Soluble Starch Synthase Enzymes in Cereals: An Updated Review
Abstract
1. Introduction
2. Mode of Action and Properties of Soluble SSs in Amylopectin Formation
2.1. SS Mutants Vital Roles in the Formation Amylopectin Chains: Starch Synthase SSI to SSIII
2.2. Initiation of Starch Granule Formation
2.3. Impact of Different Mutation Technologies on Soluble SSs Genes
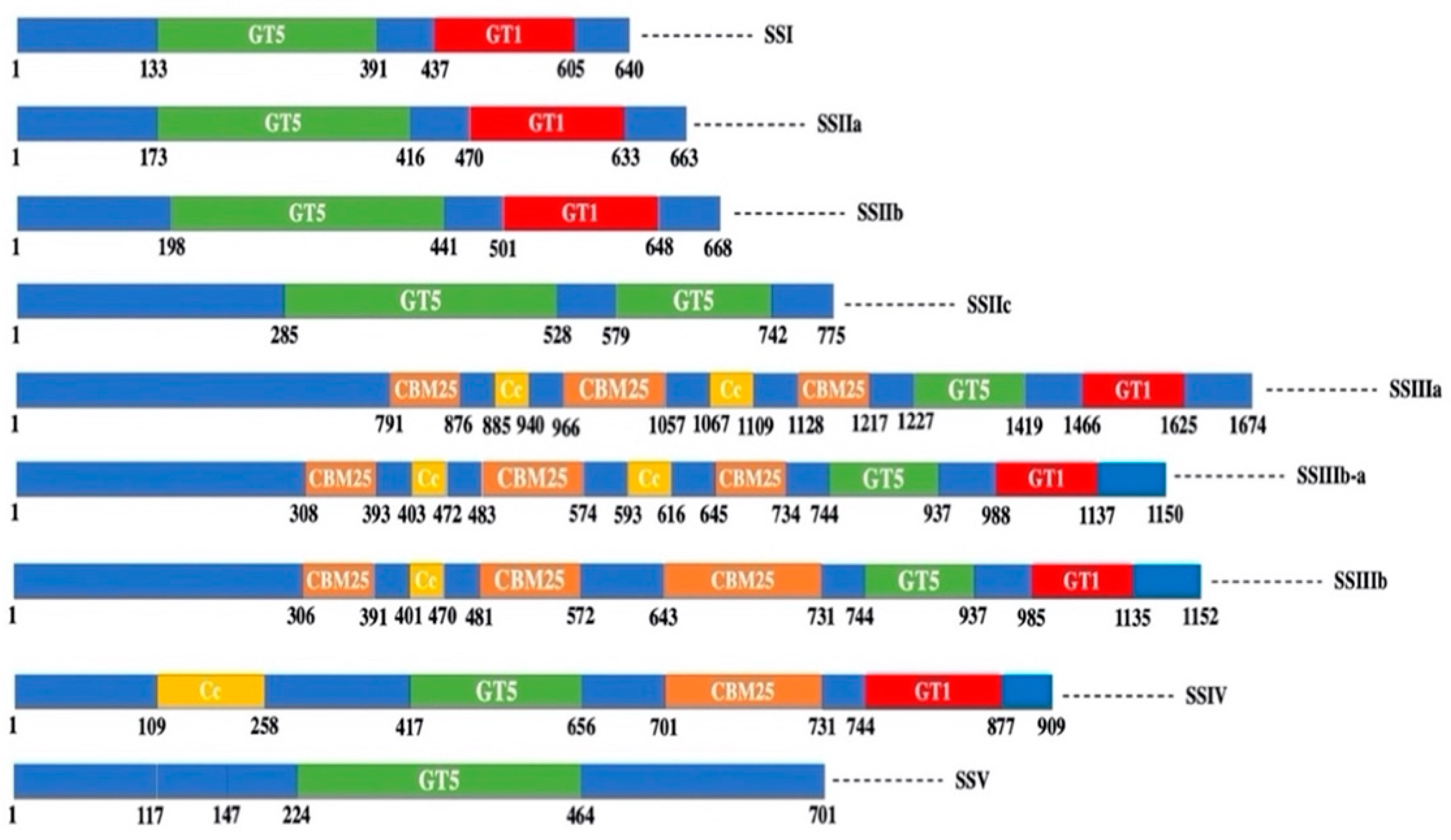
3. Structure-Function Relationships of SS in Cereals
4. Regulation of SS Activity in Cereals
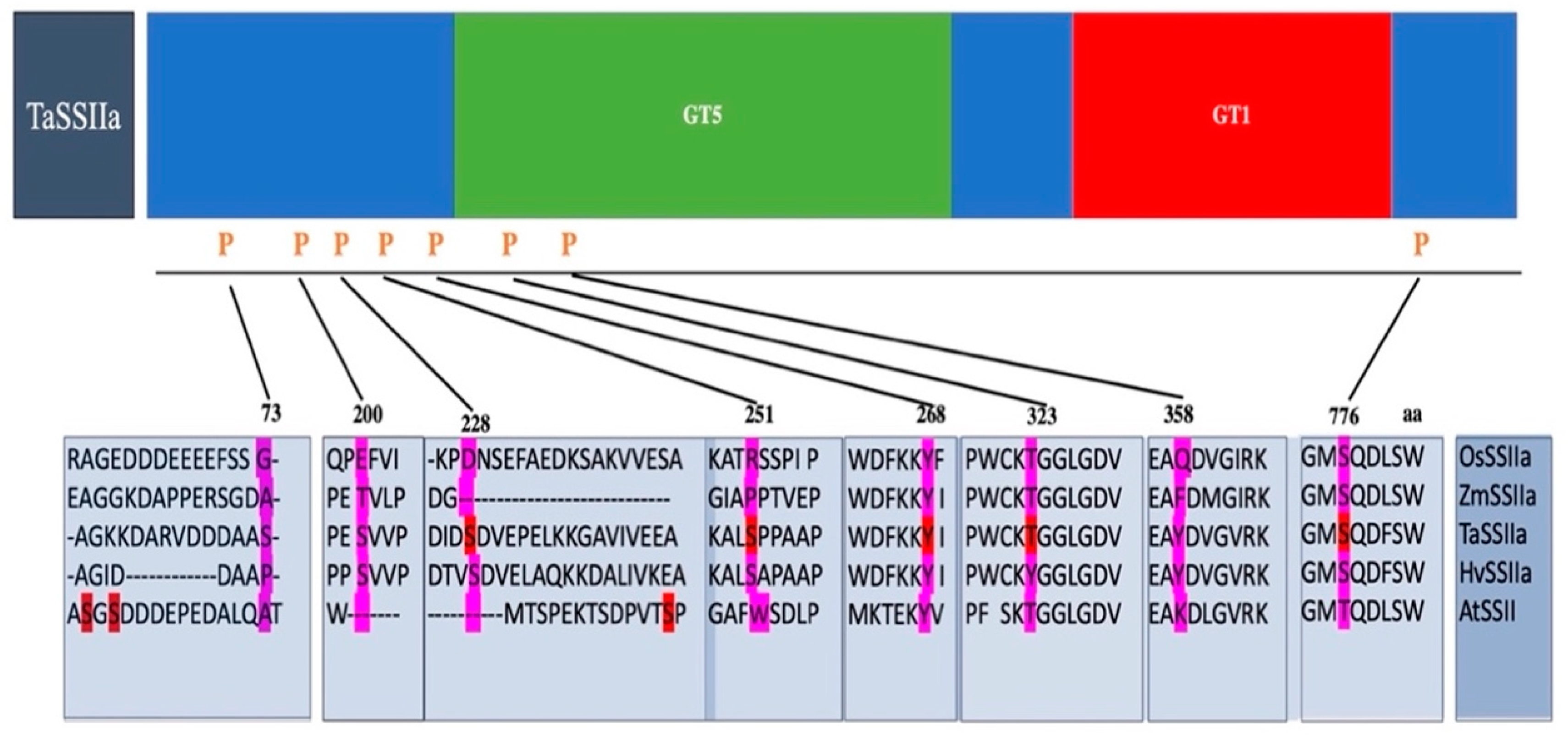
4.1. Regulation of Protein Phosphorylation
4.2. SSs form Heteromeric Protein Complexes in Amyloplasts
5. Conclusions and Future Aspects
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Polesi, L.F.; da Matta Junior, M.D.; Sarmento, S.B.S.; Canniatti-Brazaca, S.G. Starch digestibility and physicochemical and cooking properties of irradiated rice grains. Rice Sci. 2017, 24, 48–55. [Google Scholar] [CrossRef]
- Fettke, J.; Hejazi, M.; Smirnova, J.; Höchel, E.; Stage, M.; Steup, M. Eukaryotic starch degradation: Integration of plastidial and cytosolic pathways. J. Exp. Bot. 2009, 60, 2907–2922. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Yoon, M.-R.; Chun, A.; Tai, T.H. Identification of novel mutations in the rice starch branching enzyme I gene via TILLING by sequencing. Euphytica 2018, 214, 94. [Google Scholar] [CrossRef]
- Aoki, N.; Whitfeld, P.; Hoeren, F.; Scofield, G.; Newell, K.; Patrick, J. Three sucrose transporter genes are expressed in the developing grain of hexaploid wheat. Plant Mol. Biol. 2002, 50, 453–462. [Google Scholar] [CrossRef]
- Kittisuban, P.; Lee, B.H.; Suphantharika, M.; Hamaker, B.R. Slow glucose release property of enzyme-synthesized highly branched maltodextrins differs among starch sources. Carbohydr. Polym. 2014, 107, 182–191. [Google Scholar] [CrossRef]
- Pfister, B.; Zeeman, S.C. Formation of starch in plant cells. Mol. Life Sci. 2016, 73, 2781–2807. [Google Scholar] [CrossRef]
- Cenci, U.; Nitschke, F.; Steup, M.; Minassian, B.A.; Colleoni, C.; Ball, S.G. Transition from glycogen to starch metabolism in Archaeplastida. Trends Plant Sci. 2014, 19, 18–28. [Google Scholar] [CrossRef]
- Coutinho, P.M.; Deleury, E.; Davies, G.J.; Henrissat, B. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 2003, 328, 307–317. [Google Scholar] [CrossRef]
- Nougué, O.; Corbi, J.; Ball, S.G.; Manicacci, D.; Tenaillon, M.I. Molecular evolution accompanying functional divergence of duplicated genes along the plant starch biosynthesis pathway. BMC Evol. Biol. 2014, 14, 103. [Google Scholar] [CrossRef] [PubMed]
- Abt, M.; Pfister, B.; Sharma, M.; Eicke, S.; Bürgy, L.; Neale, I.; Seung, D.; Zeeman, S.C. Starch Synthase 5, a Noncanonical Starch Synthase-Like Protein, Promotes Starch Granule Initiation in Arabidopsis. Plant Cell. 2020. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F. Interactions between starch and phenolic compound. Trends Food Sci. Technol. 2015, 43, 129–143. [Google Scholar] [CrossRef]
- Gous, P.W.; Fox, G.P. Amylopectin synthesis and hydrolysis—Understanding isoamylase and limit dextrinase and their impact on starch structure on barley (Hordeum vulgare) quality. J. Trends Food Sci. 2017, 62, 23–32. [Google Scholar] [CrossRef]
- Pandey, M.K.; Rani, N.S.; Madhav, M.S.; Sundaram, R.; Varaprasad, G.; Sivaranjani, A.; Bohra, A.; Kumar, G.R.; Kumar, A. Different isoforms of starch-synthesizing enzymes controlling amylose and amylopectin content in rice (Oryza sativa L.). Biotechnol. Adv. 2012, 30, 1697–1706. [Google Scholar] [CrossRef]
- Pan, X.; Yan, H.; Li, M.; Wu, G.; Jiang, H. Evolution of the genes encoding starch synthase in Sorghum and common wheat. Mol. Plant Breed. 2011, 2, 60–67. [Google Scholar] [CrossRef]
- Wang, Z.; Li, W.; Qi, J.; Shi, P.; Yin, Y. Starch accumulation, activities of key enzyme and gene expression in starch synthesis of wheat endosperm with different starch contents. J. Food Sci. Technol. 2014, 51, 419–429. [Google Scholar] [CrossRef]
- Qu, J.; Ma, C.; Feng, J.; Xu, S.; Wang, L.; Li, F.; Li, Y.; Zhang, R.; Zhang, X.; Xue, J. Transcriptome dynamics during maize endosperm development. PLoS ONE 2016, 3, e0163814. [Google Scholar] [CrossRef]
- Crofts, N.; Abe, N.; Oitome, N.F.; Matsushima, R.; Hayashi, M.; Tetlow, I.J. Amylopectin biosynthetic enzymes from developing rice seed form enzymatically active protein complexes. J. Exp. Bot. 2015, 66, 4469–4482. [Google Scholar] [CrossRef] [PubMed]
- Edwards, A.; Fulton, D.C.; Hylton, C.M.; Jobling, S.A.; Gidley, M.; Rössner, U. A combined reduction in activity of starch synthases II and III of potato has novel effects on the starch of tubers. Plant J. 1999, 17, 251–261. [Google Scholar] [CrossRef]
- Cuesta-Seijo, J.A.; Nielsen, M.M.; Ruzanski, C.; Krucewicz, K.; Beeren, S.R.; Rydhal, M.G. In vitro biochemical characterization of all barley endosperm starch synthases. Front. Plant Sci. 2016, 6, 1265. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Keeling, P.L.; Hennen-Bierwagen, T.A.; Myers, A.M. Comparative in vitro analyses of recombinant maize starch synthases SSI, SSIIa, and SSIII reveal direct regulatory interactions and thermosensitivity. Arch. Biochem. Biophys. 2016, 596, 63–72. [Google Scholar] [CrossRef]
- Lin, Q.; Qian, S.; Li, C.; Pan, H.; Wu, Z.; Liu, G. Synthesis, flocculation and adsorption performance of amphoteric starch. Carbohydr. Polym. 2012, 90, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Tetlow, I.J.; Emes, M.J. Starch biosynthesis in the developing endosperms of grasses and cereals. Agronomy 2017, 7, 81. [Google Scholar] [CrossRef]
- Šárka, E.; Dvořáček, V. New processing and applications of waxy starch (a review). J. Food Eng. 2017, 206, 77–87. [Google Scholar] [CrossRef]
- Hanashiro, I.; Higuchi, T.; Aihara, S.; Nakamura, Y.; Fujita, N. Structures of starches from rice mutants deficient in the starch synthase isozyme SSI or SSIIIa. J. Biomacromol. 2011, 12, 1621–1628. [Google Scholar] [CrossRef] [PubMed]
- Brust, H.; Lehmann, T.; D’Hulst, C.; Fettke, J. Analysis of the functional interaction of Arabidopsis starch synthase and branching enzyme isoforms reveals that the cooperative action of SSI and BEs results in glucans with polymodal chain length distribution similar to amylopectin. PLoS ONE 2014, 9, e102364. [Google Scholar]
- Fujita, N.; Yoshida, M.; Asakura, N.; Ohdan, T.; Miyao, A.; Hirochika, H.; Nakamura, Y. Function and characterization of starch synthase I using mutants in rice. Plant Physiol. 2006, 140, 1070–1084. [Google Scholar] [CrossRef] [PubMed]
- Wilkens, C.; Svensson, B.; Møller, M.S. Functional roles of starch binding domains and surface binding sites in enzymes involved in starch biosynthesis. Front. Plant Sci. 2018, 9, 1652. [Google Scholar] [CrossRef]
- Fujita, N.; Satoh, R.; Hayashi, A.; Kodama, M.; Itoh, R.; Aihara, S.; Nakamura, Y. Starch biosynthesis in rice endosperm requires the presence of either starch synthase I or IIIa. J. Exp. Bot. 2011, 62, 4819–4831. [Google Scholar] [CrossRef]
- Nakata, M.; Miyashita, T.; Kimura, R.; Nakata, Y.; Takagi, H.; Kuroda, M.; Yamaguchi, T.; Umemoto, T.; Yamakawa, H. MutMapPlus identified novel mutant alleles of a rice starch branching enzyme II b gene for fine-tuning of cooked rice texture. Plant Biotechnol. J. 2018, 16, 111–123. [Google Scholar] [CrossRef]
- Guzmán, C.; Alvarez, J.B. Wheat waxy proteins: Polymorphism, molecular characterization and effects on starch properties. Theor. Appl. Genet. 2016, 129, 1–16. [Google Scholar] [CrossRef]
- Sparla, F.; Falini, G.; Botticella, E.; Pirone, C.; Talamè, V.; Bovina, R.; Salvi, S.; Tuberosa, R.; Sestili, F.; Trost, P. New starch phenotypes produced by TILLING in barley. PLoS ONE 2014, 9, e107779. [Google Scholar]
- Niu, L.; Ding, H.; Zhang, J.; Wang, W. Proteomic analysis of starch biosynthesis in maize seeds. Starch Starke 2019, 71, 1800294. [Google Scholar] [CrossRef]
- Szydlowski, N.; Ragel, P.; Hennen-Bierwagen, T.A.; Planchot, V.; Myers, A.M.; Mérida, A.; d’Hulst, C.; Wattebled, F. Integrated functions among multiple starch synthases determine both amylopectin chain length and branch linkage location in Arabidopsis leaf starch. J. Exp. Bot. 2011, 62, 4547–4559. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Ahmed, R.; Kosar-Hashemi, B.; Larroque, O.; Butardo, V.M.; Tanner, G.J.; Colgrave, M.L.; Upadhyaya, N.M.; Tetlow, I.J.; Emes, M.J.; et al. The different effects of starch synthase IIa mutations or variation on endosperm amylose content of barley, wheat and rice are determined by the distribution of starch synthase I and starch branching enzyme IIb between the starch granule and amyloplast stroma. Theor. Appl. Genet. 2015, 128, 1407–1419. [Google Scholar] [PubMed]
- Subasinghe, R.M.; Liu, F.; Polack, U.C.; Lee, E.A.; Emes, M.J.; Tetlow, I.J. Multimeric states of starch phosphorylase determine protein–protein interactions with starch biosynthetic enzymes in amyloplasts. Plant Physiol. Biochem. 2014, 83, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Cheng, Z.; Zhang, X.; Guo, X.; Su, N.; Jiang, L.; Mao, L.; Wan, J. Double repression of soluble starch synthase genes SSIIa and SSIIIa in rice (Oryza sativa L.) uncovers interactive effects on the physicochemical properties of starch. Genome 2011, 54, 448–459. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Gao, S.; Sun, Q.; Tang, Y.; Han, Y.; Zhang, J.; Li, Z. Induced splice site mutation generates alternative intron splicing in starch synthase II (SSII) gene in rice. Biotechnol. Equip. 2017, 31, 1093–1099. [Google Scholar] [CrossRef]
- Zhu, F.; Bertoft, E.; Seetharaman, K. Distribution of branches in whole starches from maize mutants deficient in starch synthase III. J. Agric. Food Chem. 2014, 62, 4577–4583. [Google Scholar] [CrossRef]
- Takahashi, T.; Fujita, N. Thermal and rheological characteristics of mutant rice starches with widespread variation of amylose content and amylopectin structure. Food Hydrocoll. 2017, 62, 83–93. [Google Scholar] [CrossRef]
- Lin, Q.; Huang, B.; Zhang, M.; Zhang, X.; Rivenbark, J.; Lappe, R.L.; James, M.G.; Myers, A.M.; Hennen-Bierwagen, T.A. Functional interactions between starch synthase III and isoamylase-type starch-debranching enzyme in maize endosperm. Plant Physiol. 2012, 158, 679–692. [Google Scholar] [CrossRef]
- Santelia, D.; Zeeman, S.C. Progress in Arabidopsis starch research and potential biotechnological applications. Curr. Opin. Biotechnol. 2011, 22, 271–280. [Google Scholar] [CrossRef]
- Zhang, X.; Szydlowski, N.; Delvallé, D.; d’Hulst, C.; James, M.G.; Myers, A.M. Overlapping functions of the starch synthases SSII and SSIII in amylopectin biosynthesis in Arabidopsis. BMC Plant Biol. 2008, 8, 96. [Google Scholar] [CrossRef]
- Hayashi, M.; Kodama, M.; Nakamura, Y.; Fujita, N. Thermal and pasting properties, morphology of starch granules, and crystallinity of endosperm starch in the rice SSI and SSIIIa double-mutant. J. Appl. Glycosci. 2015, 62, 81–86. [Google Scholar] [CrossRef]
- Yamamori, M.; Fujita, S.; Hayakawa, K.; Matsuki, J.; Yasui, T. Genetic elimination of a starch granule protein, SGP-1, of wheat generates an altered starch with apparent high amylose. Theor. Appl. Genet. 2000, 101, 21–29. [Google Scholar] [CrossRef]
- Kosar-Hashemi, B.; Li, Z.; Larroque, O.; Regina, A.; Yamamori, M.; Morell, M.K.; Rahman, S. Multiple effects of the starch synthase II mutation in developing wheat endosperm. Funct. Plant Biol. 2007, 34, 431–438. [Google Scholar] [CrossRef]
- Vrinten, P.L.; Shimbata, T.; Yanase, M.; Sunohara, A.; Saito, M.; Inokuma, T.; Takiya, T.; Takaha, T.; Nakamura, T. Properties of a novel type of starch found in the double mutant “sweet wheat”. Carbohydr. Polym. 2012, 89, 1250–1260. [Google Scholar] [CrossRef] [PubMed]
- Botticella, E.; Sestili, F.; Ferrazzano, G.; Mantovani, P.; Cammerata, A.; D’Egidio, M.G.; Lafiandra, D. The impact of the SSIIa null mutations on grain traits and composition in durum wheat. Breed. Sci. 2016, 16, 4. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.M.; Hogg, A.C.; Hofer, P.; Manthey, F.A.; Giroux, M.J. Impacts of SSIIa-A null allele on durum wheat noodle quality. Cereal Chem. 2014, 91, 176–182. [Google Scholar] [CrossRef]
- Shimbata, T.; Ai, Y.; Fujita, M.; Inokuma, T.; Vrinten, P.; Sunohara, A.; Saito, M.; Takiya, T.; Jane, J.L.; Nakamura, T. Effects of homoeologous wheat starch synthase IIa genes on starch properties. J. Agric. Food Chem. 2012, 60, 12004–12010. [Google Scholar] [CrossRef]
- Konik-Rose, C.; Thistleton, J.; Chanvrier, H.; Tan, I.; Halley, P.; Gidley, M.; Kosar-Hashemi, B.; Wang, H.; Larroque, O.; Ikea, J.; et al. Effects of starch synthase IIa gene dosage on grain, protein and starch in endosperm of wheat. Theor. Appl. Genet. 2007, 115, 1053. [Google Scholar] [CrossRef]
- Guo, H.; Liu, Y.; Li, X.; Yan, Z.; Xie, Y.; Xiong, H.; Zhao, L.; Gu, J.; Zhao, S.; Liu, L. Novel mutant alleles of the starch synthesis gene TaSSIVb-D result in the reduction of starch granule number per chloroplast in wheat. BMC Genom. 2017, 18, 358. [Google Scholar] [CrossRef]
- Zhang, S.; Guo, H.; Irshad, A.; Xie, Y.; Zhao, L.; Xiong, H.; Gu, J.; Zhao, S.; Ding, Y.; Liu, L. The synergistic effects of TaAGP. L-B1 and TaSSIVb-D mutations in wheat lead to alterations of gene expression patterns and starch content in grain development. PLoS ONE 2019, 14, e0223783. [Google Scholar]
- Fujita, N.; Hanashiro, I.; Toyosawa, Y.; Nakamura, Y. Functional study of rice starch synthase I (SSI) by using double mutant with lowered activities of SSI and isoamylase1. J. Appl. Glycosci. 2013, 60, 45–51. [Google Scholar] [CrossRef]
- Abe, N.; Asai, H.; Yago, H.; Oitome, N.F.; Itoh, R.; Crofts, N.; Nakamura, Y.; Fujita, N. Relationships between starch synthase I and branching enzyme isozymes determined using double mutant rice lines. BMC Plant Biol. 2014, 14, 80. [Google Scholar] [CrossRef] [PubMed]
- Crofts, N.; Iizuka, Y.; Abe, N.; Miura, S.; Kikuchi, K.; Matsushima, R.; Fujita, N. Rice mutants lacking starch synthase I or branching enzyme IIb activity altered starch biosynthetic protein complexes. Front. Plant Sci. 2018, 9, 1817. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.; Crofts, N.; Oitome, N.F.; Fujita, N. Analyses of starch biosynthetic protein complexes and starch properties from developing mutant rice seeds with minimal starch synthase activities. BMC Plant Biol. 2018, 18, 59. [Google Scholar] [CrossRef] [PubMed]
- Miura, S.; Crofts, N.; Saito, Y.; Hosaka, Y.; Oitome, N.F.; Watanabe, T.; Kumamaru, T.; Fujita, N. Starch synthase IIa-deficient mutant rice line produces endosperm starch with lower gelatinization temperature than japonica rice cultivars. Front. Plant Sci. 2018, 9, 645. [Google Scholar] [CrossRef]
- Fujita, N.; Yoshida, M.; Kondo, T.; Saito, K.; Utsumi, Y.; Tokunaga, T.; Nishi, A.; Satoh, H.; Park, J.-H. Characterization of SSIIIa-deficient mutants of rice: The function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiol. 2007, 144, 2009–2023. [Google Scholar] [CrossRef]
- Xu, L.; You, H.; Zhang, O.; Xiang, X. Genetic Effects of Soluble Starch Synthase IV-2 and It with ADPglucose Pyrophorylase Large Unit and Pullulanase on Rice Qualities. Rice 2020, 13, 46. [Google Scholar] [CrossRef]
- Ryoo, N.; Yu, C.; Park, C.-S.; Baik, M.-Y.; Park, I.M.; Cho, M.-H.; Bhoo, S.H.; An, G.; Hahn, T.-R.; Jeon, J.-S. Knockout of a starch synthase gene OsSSIIIa/Flo5 causes white-core floury endosperm in rice (Oryza sativa L.). Plant Cell Rep. 2007, 26, 1083–1095. [Google Scholar] [CrossRef]
- Toyosawa, Y.; Kawagoe, Y.; Matsushima, R.; Crofts, N.; Ogawa, M.; Fukuda, M.; Kumamaru, T.; Okazaki, Y.; Kusano, M.; Saito, K. Deficiency of starch synthase IIIa and IVb alters starch granule morphology from polyhedral to spherical in rice endosperm. Plant Physiol. 2016, 170, 1255–1270. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.-J.; Nogoy, F.M.; Lee, S.-K.; Cho, Y.-G.; Kang, K.-K. Application of ZFN for site directed mutagenesis of rice SSIVa gene. Biotechnol. Bioproc. Eng. 2018, 23, 108–115. [Google Scholar] [CrossRef]
- Zhu, F.; Bertoft, E.; Källman, A.; Myers, A.M.; Seetharaman, K. Molecular structure of starches from maize mutants deficient in starch synthase III. J. Agric. Food Chem. 2013, 61, 9899–9907. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Ahmed, Z.; Lee, E.A.; Donner, E.; Liu, Q.; Ahmed, R.; Morell, M.K.; Emes, M.J.; Tetlow, I.J. Allelic variants of the amylose extender mutation of maize demonstrate phenotypic variation in starch structure resulting from modified protein–protein interactions. J. Exp.Bot. 2012, 63, 1167–1183. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Bertoft, E.; Li, G. Morphological, thermal, and rheological properties of starches from maize mutants deficient in starch synthase III. J. Agric. Food Chem. 2016, 64, 6539–6545. [Google Scholar] [CrossRef]
- Morell, M.K.; Kosar-Hashemi, B.; Cmiel, M.; Samuel, M.S.; Chandler, P.; Rahman, S.; Buleon, A.; Batey, I.L.; Li, Z. Barley sex6 mutants lack starch synthase IIa activity and contain a starch with novel properties. Plant J. 2003, 34, 173–185. [Google Scholar] [CrossRef]
- Howard, T.P.; Fahy, B.; Leigh, F.; Howell, P.; Powell, W.; Greenland, A.; Trafford, K.; Smith, A.M. Use of advanced recombinant lines to study the impact and potential of mutations affecting starch synthesis in barley. J. Cereal Sci. 2014, 59, 196–202. [Google Scholar] [CrossRef]
- Ezquer, I.; Li, J.; Ovecka, M.; Baroja-Fernández, E.; Muñoz, F.J.; Montero, M.; Díaz de Cerio, J.; Hidalgo, M.; Sesma, M.T.; Bahaji, A. Microbial volatile emissions promote accumulation of exceptionally high levels of starch in leaves in mono-and dicotyledonous plants. Plant Cell Physiol. 2010, 51, 1674–1693. [Google Scholar] [CrossRef]
- Irshad, A.; Guo, H.; Zhang, S.; Gu, J.; Zhao, L.; Xie, Y.; Xiong, H.; Zhao, S.; Ding, Y.; Ma, Y. EcoTILLING reveals natural allelic variations in starch synthesis key gene TaSSIV and Its haplotypes associated with higher thousand grain weight. Genes 2019, 10, 307. [Google Scholar] [CrossRef]
- Gámez-Arjona, F.M.; Raynaud, S.; Ragel, P.; Mérida, Á. Starch synthase 4 is located in the thylakoid membrane and interacts with plastoglobule-associated proteins in Arabidopsis. Plant J. 2014, 80, 305–316. [Google Scholar] [CrossRef]
- Schwarte, S.; Brust, H.; Steup, M.; Tiedemann, R. Intraspecific sequence variation and differential expression in starch synthase genes of Arabidopsis thaliana. BMC Res. Notes 2013, 6, 84. [Google Scholar] [CrossRef] [PubMed]
- Crumpton-Taylor, M.; Pike, M.; Lu, K.J.; Hylton, C.M.; Feil, R.; Eicke, S.; Lunn, J.E.; Zeeman, S.C.; Smith, A.M. Starch synthase 4 is essential for coordination of starch granule formation with chloroplast division during Arabidopsis leaf expansion. New Phytol. 2013, 200, 1064–1075. [Google Scholar] [CrossRef] [PubMed]
- Borel, C.; Deutsch, S.; Letourneau, A.; Migliavacca, E.; Montgomery, S.B.; Dimas, A.S.; Vejnar, C.E.; Attar, H.; Gagnebin, M.; Gehrig, C. Identification of cis-and trans-regulatory variation modulating microRNA expression levels in human fibroblasts. Genome Res. 2011, 21, 68–73. [Google Scholar] [CrossRef]
- Dian, W.; Jiang, H.; Wu, P. Evolution and expression analysis of starch synthase III and IV in rice. J. Exp. Bot. 2005, 56, 623–632. [Google Scholar] [CrossRef]
- Hawkins, E.; Chen, J.; Watson-Lazowski, A.; Ahn-Jarvis, J.; Barclay, J.E.; Fahy, B. STARCH SYNTHASE 4 is required for normal starch granule initiation in amyloplasts of wheat endosperm. New Phytol. 2021, 230–236. [Google Scholar]
- Leterrier, M.; Holappa, L.D.; Broglie, K.E.; Beckles, D.M. Cloning, characterisation and comparative analysis of a starch synthase IV gene in wheat: Functional and evolutionary implications. BMC Plant Biol. 2008, 8, 98. [Google Scholar] [CrossRef]
- Irshad, A.; Guo, H.; Zhang, S.; Liu, L. TILLING in cereal crops for allele expansion and mutation detection by using modern sequencing technologies. Agronomy 2020, 10, 405. [Google Scholar] [CrossRef]
- Gámez-Arjona, F.M.; Li, J.; Raynaud, S.; Baroja-Fernández, E.; Muñoz, F.J.; Ovecka, M.; Ragel, P.; Bahaji, A.; Pozueta-Romero, J.; Mérida, Á. Enhancing the expression of starch synthase class IV results in increased levels of both transitory and long-term storage starch. Plant Biotechnol. J. 2011, 9, 1049–1060. [Google Scholar] [CrossRef]
- Liu, H.; Yu, G.; Wei, B.; Wang, Y.; Zhang, J.; Hu, Y.; Liu, Y.; Yu, G.; Zhang, H.; Huang, Y. Identification and phylogenetic analysis of a novel starch synthase in maize. Front. Plant Sci. 2015, 6, 1013. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Jee, M.G.; Kim, J.; Nogoy, F.M.; Yu, D.A.; Kim, M.S.; Sun, M.; Kang, K.K.; Nou, I.; Cho, Y.G. Modification of starch composition using RNAi targeting soluble starch synthase I in Japonica rice. Plant Breed. Biotechnol. 2014, 2, 301–312. [Google Scholar] [CrossRef][Green Version]
- Yaling, C.; Yuehan, P.; Jinsong, B. Expression Profiles and Protein Complexes of Starch Biosynthetic Enzymes from White-Core and Waxy Mutants Induced from High Amylose Indica Rice. Rice Sci. 2020, 27, 152–161. [Google Scholar] [CrossRef]
- Qu, J.; Xu, S.; Zhang, Z.; Chen, G.; Zhong, Y.; Liu, L.; Zhang, R.; Xue, J.; Guo, D. Evolutionary, structural and expression analysis of core genes involved in starch synthesis. Sci. Rep. 2018, 8, 1–16. [Google Scholar]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Meekins, D.A.; Vander Kooi, C.W.; Gentry, M.S. Structural mechanisms of plant glucan phosphatases in starch metabolism. FEBS J. 2016, 283, 2427–2447. [Google Scholar] [CrossRef]
- Mishra, B.P.; Kumar, R.; Mohan, A.; Gill, K.S. Conservation and divergence of Starch Synthase III genes of monocots and dicots. PLoS ONE 2017, 12, e0189303. [Google Scholar] [CrossRef]
- Momma, M.; Fujimoto, Z. Interdomain disulfide bridge in the rice granule bound starch synthase I catalytic domain as elucidated by X-ray structure analysis. Biosci. Biotechnol. Biochem. 2012, 76, 1591–1595. [Google Scholar] [CrossRef]
- Xiao, Y.; Thatcher, S.; Wang, M.; Wang, T.; Beatty, M.; Zastrow-Hayes, G.; Li, L.; Li, J.; Li, B.; Yang, X. Transcriptome analysis of near-isogenic lines provides molecular insights into starch biosynthesis in maize kernel. J. Integr. Plant Biol. 2016, 58, 713–723. [Google Scholar] [CrossRef]
- Jiang, H.; Lio, J.; Blanco, M.; Campbell, M.; Jane, J.-l. Resistant-starch formation in high-amylose maize starch during kernel development. J. Agric. Food Chem. 2010, 58, 8043–8047. [Google Scholar] [CrossRef]
- Seung, D.; Lu, K.-J.; Stettler, M.; Streb, S.; Zeeman, S.C. Degradation of glucan primers in the absence of starch synthase 4 disrupts starch granule initiation in Arabidopsis. J. Biol. Chem. 2016, 291, 20718–20728. [Google Scholar] [CrossRef]
- Raynaud, S.; Ragel, P.; Rojas, T.; Mérida, Á. The N-terminal part of Arabidopsis thaliana starch synthase 4 determines the localization and activity of the enzyme. J. Biol. Chem. 2016, 291, 10759–10771. [Google Scholar] [CrossRef]
- Lu, K.-J.; Pfister, B.; Jenny, C.; Eicke, S.; Zeeman, S.C. Distinct functions of STARCH SYNTHASE 4 domains in starch granule formation. Plant Physiol. 2018, 176, 566–581. [Google Scholar] [CrossRef] [PubMed]
- You, Y.; Zhang, M.; Yang, W.; Li, C.; Liu, Y.; Li, C. Starch phosphorylation and the in vivo regulation of starch metabolism and characteristics. Int. J. Biol. Macromol. 2020, 159, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, Z.; Tetlow, I.J.; Ahmed, R.; Morell, M.K.; Emes, M.J. Protein–protein interactions among enzymes of starch biosynthesis in high-amylose barley genotypes reveal differential roles of heteromeric enzyme complexes in the synthesis of A and B granules. Plant Sci. 2015, 233, 95–106. [Google Scholar] [CrossRef]
- Nakamura, Y. Rice starch biotechnology: Rice endosperm as a model of cereal endosperms. Starch Starke 2018, 70, 1600375. [Google Scholar] [CrossRef]
- Blennow, A.; Jensen, S.L.; Shaik, S.S.; Skryhan, K.; Carciofi, M.; Holm, P.B.; Hebelstrup, K.H.; Tanackovic, V. Future cereal starch bioengineering: Cereal ancestors encounter gene technology and designer enzymes. Cereal Chem. 2013, 90, 274–287. [Google Scholar] [CrossRef]
- Xia, J.; Zhu, D.; Wang, R.; Cui, Y.; Yan, Y. Crop resistant starch and genetic improvement: A review of recent advances. BMC Plant Biol. 2018, 131, 2495–2511. [Google Scholar] [CrossRef]
- Carpenter, M.A.; Joyce, N.I.; Genet, R.A.; Cooper, R.D.; Murray, S.R.; Noble, A.D.; Butler, R.C.; Timmerman-Vaughan, G.M. Starch phosphorylation in potato tubers is influenced by allelic variation in the genes encoding glucan water dikinase, starch branching enzymes I and II, and starch synthase III. Front. Plant Sci. 2015, 6, 143. [Google Scholar] [CrossRef]
- Song, Y.; Luo, G.; Shen, L.; Yu, K.; Yang, W.; Li, X. TubZIP28, a novel bZIP family transcription factor from Triticum urartu, and TabZIP28, its homologue from Triticum aestivum, enhance starch synthesis in wheat. New Phytol. 2020, 226, 1384–1398. [Google Scholar] [CrossRef]
- Waterschoot, J.; Gomand, S.V.; Fierens, E.; Delcour, J.A. Production, structure, physicochemical and functional properties of maize, cassava, wheat, potato and rice starches. Starch Starke 2015, 67, 14–29. [Google Scholar] [CrossRef]
- Crofts, N.; Abe, K.; Aihara, S.; Itoh, R.; Nakamura, Y.; Itoh, K.; Fujita, N.J.A.-B.M. Lack of starch synthase IIIa and high expression of granule-bound starch synthase I synergistically increase the apparent amylose content in rice endosperm. Plant Sci. 2012, 193, 62–69. [Google Scholar] [CrossRef] [PubMed]



| Species | No. of SS Genes | Gene Names with Accession No./ID | Reference |
|---|---|---|---|
| Hordeum vulgare | 6 | GBSSI (AAM560327.2), SSI (AAF37876), SSII (AAN28307), SSIIIa (AAF87999), SSIIIb (AAL40942), SSIV (AAK97773) | [12] |
| Oryza sativa | 11 | GBSSI (AB425323), GBSSII (AY069940), SSI (AY299404), SSIIa (AF419099), SSIIb (AF395537), SSIIc (AF383878), SSIIIa (AY100469), SSIIIb (AF432915), SSIVa (AY373257), SSIVb (AY373258), SSV (EU621837.1) | [13] |
| Sorghum bicolor | 10 | GBSSI (LOC8068390), GBSSII, SSI (NC054143), SSIIa (EU620718), SSIIb (EU620719), SSIIIa (EU620720), SSIIIb (EU620721), SSIV, SSV (HQ661801) | [14] |
| Triticum aestivum | 7 | GBSSI (AF286320), GBSSII (AF109395), SSI (AJ269503), SSII (AJ269503), SSIIIa (AF258608), SSIIIb (EU333946), SSIV (AY044844) | [15] |
| Zea mays | 10 | GBSSI (AY109531), GBSSII (EF471312), SSI (AF036891), SSIIa (AF019296), SSIIb (EF472249), SSIIc (EU284113), SSIIIa (AF023159), SSIIIb (EF472250), SSIV (EU599036), SSV (NM_001 130131.1) | [16] |
| Cereals | Amylose Content (%) | Inactivated Genes | Mutant Lines | Structural and Functional Changes in Mutant | Reference |
|---|---|---|---|---|---|
| Wheat | 22.9–32.3 | SSSII | sgp-1 | Alteration in amylopectin structure, high amylose contents | [44] |
| SSII | sgp-1, a7, a63 | Increase in short chains, decrease in starch branching enzyme | [45] | ||
| SSIIa | Increase in proportion of short chains, difference in gelatinization, retrogradation and pasting | [46] | |||
| SSIIa | svevo, semolina | Increased in dietary fiber of contents, change in total starch content, improved quality traits | [47] | ||
| SSIIa | ssIIa-Ab | Amylose contents increased 3%, cooked noodles firmness increased | [48] | ||
| SSIIa, GBSS | sw | Changes in seed size, starch granules and starch content, shrunken seed during maturity | [49] | ||
| SSIIa | abd null line | Grain properties (change in 1000 grain weight, grain size) and starch properties (fluctuation in amylose content, increased in resistant content) changed in null line | [50] | ||
| SSIV-D | e054-13, e1137 | Altered granule number/chloroplast | [51] | ||
| SSIV | e3-1-3, e1137 | Total starch and amylopectin content decreased | [52] | ||
| Rice | 15.4–25 | SSI | e7, i2-1, i2-2, i4 | Decrease in chains with DP 8 to 12, Increase in chains with DP 6 to 7 | [26] |
| SSI, SSIIIa | np | Higher amylose content, internal chain length of B2 and B3 fractions observed | [24] | ||
| SSI | ss1, isa1 | Take part in chain length distribution, outer chain elongation with little effect on branch position distribution | [53] | ||
| SSI, BEI | ss1/be1, ss1/be2b | Seed weight of mutant was higher than WT Number of short chains of amylopectin decrease, Amylose content almost same to WT | [54] | ||
| SSI | ssI, be2b | Subtle difference in protein profile, reduced association of SSI and BEIIb in ssI mutant | [55] | ||
| SSI, SSIIa, SSIIIa | ss1L/ss2aL/ss3a | Increase amylose, decrease grain weight, increase in level of ADP-glucose pyrophosphorylases | [56] | ||
| SSII | zhonghua-15 | GC-AG intron splicing offer more variants for genetic divergence in rice | [37] | ||
| SSIIa | ss2a(em204) | SSIIa protein was totally absent in seeds, higher amylose content, Number of short chains formation increased in amylopectin | [57] | ||
| SSIIIa | ss3a-1, ss3a-2 | Chains with DP 6 to 9 and DP 16 to 19 decreased, chains with DP 10 to 15 and DP 20 to 25 increased, amylose and amylopectin content increased | [58] | ||
| SSIV-2 | allelic variation | Affected gel consistency, percent of retrogradation, | [59] | ||
| SSIIIa | flo5-1, flo5-2 | Starch granules smaller and round as compared to WT, reduced contents of long chains | [60] | ||
| SSIIIa, SSIVb | ss3a, ss4b | Produced compound type starch granules in the early stages, glucan chain length distribution identified overlapping roles for SSIIIa and SSIVb in amylopectin chain synthesis | [61] | ||
| SSIVb | Transgenic plant contains premature codons, no mRNA expression, low starch contents, dwarf phenotype | [62] | |||
| Maize | 25–30 | SSIII | dull1 | Lager clusters of chain with more branched building blocks, average cluster contained 5.4 blocks in mutant and 4.2 blocks in WT. | [63] |
| SSIII, ISA2 | du1-R4059 | Starch deficient, accumulation of phytoglycogen | [21] | ||
| SSIIa | sugary-2 | Loss of activity of endosperm specific SS, impact on the SSI and SBEIIb | [64] | ||
| SSIII | w64a | Reduced granule size, decreased the enthalpy change of starch gelatinization | [65] | ||
| Barley | 29.9–31.6 | SSII | m292, m342 | Decrease in amylopectin synthesis, pleiotropic effect on other enzymes of starch biosynthesis | [66] |
| GBSS, ISA1, SSIIa | Sex6, wax, lys5fisa1 | SSIIa mutation caused low seed weight and starch content | [67] | ||
| SSI, SSIIa, GBSS | TILLING | SSI mutant increased A and B granules, SSIIa mutant caused shrunken seed | [31] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Irshad, A.; Guo, H.; Rehman, S.U.; Wang, X.; Wang, C.; Raza, A.; Zhou, C.; Li, Y.; Liu, L. Soluble Starch Synthase Enzymes in Cereals: An Updated Review. Agronomy 2021, 11, 1983. https://doi.org/10.3390/agronomy11101983
Irshad A, Guo H, Rehman SU, Wang X, Wang C, Raza A, Zhou C, Li Y, Liu L. Soluble Starch Synthase Enzymes in Cereals: An Updated Review. Agronomy. 2021; 11(10):1983. https://doi.org/10.3390/agronomy11101983
Chicago/Turabian StyleIrshad, Ahsan, Huijun Guo, Shoaib Ur Rehman, Xueqing Wang, Chaojie Wang, Ali Raza, Chunyun Zhou, Yuting Li, and Luxiang Liu. 2021. "Soluble Starch Synthase Enzymes in Cereals: An Updated Review" Agronomy 11, no. 10: 1983. https://doi.org/10.3390/agronomy11101983
APA StyleIrshad, A., Guo, H., Rehman, S. U., Wang, X., Wang, C., Raza, A., Zhou, C., Li, Y., & Liu, L. (2021). Soluble Starch Synthase Enzymes in Cereals: An Updated Review. Agronomy, 11(10), 1983. https://doi.org/10.3390/agronomy11101983








