High-Throughput Root Network System Analysis for Low Phosphorus Tolerance in Maize at Seedling Stage
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Site
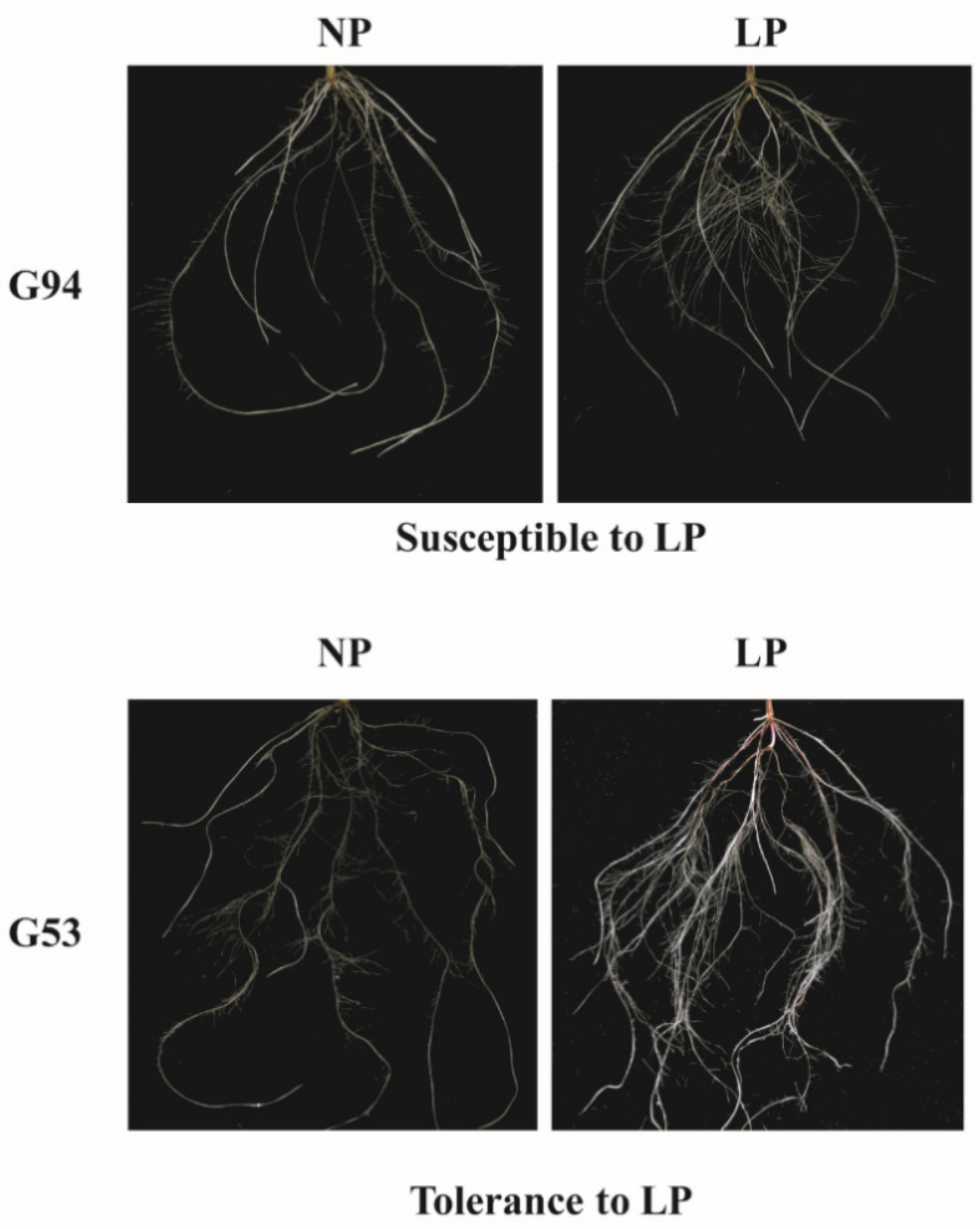
2.2. Plant Materials
2.3. Plant Growth Condition
2.4. Camera Arrangements
2.5. Imaging System
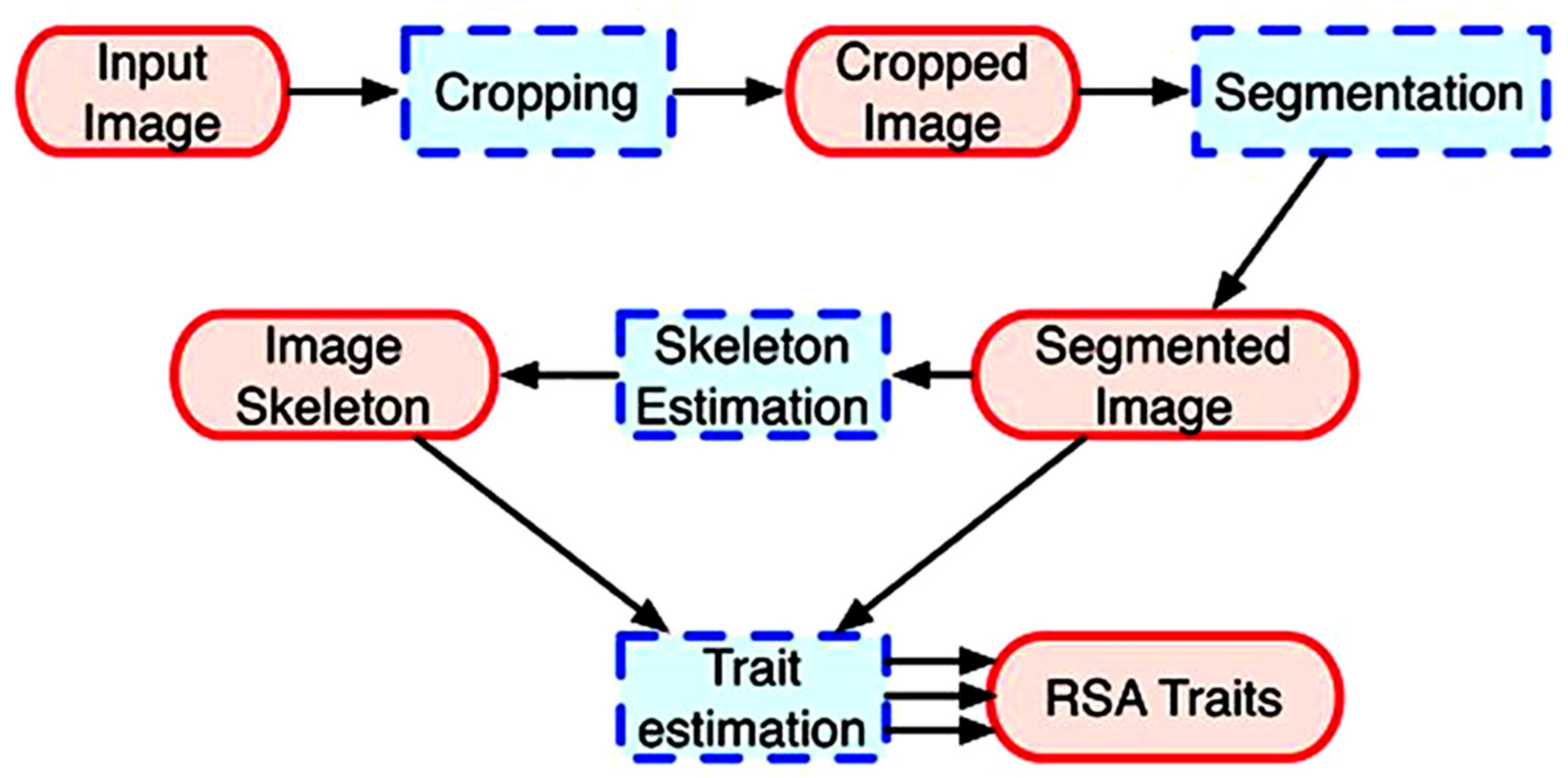
2.6. Image Acquisition, Analysis and Output
2.7. Recorded Data
2.8. Statistical Analysis
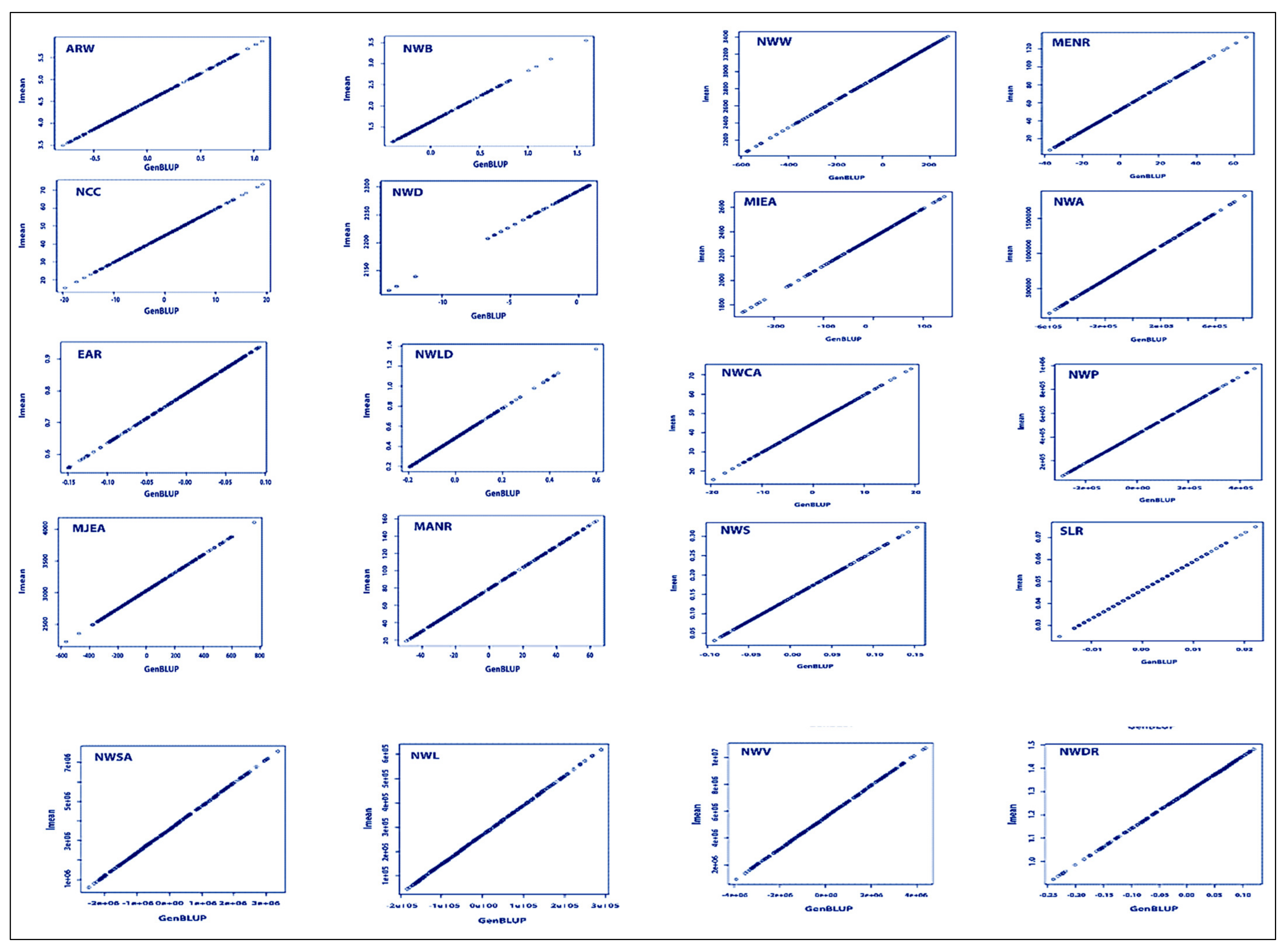
2.8.1. Best Linear Unbiased Predictors (BLUPs)
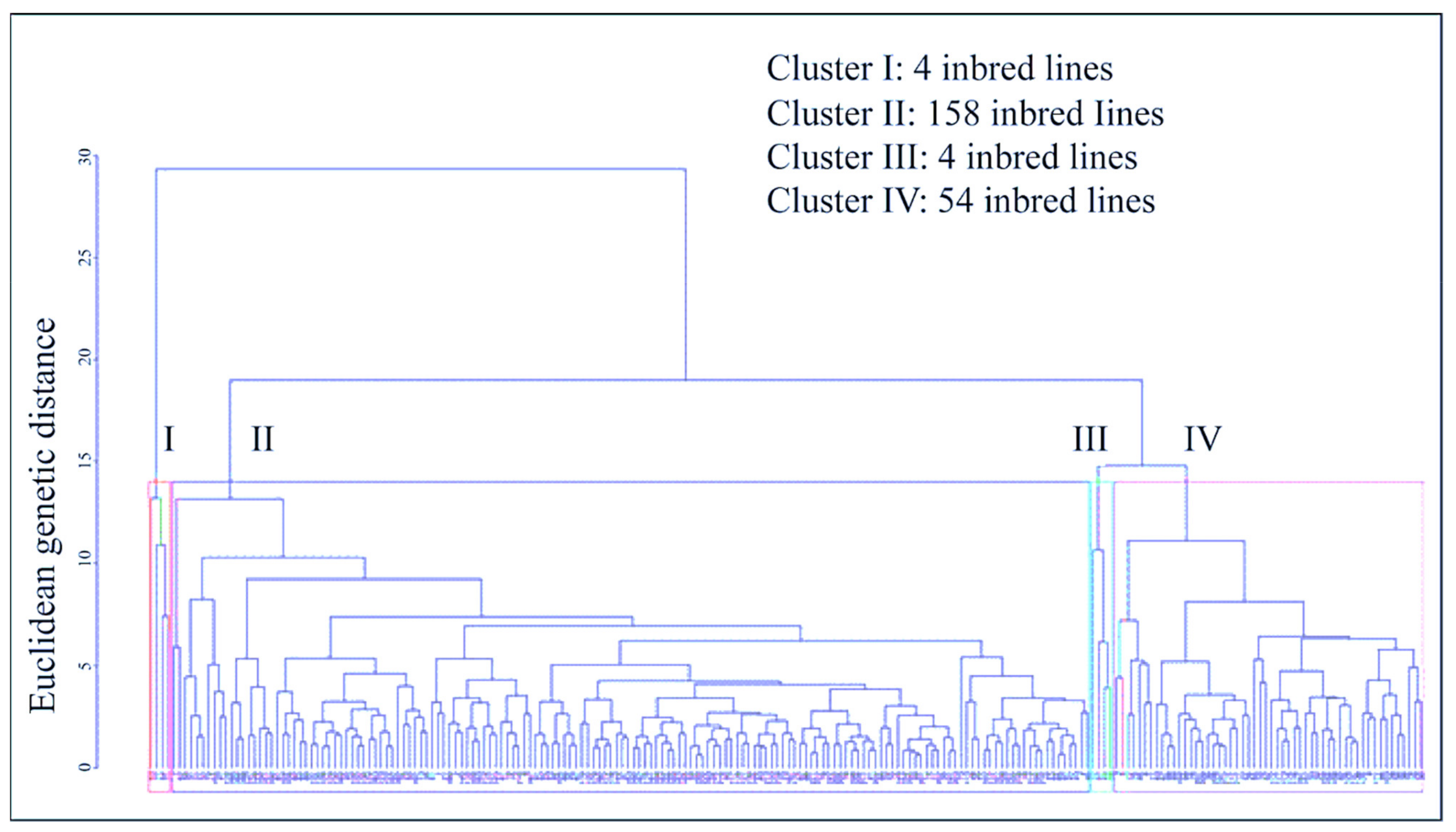
2.8.2. Multivariate Analysis
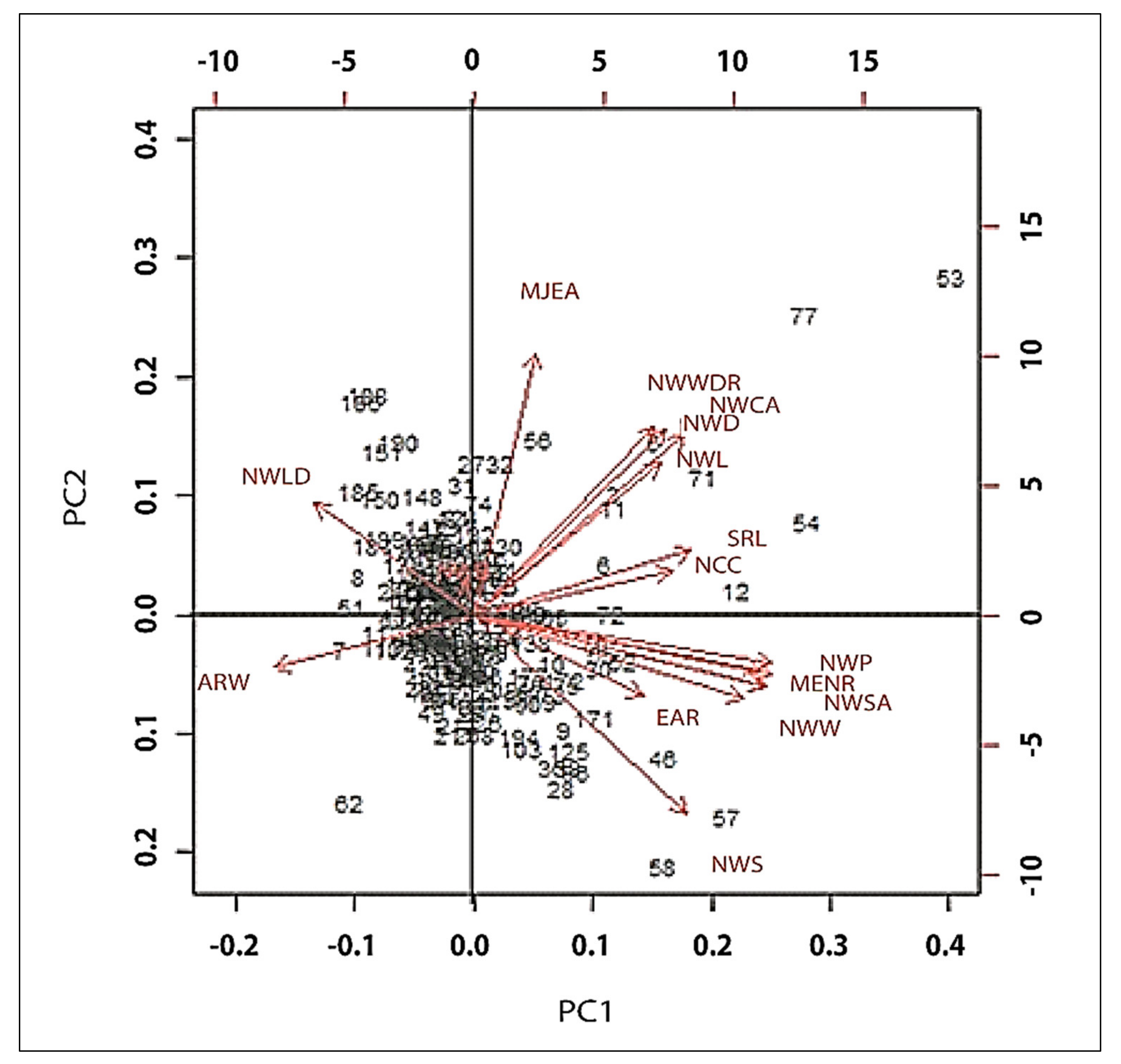
2.8.3. Genotype by Trait (GT) Interactions Biplot
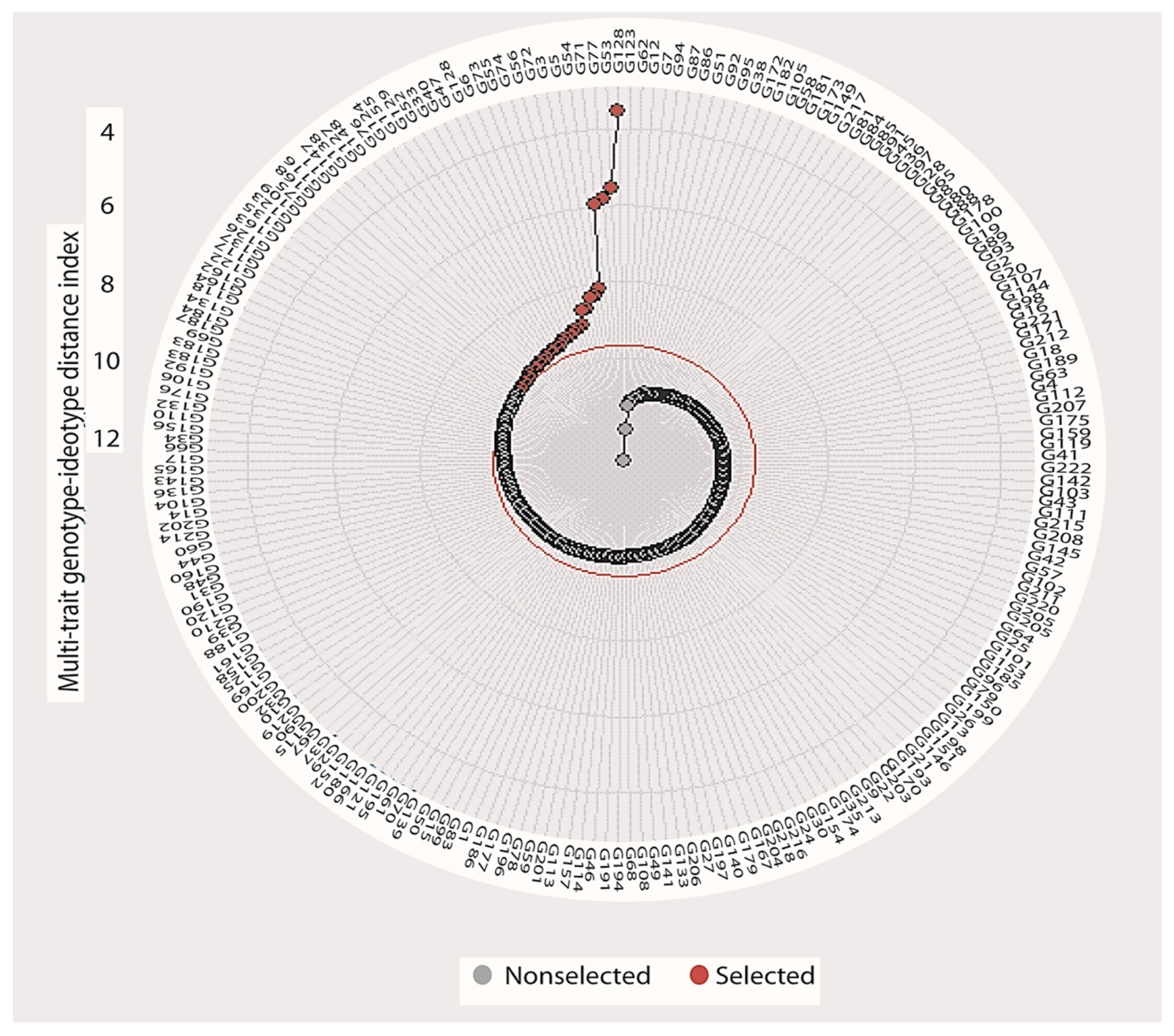
2.8.4. Multi-Trait Index Based on Factor Analysis and Genotype-Ideotypes Distance (MGIDI)
3. Results
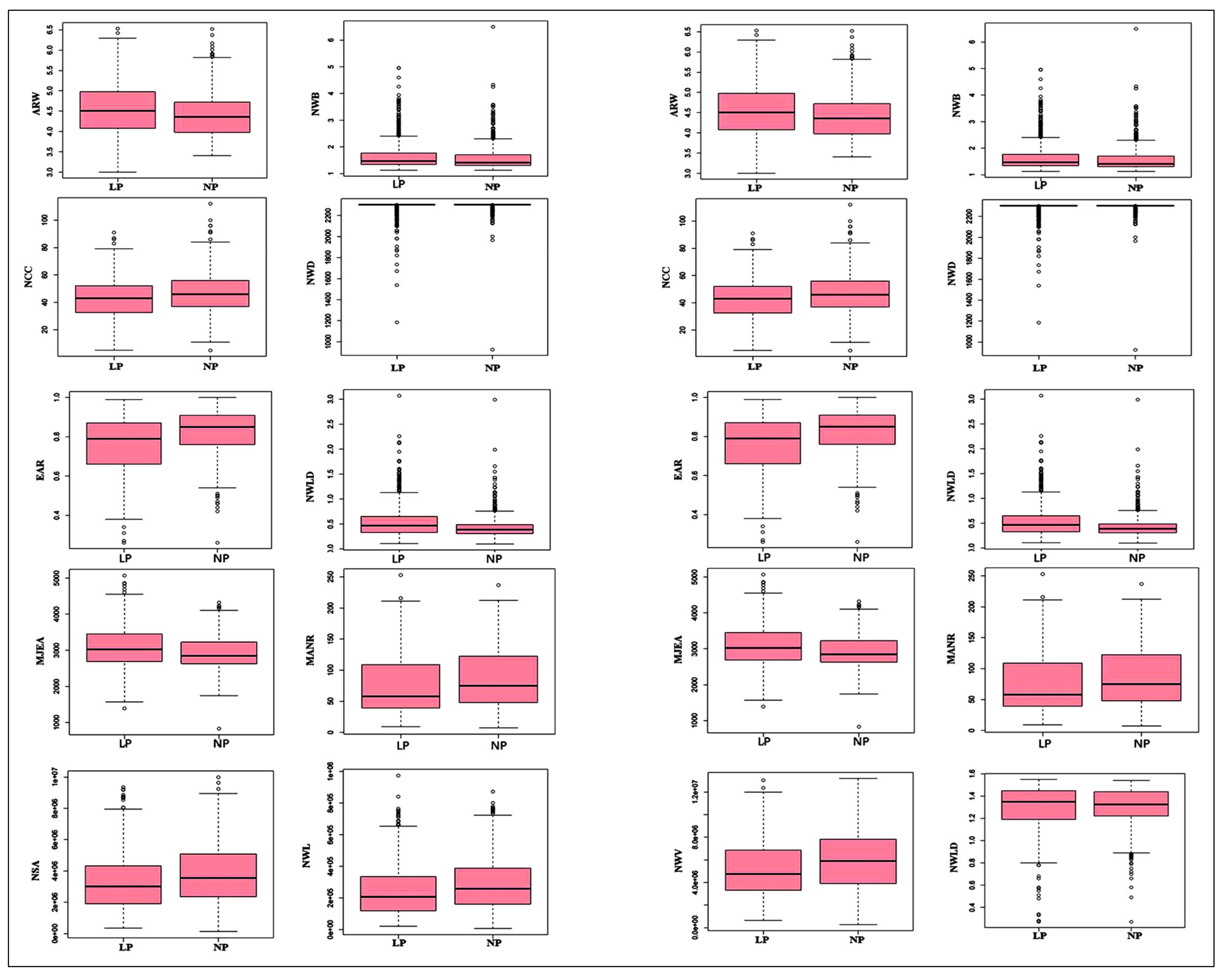
3.1. Descriptive Statistic and Analysis of Variance
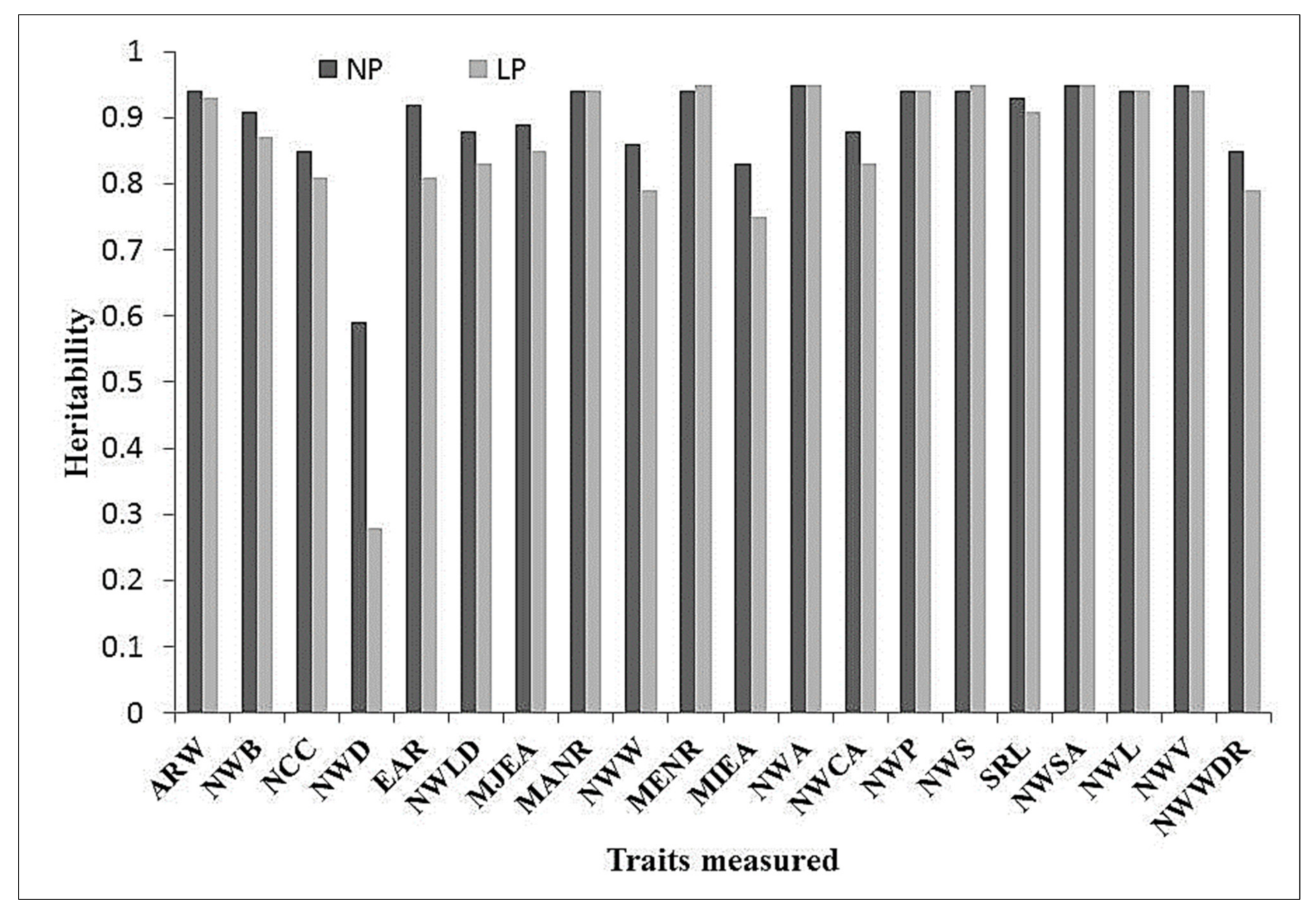
3.2. Analysis of Variance and Broad-Sense Heritability Estimated
3.3. Variance Components
3.4. BLUP Analysis
3.5. Multivariate Analysis
3.6. Genotype by Trait Interactions
3.7. Multi-Trait Index Based on Factor Analysis and Genotype-Ideotype Distance (MGIDI)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bailey-Serres, J.; Parker, J.E.; Ainsworth, E.A.; Oldroyd, G.E.D.; Schroeder, J.I. Genetic strategies for improving crop yields. Nat. Cell Biol. 2019, 575, 109–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raza, A.; Razzaq, A.; Mehmood, S.S.; Zou, X.; Zhang, X.; Lv, Y.; Xu, J. Impact of Climate Change on Crops Adaptation and Strategies to Tackle Its Outcome: A Review. Plants 2019, 8, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yee, M.O.; Kim, P.; Li, Y.; Singh, A.K.; Northen, T.R.; Chakraborty, R. Specialized Plant Growth Chamber Designs to Study Complex Rhizosphere Interactions. Front. Microbiol. 2021, 12, 625752. [Google Scholar] [CrossRef] [PubMed]
- FAO. The Future of Food and Agriculture—Trends and Challenges; Annual Report; FAO: Rome, Italy, 2017; p. 296. [Google Scholar]
- Jia, X.; Liu, P.; Lynch, J.P. Greater lateral root branching density in maize improves phosphorus acquisition from low phosphorus soil. J. Exp. Bot. 2018, 69, 4961–4970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Yan, C.; Guo, Q.; Zhang, J.; Ruiz-Menjivar, J. The impact of agricultural chemical inputs on environment: Global evidence from informetrics analysis and visualization. Int. J. Low-Carbon Technol. 2018, 13, 338–352. [Google Scholar] [CrossRef] [Green Version]
- Cordell, D.; White, S. Tracking phosphorus security: Indicators of phosphorus vulnerability in the global food system. Food Secur. 2015, 7, 337–350. [Google Scholar] [CrossRef]
- Pavinato, P.S.; Rodrigues, M.; Soltangheisi, A.; Sartor, L.R.; Withers, P.J.A. Effects of Cover Crops and Phosphorus Sources on Maize Yield, Phosphorus Uptake, and Phosphorus Use Efficiency. Agron. J. 2017, 109, 1039–1047. [Google Scholar] [CrossRef]
- Liao, D.; Zhang, C.; Li, H.; Lambers, H.; Zhang, F. Changes in soil phosphorus fractions following sole cropped and intercropped maize and faba bean grown on calcareous soil. Plant Soil 2020, 448, 587–601. [Google Scholar] [CrossRef]
- Miguel, M.A.; Postma, J.A.; Lynch, J.P. Phene Synergism between Root Hair Length and Basal Root Growth Angle for Phosphorus Acquisition. Plant Physiol. 2015, 167, 1430–1439. [Google Scholar] [CrossRef]
- Lynch, J.P. Root Phenes for Enhanced Soil Exploration and Phosphorus Acquisition: Tools for Future Crops. Plant Physiol. 2011, 156, 1041–1049. [Google Scholar] [CrossRef] [Green Version]
- Galindo-Castañeda, T.; Brown, K.M.; Lynch, J.P. Reduced root cortical burden improves growth and grain yield under low phosphorus availability in maize. Plant Cell Environ. 2018, 41, 1579–1592. [Google Scholar] [CrossRef]
- Klamer, F.; Vogel, F.; Li, X.; Bremer, H.; Neumann, G.; Neuhäuser, B.; Hochholdinger, F.; Ludewig, U. Estimating the importance of maize root hairs in low phosphorus conditions and under drought. Ann. Bot. 2019, 124, 961–968. [Google Scholar] [CrossRef]
- Miller, C.R.; Ochoa, I.; Nielsen, K.L.; Beck, D.; Lynch, J.P. Genetic variation for adventitious rooting in response to low phosphorus availability: Potential utility for phosphorusacquisition from stratified soils. Funct. Plant Biol. 2003, 30, 973–985. [Google Scholar] [CrossRef] [Green Version]
- Perkins, A.C.; Lynch, J.P. Increased seminal root number associated with domestication improves nitrogen and phosphorus acquisition in maize seedlings. Ann. Bot. 2021, 128, 453–468. [Google Scholar] [CrossRef]
- Richardson, A.E.; Lynch, J.P.; Ryan, P.R.; Delhaize, E.; Smith, F.A.; Smith, S.E.; Harvey, P.R.; Ryan, M.; Veneklaas, E.J.; Lambers, H.; et al. Plant and microbial strategies to improve the phosphorus efficiency of agriculture. Plant Soil 2011, 349, 121–156. [Google Scholar] [CrossRef]
- Mäkelä, P.; Wasonga, D.; Hernandez, A.S.; Santanen, A. Seedling Growth and Phosphorus Uptake in Response to Different Phosphorus Sources. Agronomy 2020, 10, 1089. [Google Scholar] [CrossRef]
- Tang, H.; Chen, X.; Gao, Y.; Hong, L.; Chen, Y. Alteration in root morphological and physiological traits of two maize cultivars in response to phosphorus deficiency. Rhizosphere 2020, 14, 100201. [Google Scholar] [CrossRef]
- Bhatta, M.; Shamanin, V.; Shepelev, S.; Baenziger, P.S.; Pozherukova, V.; Pototskaya, I.; Morgounov, A. Marker-Trait Associations for Enhancing Agronomic Performance, Disease Resistance, and Grain Quality in Synthetic and Bread Wheat Accessions in Western Siberia. G3 Genes Genomes Genet. 2019, 9, 4209–4222. [Google Scholar] [CrossRef] [Green Version]
- Hadfield, J. The quantitative genetic theory of parental effects. In The Evolution of Parental Care; Oxford University Press: Oxford, UK, 2012; pp. 267–284. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, A.; Czauderna, T.; Hoffmann, R.; Stein, N.; Schreiber, F. HTPheno: An image analysis pipeline for high-throughput plant phenotyping. BMC Bioinform. 2011, 12, 148. [Google Scholar] [CrossRef] [Green Version]
- Kaspar, T.C.; Ewing, R.P. ROOTEDGE: Software for Measuring Root Length from Desktop Scanner Images. Agron. J. 1997, 89, 932–940. [Google Scholar] [CrossRef]
- Le Bot, J.; Serra, V.; Fabre, J.; Draye, X.; Adamowicz, S.; Pagès, L. DART: A software to analyse root system architecture and development from captured images. Plant Soil 2009, 326, 261–273. [Google Scholar] [CrossRef]
- Leitner, D.; Felderer, B.; Vontobel, P.; Schnepf, A. Recovering Root System Traits Using Image Analysis Exemplified by Two-Dimensional Neutron Radiography Images of Lupine. Plant Physiol. 2013, 164, 24–35. [Google Scholar] [CrossRef] [Green Version]
- Galkovskyi, T.; Mileyko, Y.; Bucksch, A.; Moore, B.; Symonova, O.; A Price, C.; Topp, C.N.; Iyer-Pascuzzi, A.S.; Zurek, P.R.; Fang, S.; et al. GiA Roots: Software for the high throughput analysis of plant root system architecture. BMC Plant Biol. 2012, 12, 116. [Google Scholar] [CrossRef] [Green Version]
- Messmer, R.; Fracheboud, Y.; Bänziger, M.; Stamp, P.; Ribaut, J.-M. Drought stress and tropical maize: QTLs for leaf greenness, plant senescence, and root capacitance. Field Crop. Res. 2011, 124, 93–103. [Google Scholar] [CrossRef]
- R Core Team. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Olivoto, T.; Nardino, M. MGIDI: Toward an effective multivariate selection in biological experiments. Bioinformatics 2020, 37, 1383–1389. [Google Scholar] [CrossRef]
- Smith, H.F. A Discriminant function for plant selection. Ann. Eugen. 1936, 7, 240–250. [Google Scholar] [CrossRef]
- Hazel, L.N. The genetic basis for constructing selection indexes. Genetics 1943, 28, 476–490. [Google Scholar] [CrossRef]
- Rocha, J.R.d.A.S.d.C.; Nunes, K.V.; Carneiro, A.L.N.; Marçal, T.d.S.; Salvador, F.V.; Carneiro, P.C.S.; Carneiro, J.E.S. Selection of superior inbred progenies toward the common bean ideotype. Agron. J. 2019, 111, 1181–1189. [Google Scholar] [CrossRef]
- Deng, Y.; Chen, K.; Teng, W.; Zhan, A.; Tong, Y.; Feng, G.; Cui, Z.; Zhang, F.; Chen, X. Is the Inherent Potential of Maize Roots Efficient for Soil Phosphorus Acquisition? PLoS ONE 2014, 9, e90287. [Google Scholar] [CrossRef]
- Jiang, H.; Zhang, J.; Han, Z.; Yang, J.; Ge, C.; Wu, Q. Revealing new insights into different phosphorus-starving responses between two maize (Zea mays) inbred lines by transcriptomic and proteomic studies. Sci. Rep. 2017, 7, 44294. [Google Scholar] [CrossRef] [Green Version]
- Ruiz, S.; Koebernick, N.; Duncan, S.; Fletcher, D.M.; Scotson, C.; Boghi, A.; Marin, M.; Bengough, A.G.; George, T.S.; Brown, L.K.; et al. Significance of root hairs at the field scale–modelling root water and phosphorus uptake under different field conditions. Plant Soil 2020, 447, 281–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, Q.-L.; Yang, K.-C.; Pan, G.-T.; Rong, T.-Z. The Effects of Low Phosphorus Stress on Morphological and Physiological Characteristics of Maize (Zea mays L.) Landraces. Agric. Sci. China 2007, 6, 559–566. [Google Scholar] [CrossRef]
- Zhang, Y.-K.; Chen, F.-J.; Chen, X.-C.; Long, L.-Z.; Gao, K.; Yuan, L.-X.; Zhang, F.-S.; Mi, G.-H. Genetic Improvement of Root Growth Contributes to Efficient Phosphorus Acquisition in maize (Zea mays L.). J. Integr. Agric. 2013, 12, 1098–1111. [Google Scholar] [CrossRef]
- Bayuelo-Jiménez, J.S.; Ochoa-Cadavid, I. Phosphorus acquisition and internal utilization efficiency among maize landraces from the central Mexican highlands. Field Crop. Res. 2014, 156, 123–134. [Google Scholar] [CrossRef]
- Haling, R.E.; Brown, L.K.; Stefanski, A.; Kidd, D.R.; Ryan, M.; Sandral, G.A.; George, T.S.; Lambers, H.; Simpson, R.J. Differences in nutrient foraging among Trifolium subterraneum cultivars deliver improved P-acquisition efficiency. Plant Soil 2018, 424, 539–554. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.; Yang, J.; Zhang, J.; Hou, Y. Screening of tolerant maize genotypes in the low phosphorus field soil. In Proceedings of the 19th World Congress of Soil Science: Soil Solutions for a Changing World, Symposium 3.1.2 Farm System and Environment Impacts, Brisbane, Australia, 1–6 August 2010; pp. 214–217. [Google Scholar]
- Li, H.; Zhang, D.; Wang, X.; Li, H.; Rengel, Z.; Shen, J. Competition between Zea mays genotypes with different root morphological and physiological traits is dependent on phosphorus forms and supply patterns. Plant Soil 2018, 434, 125–137. [Google Scholar] [CrossRef]
- Li, X.; Mang, M.; Piepho, H.; Melchinger, A.; Ludewig, U. Decline of seedling phosphorus use efficiency in the heterotic pool of flint maize breeding lines since the onset of hybrid breeding. J. Agron. Crop Sci. 2021, 207, 857–872. [Google Scholar] [CrossRef]
- Mundim, G.B.; Viana, J.M.S.; Maia, C. Early evaluation of popcorn inbred lines for phosphorus use efficiency. Plant Breed. 2013, 132, 613–619. [Google Scholar] [CrossRef]
- Coetzee, P.-E.; Ceronio, G.M.; du Preez, C.C. Evaluation of the effects of phosphorus and nitrogen source on aerial and subsoil parameters of maize (Zea mays L.) during early growth and development. S. Afr. J. Plant Soil 2016, 33, 237–244. [Google Scholar] [CrossRef]
- Zhang, H.; Xu, R.; Xie, C.; Huang, C.; Liao, H.; Xu, Y.; Wang, J.; Li, W.-X. Large-Scale Evaluation of Maize Germplasm for Low-Phosphorus Tolerance. PLoS ONE 2015, 10, e0124212. [Google Scholar] [CrossRef]
- Jones, J.B., Jr. Complete Guide for Growing Plants Hydroponically; CRC Press: Boca Raton, FL, USA, 2014. [Google Scholar]
- Tuberosa, R.; Salvi, S.; Sanguineti, M.C.; Maccaferri, M.; Giuliani, S.; Landi, P. Searching for quantitative trait loci controlling root traits in maize: A critical appraisal. Plant Soil 2003, 255, 35–54. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, X.; Craft, E.J.; Yuan, L.; Cheng, L.; Mi, G.; Chen, F. Physiological and genetic analysis for maize root characters and yield in response to low phosphorus stress. Breed. Sci. 2018, 68, 268–277. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Li, H.; Wang, J.; Zhang, H.; Hu, Z.; Chu, S.; Lv, H.; Yu, D. High-Density Genetic Mapping Identifies New Major Loci for Tolerance to Low-Phosphorus Stress in Soybean. Front. Plant Sci. 2016, 7, 372. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Uddin, M.S.; Zou, C.; Xie, C.; Xu, Y.; Li, W.-X. Meta-analysis and candidate gene mining of low-phosphorus tolerance in maize. J. Integr. Plant Biol. 2014, 56, 262–270. [Google Scholar] [CrossRef] [Green Version]
- Gu, R.; Chen, F.; Long, L.; Cai, H.; Liu, Z.; Yang, J.; Wang, L.; Li, H.; Li, J.; Liu, W.; et al. Enhancing phosphorus uptake efficiency through QTL-based selection for root system architecture in maize. J. Genet. Genom. 2016, 43, 663–672. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Kaeppler, S.; Lynch, J.P. Mapping of QTL controlling root hair length in maize (Zea mays L.) under phosphorus deficiency. Plant Soil 2005, 270, 299–310. [Google Scholar] [CrossRef]
- Liu, Z.; Gao, K.; Shan, S.; Gu, R.; Wang, Z.; Craft, E.J.; Mi, G.; Yuan, L.; Chen, F. Comparative analysis of root traits and the associated QTLs for maize seedlings grown in paper roll, hydroponics and vermiculite culture system. Front. Plant Sci. 2017, 8, 436. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Mickelson, S.M.; Kaeppler, S.M.; Lynch, J.P. Detection of quantitative trait loci for seminal root traits in maize (Zea mays L.) seedlings grown under differential phosphorus levels. Theor. Appl. Genet. 2006, 113, 1–10. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, H.; Chu, S.; Li, H.; Chi, Y.; Triebwasser-Freese, D.; Lv, H.; Yu, D. Integrating QTL mapping and transcriptomics identifies candidate genes underlying QTLs associated with soybean tolerance to low-phosphorus stress. Plant Mol. Biol. 2016, 93, 137–150. [Google Scholar] [CrossRef]
- Ouma, E.O. Evaluating Heritability and Relationships among Phosphorus Efficiency Traits in Maize under Low P Soils of Western Kenya. Curr. J. Appl. Sci. Technol. 2021, 83–96. [Google Scholar] [CrossRef]
- Tuberosa, R.; Salvi, S.; Giuliani, S.; Sanguineti, M.C.; Frascaroli, E.; Conti, S.; Landi, P. Genomics of root architecture and functions in maize. In Root Genomics; Springer: Berlin/Heidelberg, Germany, 2010; pp. 179–204. [Google Scholar] [CrossRef]
- Zhang, L.-T.; Li, J.; Rong, T.-Z.; Gao, S.-B.; Wu, F.-K.; Xu, J.; Li, M.-L.; Cao, M.-J.; Wang, J.; Hu, E.-L.; et al. Large-scale screening maize germplasm for low-phosphorus tolerance using multiple selection criteria. Euphytica 2014, 197, 435–446. [Google Scholar] [CrossRef]
- Bayuelo-Jiménez, J.S.; Gallardo-Valdéz, M.; Pérez-Decelis, V.A.; Magdaleno-Armas, L.; Ochoa, I.; Lynch, J.P. Genotypic variation for root traits of maize (Zea mays L.) from the Purhepecha Plateau under contrasting phosphorus availability. Field Crop. Res. 2011, 121, 350–362. [Google Scholar] [CrossRef]
- Bayuelo-Jiménez, J.S.; Ochoa-Cadavid, I. Interacción genotipo × ambiente para eficiencia en el uso de fósforo en maíz nativo de la meseta p’urhépecha. Rev. Fitotec. Mex. 2018, 41, 39–47. [Google Scholar] [CrossRef] [Green Version]
- Gaggion, N.; Ariel, F.; Daric, V.; Lambert, É.; Legendre, S.; Roulé, T.; Camoirano, A.; Milone, D.H.; Crespi, M.; Blein, T.; et al. ChronoRoot: High-throughput phenotyping by deep segmentation networks reveals novel temporal parameters of plant root system architecture. GigaScience 2021, 10, giab052. [Google Scholar] [CrossRef]
- Bhandari, H.; Bhanu, A.; Srivastava, K.; Singh, M.; Shreya, H.A. Assessment of genetic diversity in crop plants-an overview. Adv. Plants Agric. Res. 2017, 7, 00255. [Google Scholar]
- Olivoto, T.; Diel, M.I.; Schmidt, D.; Lúcio, A.D.C. Multivariate analysis of strawberry experiments: Where are we now and where can we go? BioRxiv 2021. [Google Scholar]

| Sl. No | Name of the Chemical | Net Weight (g)/10 L | Volume Needs for 50 L Stock Solution |
|---|---|---|---|
| 01 | Calcium Nitrate {Ca(NO3)2·4H2O} | 944.6 | 250 mL |
| 02 | Potassium Sulfate (K2SO4) + Magnesium Sulfate (MgSO4) | 261.39 + 324.12 | 250 mL |
| 03 | Potassium Chloride (KCl) | 14.9 | 250 mL |
| 04 | Potassium dihydrogen phosphate (KH2PO4) | 68.046 | 2.5 mL |
| 05 | Boric Acid (H3BO4) | 0.6108 | 5 mL |
| 06 | Manganese sulfate (MnSO4) + Copper sulfate (CuSO4·5H2O) + Zinc sulfate (ZnSO4·5H2O) + {(NH4)6·Mo7·O24·4H2O} | 1.690 + 0.25 + 2.87 + 0.062 | 5 mL |
| 07 | Iron sodium salt Fe-EDTA | 146.82 | 250 mL |
| Sl. No | Name of the Chemical | Net Weight (g)/10 L | Volume Needs for 50 L Stock Solution |
|---|---|---|---|
| 01 | Calcium Nitrate {Ca(NO3)2·4H2O} | 944.6 | 250 mL |
| 02 | Potassium Sulfate (K2SO4) + Magnesium Sulfate (MgSO4) | 261.39 + 324.12 | 250 mL |
| 03 | Potassium Chloride (KCl) | 14.9 | 250 mL |
| 04 | Potassium dihydrogen phosphate (KH2PO4) | 68.046 | 250 mL |
| 05 | Boric Acid (H3BO4) | 0.618 | 5 mL |
| 06 | Manganese sulfate (MnSO4) + Copper sulfate (CuSO4·5H2O) + Zinc sulfate (ZnSO4·5H2O) + {(NH4)6·Mo7·O24·4H2O} | 1.690 + 0.25 + 2.87 + 0.062 | 5 mL |
| 07 | Iron sodium salt Fe-EDTA | 146.82 | 250 mL |
| Traits | Treat. | Mean | SE | Minimum | Maximum | Median | Mode | Kurtosis | Skewness |
|---|---|---|---|---|---|---|---|---|---|
| ARW | NP | 4.52 | 0.02 | 3.00 | 6.53 | 4.44 | 6.01 | −0.38 | 0.46 |
| LP | 4.45 | 0.02 | 3.41 | 6.52 | 4.40 | 4.12 | −0.05 | 0.62 | |
| NWB | NP | 1.62 | 0.01 | 1.12 | 4.96 | 1.45 | 1.50 | 7.91 | 2.50 |
| LP | 1.54 | 0.01 | 1.10 | 6.50 | 1.39 | 1.33 | 20.68 | 3.40 | |
| NCC | NP | 43.89 | 0.42 | 3.00 | 91.00 | 44.00 | 43.00 | −0.11 | 0.13 |
| LP | 46.15 | 0.37 | 5.00 | 112.00 | 45.00 | 44.00 | 0.64 | 0.38 | |
| NWD | NP | 2290.35 | 2.17 | 910.00 | 2303.00 | 2303.00 | 2303.00 | 158.25 | −11.45 |
| LP | 2297.84 | 1.25 | 926.00 | 2303.00 | 2303.00 | 2303.00 | 634.59 | −22.55 | |
| EAR | NP | 0.76 | 0.00 | 0.11 | 0.99 | 0.79 | 0.84 | −0.24 | −0.56 |
| LP | 0.82 | 0.00 | 0.26 | 1.00 | 0.85 | 0.84 | 0.24 | −0.87 | |
| NWLD | NP | 0.53 | 0.01 | 0.11 | 3.07 | 0.46 | 0.24 | 8.27 | 2.20 |
| LP | 0.44 | 0.01 | 0.06 | 2.99 | 0.41 | 0.61 | 26.59 | 3.20 | |
| MJEA | NP | 3105.60 | 15.08 | 823.57 | 5072.72 | 3001.58 | 2632.98 | 0.05 | 0.47 |
| LP | 2972.72 | 11.85 | 830.81 | 4465.25 | 2856.90 | 2679.85 | 0.33 | 0.66 | |
| MANR | NP | 77.86 | 1.25 | 5.00 | 253.00 | 62.00 | 56.00 | −0.51 | 0.64 |
| LP | 83.14 | 1.24 | 7.00 | 237.00 | 69.00 | 44.00 | −0.55 | 0.70 | |
| NWW | NP | 2956.82 | 13.59 | 179.00 | 3455.00 | 3116.00 | 3455.00 | 3.19 | −1.54 |
| LP | 2979.72 | 11.07 | 249.00 | 3455.00 | 3046.00 | 3455.00 | 2.48 | −1.22 | |
| MENR | NP | 52.43 | 0.97 | 2.00 | 223.00 | 42.00 | 21.00 | 0.52 | 1.02 |
| LP | 57.32 | 0.97 | 4.00 | 177.00 | 45.00 | 37.00 | 0.48 | 1.08 | |
| MIEA | NP | 2310.48 | 7.83 | 239.24 | 2972.45 | 2353.58 | 2201.84 | 8.46 | −1.97 |
| LP | 2386.18 | 6.64 | 213.02 | 3162.82 | 2398.86 | 2255.16 | 7.44 | −1.09 | |
| NWA | NP | 85,2169.61 | 12,698.0 | 28,448.00 | 2,306,467.00 | 766,588.00 | 425,187.00 | −0.21 | 0.67 |
| LP | 927,120.77 | 12,626.3 | 33,913.00 | 2,380,819.00 | 828,074.00 | 1,399,665.00 | −0.15 | 0.72 | |
| NWCA | NP | 6,256,640.0 | 33,659.7 | 331,902.00 | 7,899,088.00 | 6,517,483.75 | 4,864,456.50 | 2.12 | −1.20 |
| LP | 6,326,244.7 | 26,555.5 | 148,855.50 | 7,907,468.00 | 6,493,552.25 | 4,333,747.00 | 2.02 | −0.99 | |
| NWP | NP | 419,776.22 | 7310.85 | 10,665.00 | 1,449,259.00 | 346,500.50 | 149,445.00 | 0.20 | 0.88 |
| LP | 455,401.13 | 7309.86 | 11,192.00 | 1,372,171.00 | 375,264.00 | 709,723.00 | 0.21 | 0.95 | |
| NWS | NP | 0.14 | 0.00 | 0.02 | 0.47 | 0.12 | 0.09 | 0.84 | 1.11 |
| LP | 0.15 | 0.00 | 0.03 | 0.46 | 0.13 | 0.32 | 0.23 | 1.00 | |
| SRL | NP | 0.05 | 0.00 | 0.02 | 0.10 | 0.04 | 0.03 | 0.43 | 0.70 |
| LP | 0.05 | 0.00 | 0.02 | 0.08 | 0.05 | 0.05 | −0.61 | 0.27 | |
| NWSA | NP | 3,523,843.5 | 53,442.4 | 109,925.66 | 9,490,765.02 | 3,153,257.77 | 1,669,954.28 | −0.20 | 0.69 |
| LP | 3,832,697.7 | 53,021.9 | 127,452.83 | 10,048,319.6 | 3,407,774.88 | 5,810,109.73 | −0.12 | 0.73 | |
| NWL | NP | 265,436.89 | 4685.19 | 6132.00 | 975,158.00 | 219,807.50 | 88,374.00 | 0.23 | 0.89 |
| LP | 287,929.75 | 4665.79 | 6225.00 | 879,854.00 | 238,309.00 | 449,101.00 | 0.24 | 0.96 | |
| NWV | NP | 5,462,435.1 | 71,955.11 | 204,831.84 | 13,046,946.3 | 5,082,050.76 | 3,397,327.72 | −0.53 | 0.43 |
| LP | 5,916,641.1 | 70,714.1 | 256,796.93 | 13,200,212.0 | 5,630,054.48 | 8,591,461.09 | −0.62 | 0.39 | |
| NWWDR | NP | 1.29 | 0.01 | 0.12 | 1.57 | 1.35 | 1.50 | 2.52 | −1.41 |
| LP | 1.30 | 0.00 | 0.27 | 1.68 | 1.32 | 1.50 | 1.79 | −1.13 |
| Traits | Treatment | Mean Square | F-Value (Gen) | Significance Level | ||
|---|---|---|---|---|---|---|
| Replication | Genotype | Error | ||||
| ARW | NP | 0.04 | 1.45 | 0.09 | 15.69 | *** |
| LP | 0.02 | 0.93 | 0.07 | 14.2 | *** | |
| NWB | NP | 0.04 | 0.78 | 0.07 | 10.83 | *** |
| LP | 0.07 | 0.61 | 0.08 | 7.78 | *** | |
| NCC | NP | 13.07 | 648.64 | 95.46 | 6.80 | *** |
| LP | 83.32 | 482.43 | 93.30 | 5.17 | *** | |
| NWD | NP | 161.82 | 11,386.40 | 4695.10 | 2.43 | *** |
| LP | 838.02 | 2807.12 | 2030.71 | 1.38 | * | |
| EAR | NP | 0.01 | 0.06 | 0.01 | 12.7 | * |
| LP | 0.01 | 0.03 | 0.01 | 5.30 | *** | |
| NWLD | NP | 0.03 | 0.27 | 0.03 | 8.11 | *** |
| LP | 0.01 | 0.11 | 0.02 | 6.03 | *** | |
| MJEA | NP | 76,230.1 | 916,523 | 103,873 | 8.82 | *** |
| LP | 99,507.1 | 486,751 | 71,573.5 | 6.80 | *** | |
| MANR | NP | 12.857 | 7172.570 | 442.848 | 16.2 | *** |
| LP | 229.32 | 7031.23 | 456.18 | 15.41 | *** | |
| NWW | NP | 64,219.4 | 696,475 | 100,280 | 6.95 | *** |
| LP | 57,281.70 | 407,323.00 | 85,124.00 | 4.79 | *** | |
| MENR | NP | 12.86 | 7172.57 | 442.85 | 16.2 | *** |
| LP | 229.32 | 7031.23 | 456.18 | 15.41 | *** | |
| MIEA | NP | 76,230.1 | 916,523 | 103,873 | 8.82 | *** |
| LP | 99,507.10 | 486,751.00 | 71,573.50 | 6.80 | *** | |
| NWA | NP | 851,326,000.00 | 756,240,000,000.00 | 38,367,000,000.00 | 19.71 | *** |
| LP | 27,187,900,000.00 | 761,409,000,000.00 | 40,363,800,000.00 | 18.86 | *** | |
| NWCA | NP | 663,782,000,000.00 | 4,530,370,000,000.00 | 528,786,000,000.00 | 8.57 | *** |
| LP | 217,830,000,000.00 | 2,475,580,000,000.00 | 428,953,000,000.00 | 5.77 | *** | |
| NWP | NP | 120,272,000.00 | 246,041,000,000.00 | 14,265,200,000.00 | 17.25 | *** |
| LP | 9,820,570,000.00 | 252,527,000,000.00 | 14,005,400,000.00 | 18.03 | *** | |
| NWS | NP | 0.001 | 0.025 | 0.001 | 16.83 | *** |
| LP | 0.001 | 0.026 | 0.001 | 19.64 | *** | |
| SRL | NP | 0.00003 | 0.00055 | 0.00004 | 14.78 | *** |
| LP | 0.00001 | 0.00036 | 0.00003 | 13.06 | *** | |
| NWSA | NP | 12,716,500,000.00 | 13,361,400,000,000.00 | 690,971,000,000.00 | 19.34 | *** |
| LP | 523,286,000,000.00 | 13,377,900,000,000.00 | 721,558,000,000.00 | 18.54 | *** | |
| NWL | NP | 65,982,000.00 | 100,591,000,000.00 | 6,010,840,000.00 | 16.73 | *** |
| LP | 4,479,120,000.00 | 102,500,000,000.00 | 5,795,460,000.00 | 17.69 | *** | |
| NWV | NP | 120,369,000,000.00 | 24,219,600,000,000.00 | 1,252,990,000,000.00 | 19.33 | *** |
| LP | 902,056,000,000.00 | 23,414,500,000,000.00 | 1,350,090,000,000.00 | 17.34 | *** | |
| NWWDR | NP | 0.01 | 0.12 | 0.02 | 6.72 | *** |
| LP | 0.01 | 0.08 | 0.02 | 4.71 | *** | |
| Traits | Genotype | Genotype × Treatment | Treatment | Residual |
|---|---|---|---|---|
| ARW | 0.53 | 0.24 | 0.03 | 0.21 |
| NWB | 0.53 | 0.14 | 0.01 | 0.33 |
| NCC | 0.32 | 0.21 | 0.05 | 0.41 |
| NWD | 0.01 | 0.14 | 0.01 | 0.84 |
| EAR | 0.30 | 0.26 | 0.11 | 0.33 |
| NWLD | 0.34 | 0.24 | 0.08 | 0.34 |
| MJEA | 0.37 | 0.22 | 0.04 | 0.37 |
| MANR | 0.55 | 0.20 | 0.03 | 0.22 |
| NWW | 0.29 | 0.20 | 0.00 | 0.50 |
| MENR | 0.56 | 0.19 | 0.03 | 0.21 |
| MIEA | 0.16 | 0.31 | 0.05 | 0.48 |
| NWA | 0.60 | 0.18 | 0.04 | 0.18 |
| NWCA | 0.39 | 0.18 | 0.01 | 0.42 |
| NWP | 0.57 | 0.19 | 0.04 | 0.20 |
| NWS | 0.58 | 0.18 | 0.03 | 0.21 |
| SRL | 0.50 | 0.24 | 0.01 | 0.26 |
| NWSA | 0.59 | 0.18 | 0.04 | 0.19 |
| NWL | 0.56 | 0.19 | 0.04 | 0.21 |
| NWV | 0.60 | 0.17 | 0.04 | 0.19 |
| NWWDR | 0.29 | 0.19 | 0.00 | 0.51 |
| Parameter | PC1 | PC2 | PC3 | PC4 |
|---|---|---|---|---|
| ARW | −0.1955 | −0.0387 | 0.2908 | −0.3165 |
| NWB | 0.0163 | 0.0311 | −0.2666 | 0.6438 |
| NCC | 0.2166 | 0.0606 | 0.2587 | −0.0784 |
| NWD | 0.2171 | 0.2043 | 0.1941 | 0.1486 |
| EAR | 0.1465 | −0.1705 | 0.4498 | 0.3054 |
| NWLD | −0.1413 | 0.2499 | −0.0488 | −0.1898 |
| MJEA | 0.1008 | 0.4366 | −0.2509 | −0.2223 |
| MANR | 0.2823 | −0.098 | −0.1934 | −0.0069 |
| NWW | 0.2087 | 0.2918 | 0.2853 | 0.1004 |
| MENR | 0.2823 | −0.098 | −0.1934 | −0.0069 |
| MIEA | 0.1008 | 0.4366 | −0.2509 | −0.2223 |
| NWA | 0.2952 | −0.1247 | −0.0122 | −0.1458 |
| NWCA | 0.2387 | 0.2501 | 0.2458 | 0.0917 |
| NWP | 0.2999 | −0.0692 | −0.0912 | −0.0696 |
| NWS | 0.1752 | −0.3885 | −0.0632 | −0.16 |
| SRL | 0.2249 | 0.046 | −0.3095 | 0.2565 |
| NWSA | 0.2957 | −0.1204 | −0.016 | −0.1436 |
| NWL | 0.2999 | −0.0651 | −0.0875 | −0.074 |
| NWV | 0.2685 | −0.1737 | 0.0867 | −0.2503 |
| NWWDR | 0.1947 | 0.3003 | 0.2624 | 0.0654 |
| Cumulative% of total variance | 52.18 | 69.24 | 79.84 | 85.71 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uddin, M.S.; Azam, M.G.; Billah, M.; Bagum, S.A.; Biswas, P.L.; Khaldun, A.B.M.; Hossain, N.; Gaber, A.; Althobaiti, Y.S.; Abdelhadi, A.A.; et al. High-Throughput Root Network System Analysis for Low Phosphorus Tolerance in Maize at Seedling Stage. Agronomy 2021, 11, 2230. https://doi.org/10.3390/agronomy11112230
Uddin MS, Azam MG, Billah M, Bagum SA, Biswas PL, Khaldun ABM, Hossain N, Gaber A, Althobaiti YS, Abdelhadi AA, et al. High-Throughput Root Network System Analysis for Low Phosphorus Tolerance in Maize at Seedling Stage. Agronomy. 2021; 11(11):2230. https://doi.org/10.3390/agronomy11112230
Chicago/Turabian StyleUddin, Md. Shalim, Md. Golam Azam, Masum Billah, Shamim Ara Bagum, Priya Lal Biswas, Abul Bashar Mohammad Khaldun, Neelima Hossain, Ahmed Gaber, Yusuf S. Althobaiti, Abdelhadi A. Abdelhadi, and et al. 2021. "High-Throughput Root Network System Analysis for Low Phosphorus Tolerance in Maize at Seedling Stage" Agronomy 11, no. 11: 2230. https://doi.org/10.3390/agronomy11112230
APA StyleUddin, M. S., Azam, M. G., Billah, M., Bagum, S. A., Biswas, P. L., Khaldun, A. B. M., Hossain, N., Gaber, A., Althobaiti, Y. S., Abdelhadi, A. A., & Hossain, A. (2021). High-Throughput Root Network System Analysis for Low Phosphorus Tolerance in Maize at Seedling Stage. Agronomy, 11(11), 2230. https://doi.org/10.3390/agronomy11112230










