Srivastava, N.; Mansimov, E.; Salakhudinov, R. Unsupervised learning of video representations using lstms. In Proceedings of the International Conference on Machine Learning, PMLR, Lile, France, 6–11 July 2015; pp. 843–852. [Google Scholar]
- Xingjian, S.; Chen, Z.; Wang, H.; Yeung, D.Y.; Wong, W.K.; Woo, W.c. Convolutional LSTM network: A machine learning approach for precipitation nowcasting. In Proceedings of the Advances in Neural Information Processing Systems, Montreal, QC, Canada, 7–12 December 2015; pp. 802–810. [Google Scholar]
- Lotter, W.; Kreiman, G.; Cox, D. Deep predictive coding networks for video prediction and unsupervised learning. arXiv 2016, arXiv:1605.08104. [Google Scholar]
- Wang, Y.; Long, M.; Wang, J.; Gao, Z.; Yu, P.S. Predrnn: Recurrent neural networks for predictive learning using spatiotemporal lstms. In Proceedings of the International Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 879–888. [Google Scholar]
- Wang, Y.; Gao, Z.; Long, M.; Wang, J.; Philip, S.Y. Predrnn++: Towards a resolution of the deep-in-time dilemma in spatiotemporal predictive learning. In Proceedings of the International Conference on Machine Learning, PMLR, Stockholm, Sweden, 10–15 July 2018; pp. 5123–5132. [Google Scholar]
- Koenig, N.; Howard, A. Design and use paradigms for gazebo, an open-source multi-robot simulator. In Proceedings of the 2004 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS)(IEEE Cat. No. 04CH37566), Sendai, Japan, 28 September 2004; Volume 3, pp. 2149–2154. [Google Scholar]
- Sutskever, I.; Vinyals, O.; Le, Q.V. Sequence to sequence learning with neural networks. In Proceedings of the Advances in Neural Information Processing Systems, Montreal, QC, Canada, 12–13 December 2014; pp. 3104–3112. [Google Scholar]
- Gal, Y.; Ghahramani, Z. Dropout as a bayesian approximation: Representing model uncertainty in deep learning. In Proceedings of the International Conference on Machine Learning, PMLR, New York, NY, USA, 19–24 June 2016; pp. 1050–1059. [Google Scholar]
- Archontoulis, S.V.; Miguez, F.E.; Moore, K.J. Evaluating APSIM Maize, Soil Water, Soil Nitrogen, Manure, and Soil Temperature Modules in the Midwestern United States. Agron. J. 2014, 106, 1025–1040. [Google Scholar] [CrossRef]
- Li, F.; Newton, P.; Lieffering, M. Testing simulations of intra- and inter-annual variation in the plant production response to elevated CO2 against measurements from an 11-year FACE experiment on grazed pasture. Glob. Chang. Biol. 2013, 20, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhou, L.; Tokekar, P.; Williams, R.K. Distributed Resilient Submodular Action Selection in Adversarial Environments. IEEE Robot. Autom. Lett. 2021, 6, 5832–5839. [Google Scholar] [CrossRef]
- Wehbe, R.; Williams, R.K. Probabilistic Resilience of Dynamic Multi-Robot Systems. IEEE Robot. Autom. Lett. 2021, 6, 1777–1784. [Google Scholar]
- Heintzman, L.; Williams, R.K. Nonlinear observability of unicycle multi-robot teams subject to nonuniform environmental disturbances. Auton. Robot. 2020, 44, 1149–1166. [Google Scholar] [CrossRef]
- Wehbe, R.; Williams, R.K. Probabilistic Security for Multirobot Systems. IEEE Trans. Rob. 2021, 37, 146–165. [Google Scholar] [CrossRef]
- Innamorati, C.; Ritschel, T.; Weyrich, T.; Mitra, N.J. Learning on the edge: Explicit boundary handling in cnns. arXiv 2018, arXiv:1805.03106. [Google Scholar]
- Innamorati, C.; Ritschel, T.; Weyrich, T.; Mitra, N.J. Learning on the edge: Investigating boundary filters in cnns. Int. J. Comput. Vis. 2019, 128, 773–782. [Google Scholar] [CrossRef] [Green Version]
- Hashemi, M. Enlarging smaller images before inputting into convolutional neural network: Zero-padding vs. interpolation. J. Big Data 2019, 6, 1–13. [Google Scholar] [CrossRef]
- Albawi, S.; Mohammed, T.A.; Al-Zawi, S. Understanding of a convolutional neural network. In Proceedings of the International Conference on Engineering and Technology, Antalya, Turkey, 21–23 August 2017; pp. 1–6. [Google Scholar]
- Tang, H.; Ortis, A.; Battiato, S. The impact of padding on image classification by using pre-trained convolutional neural networks. In Proceedings of the International Conference on Image Analysis and Processing, Trento, Italy, 9–13 September 2019; Springer: Berlin/Heidelberg, Germany, 2019; pp. 337–344. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar]
- Graves, A. Generating sequences with recurrent neural networks. arXiv 2013, arXiv:1308.0850. [Google Scholar]
- Cho, K.; Van Merriënboer, B.; Gulcehre, C.; Bahdanau, D.; Bougares, F.; Schwenk, H.; Bengio, Y. Learning phrase representations using RNN encoder–decoder for statistical machine translation. arXiv 2014, arXiv:1406.1078. [Google Scholar]
- Donahue, J.; Anne Hendricks, L.; Guadarrama, S.; Rohrbach, M.; Venugopalan, S.; Saenko, K.; Darrell, T. Long-term recurrent convolutional networks for visual recognition and description. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 8–10 June 2015; pp. 2625–2634. [Google Scholar]
- Karpathy, A.; Fei-Fei, L. Deep visual-semantic alignments for generating image descriptions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 8–10 June 2015; pp. 3128–3137. [Google Scholar]
- Ranzato, M.; Szlam, A.; Bruna, J.; Mathieu, M.; Collobert, R.; Chopra, S. Video (language) modeling: A baseline for generative models of natural videos. arXiv 2014, arXiv:1412.6604. [Google Scholar]
- Xu, K.; Ba, J.; Kiros, R.; Cho, K.; Courville, A.; Salakhudinov, R.; Zemel, R.; Bengio, Y. Show, attend and tell: Neural image caption generation with visual attention. In Proceedings of the International Conference on Machine Learning, PMLR, Lille, France, 6–11 July 2015; pp. 2048–2057. [Google Scholar]
- Horn, R.A. The hadamard product. Proc. Symp. Appl. Math. 1990, 40, 87–169. [Google Scholar]
- Song, H.; Wang, W.; Zhao, S.; Shen, J.; Lam, K.M. Pyramid dilated deeper convlstm for video salient object detection. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 715–731. [Google Scholar]
- Graves, A.; Schmidhuber, J. Framewise phoneme classification with bidirectional LSTM and other neural network architectures. Neural Netw. 2005, 18, 602–610. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Zhang, J.; Zheng, Y.; Qi, D.; Li, R.; Yi, X. DNN-based prediction model for spatio-temporal data. In Proceedings of the International Conference on Advances in Geographic Information Systems, Burlingame, CA, USA, 31 October 2016; pp. 1–4. [Google Scholar]
- Srivastava, R.K.; Greff, K.; Schmidhuber, J. Training very deep networks. arXiv 2015, arXiv:1507.06228. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 23–30 June 2016; pp. 770–778. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity mappings in deep residual networks. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 11–14 October 2016; Springer: Berlin/Heidelberg, Germany, 2016; pp. 630–645. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the International Conference on Machine Learning, PMLR, Lille, France, 6–11 July 2015; pp. 448–456. [Google Scholar]











| Model | Test Dataset (GMM) | 3D Pasture (Gazebo) | ||||||
|---|---|---|---|---|---|---|---|---|
| RMSE | MAE | MAPE | aSt. Dev. | RMSE | MAE | MAPE | aSt. Dev. | |
| + MCMC | 20.02 | 14.54 | 12.25 | 8.55 | 12.37 | 11.21 | 6.49 | 11.15 |
| + MCMC | 19.11 | 13.36 | 11.79 | 9.14 | 7.37 | 6.33 | 3.61 | 12.3 |
| + MCMC | 11.52 | 8.13 | 7.33 | 8.48 | – | – | – | – |
| + MCMC | 6.85 | 5.05 | 4.63 | 8.28 | – | – | – | – |
| 26.35 | 20.04 | 15.84 | – | 24.91 | 24.03 | 14.02 | – | |
| 24.76 | 18.81 | 15.65 | – | 19.41 | 18.13 | 10.6 | – | |
| 21.74 | 16.7 | 14.49 | – | – | - | – | – | |
| 18.66 | 14.40 | 13.15 | – | – | – | – | – | |
| Imputation | + MCMC | |||
|---|---|---|---|---|
| RMSE | MAE | MAPE | aSt. Dev. | |
| 6.85 | 5.05 | 4.63 | 8.28 | |
| 5.98 | 4.29 | 4.24 | 8.57 | |
| 6.04 | 4.44 | 4.29 | 8.73 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rangwala, M.; Liu, J.; Ahluwalia, K.S.; Ghajar, S.; Dhami, H.S.; Tracy, B.F.; Tokekar, P.; Williams, R.K. DeepPaSTL: Spatio-Temporal Deep Learning Methods for Predicting Long-Term Pasture Terrains Using Synthetic Datasets. Agronomy 2021, 11, 2245. https://doi.org/10.3390/agronomy11112245
Rangwala M, Liu J, Ahluwalia KS, Ghajar S, Dhami HS, Tracy BF, Tokekar P, Williams RK. DeepPaSTL: Spatio-Temporal Deep Learning Methods for Predicting Long-Term Pasture Terrains Using Synthetic Datasets. Agronomy. 2021; 11(11):2245. https://doi.org/10.3390/agronomy11112245
Chicago/Turabian StyleRangwala, Murtaza, Jun Liu, Kulbir Singh Ahluwalia, Shayan Ghajar, Harnaik Singh Dhami, Benjamin F. Tracy, Pratap Tokekar, and Ryan K. Williams. 2021. "DeepPaSTL: Spatio-Temporal Deep Learning Methods for Predicting Long-Term Pasture Terrains Using Synthetic Datasets" Agronomy 11, no. 11: 2245. https://doi.org/10.3390/agronomy11112245
APA StyleRangwala, M., Liu, J., Ahluwalia, K. S., Ghajar, S., Dhami, H. S., Tracy, B. F., Tokekar, P., & Williams, R. K. (2021). DeepPaSTL: Spatio-Temporal Deep Learning Methods for Predicting Long-Term Pasture Terrains Using Synthetic Datasets. Agronomy, 11(11), 2245. https://doi.org/10.3390/agronomy11112245






