Mapping Quantitative Trait Loci for Yield Potential Traits in Wheat Recombinant Inbred Lines
Abstract
1. Introduction
2. Materials and Methods
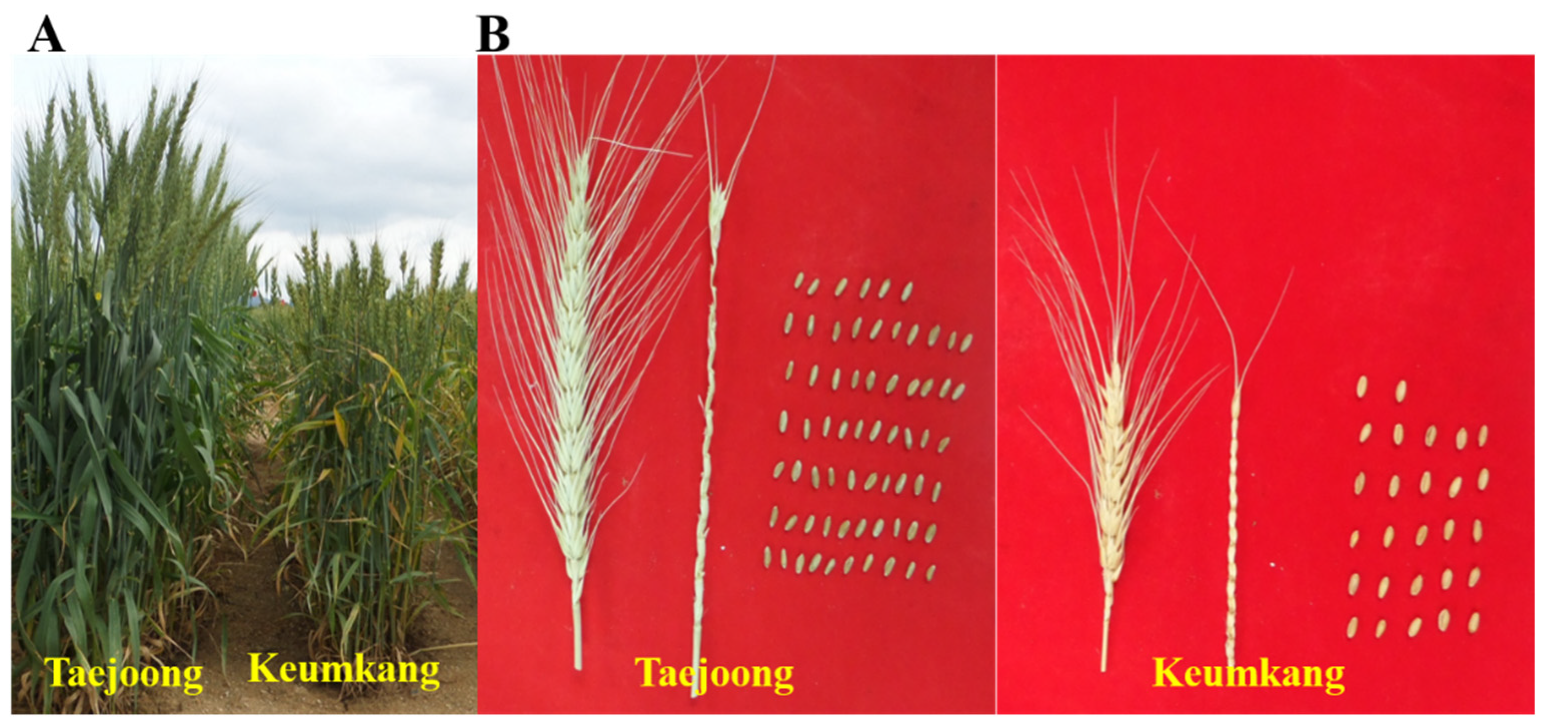
2.1. Plant Materials
2.2. Measurement of Yield Potential Traits
2.3. Genotypic Analysis
2.4. QTL Mapping and Statistical Analysis
3. Results
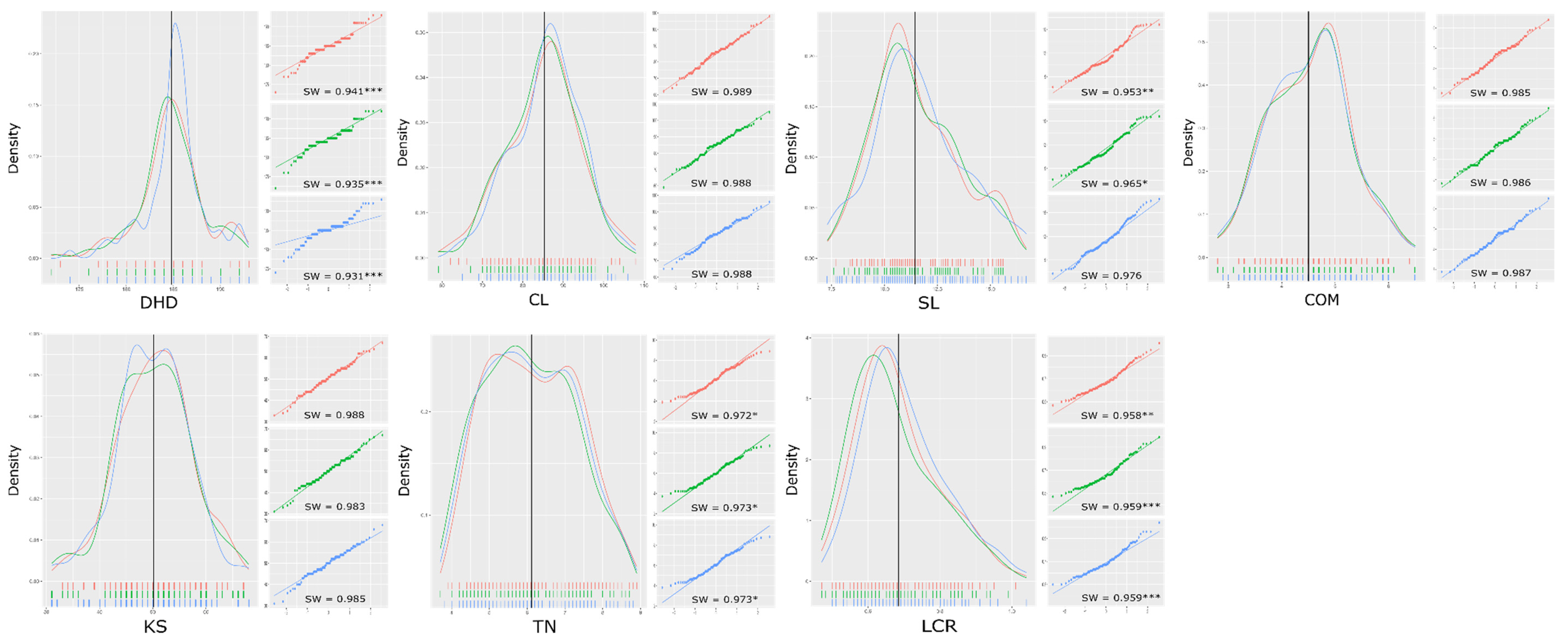
3.1. Phenotype Evaluation of the RIL Population
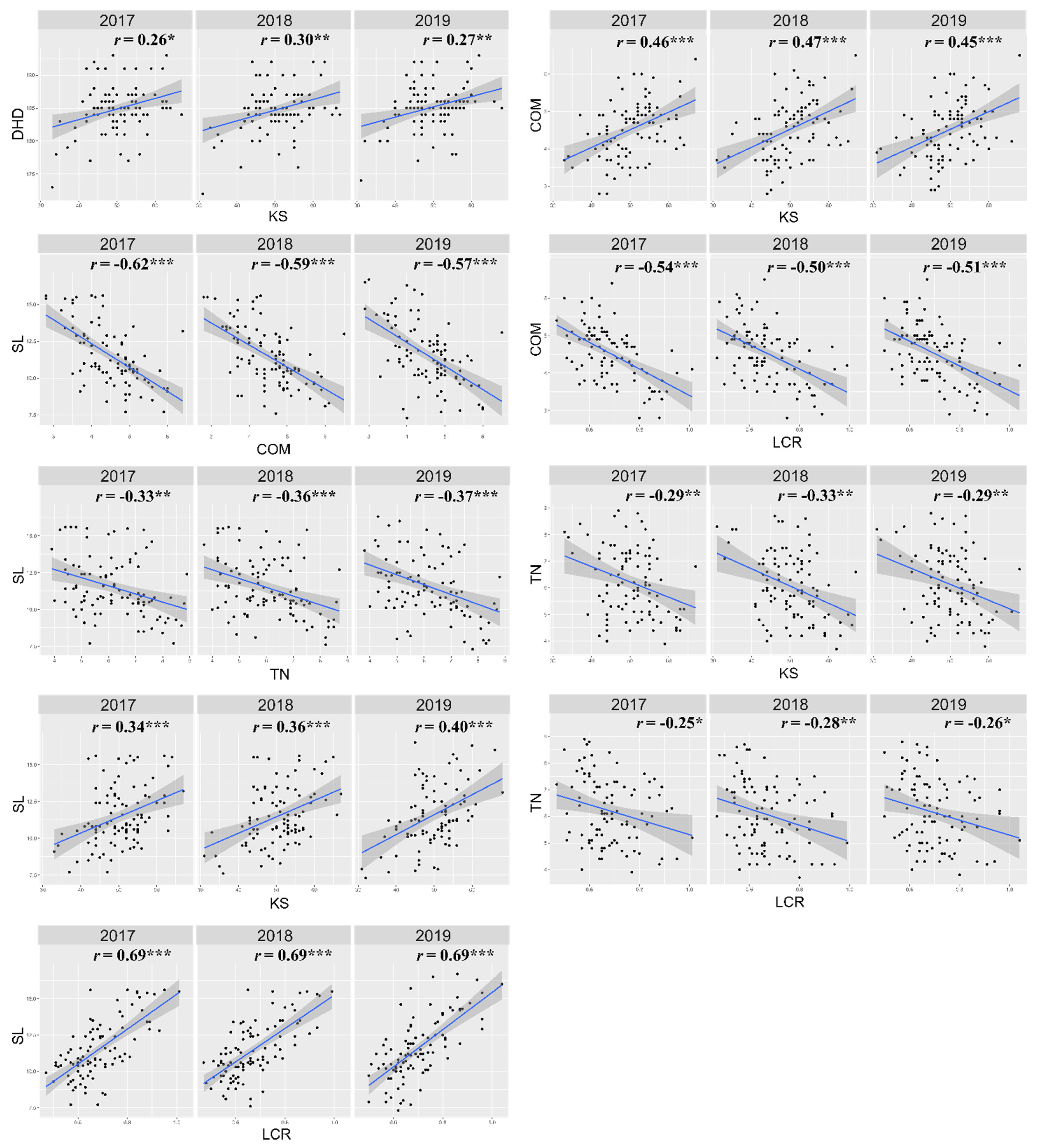
3.2. QTLs for Yield Potential Traits
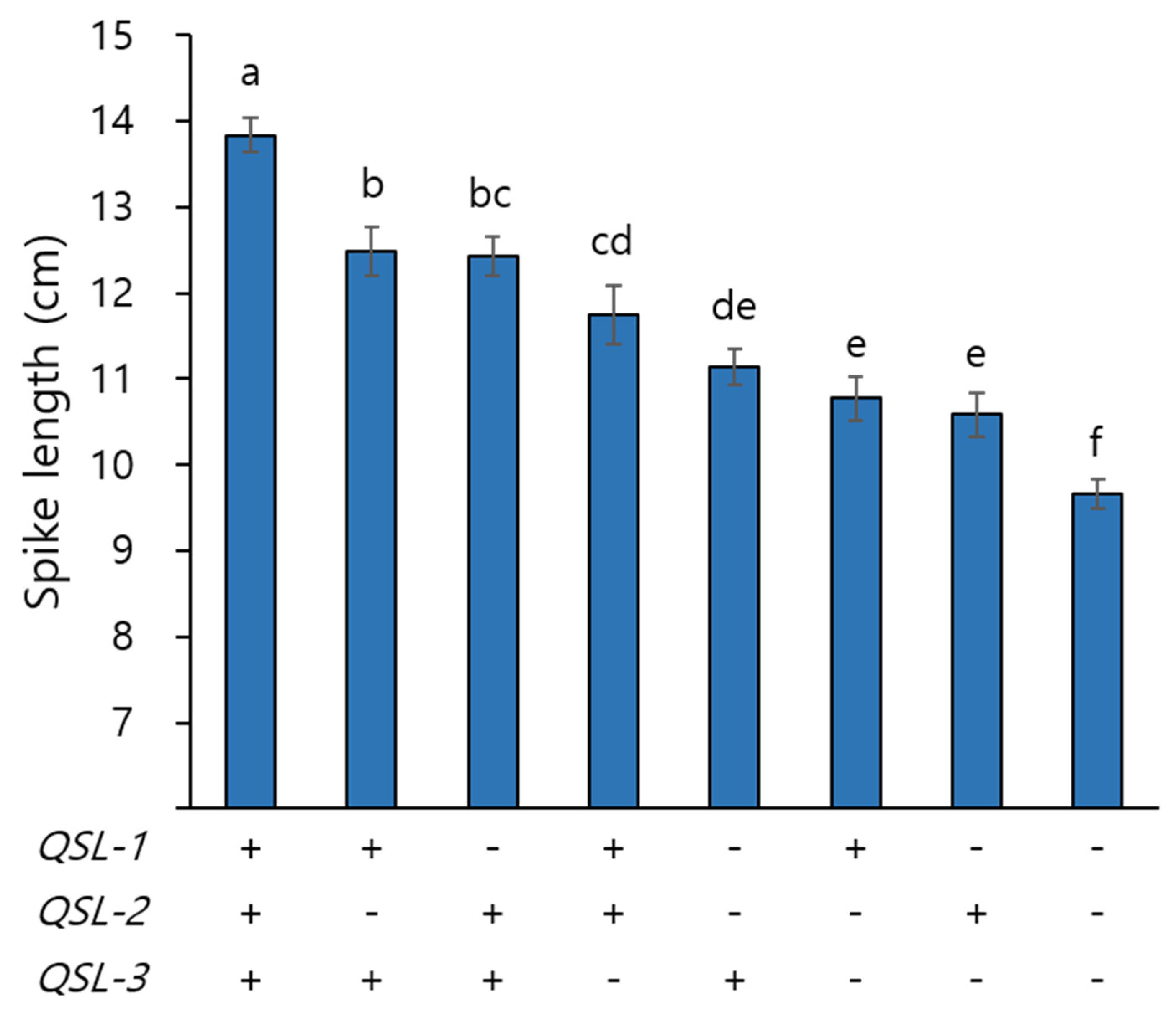
3.3. Effects of QSL-1, QSL-2, and QSL-3 on Spike Length and Other Yield Potential Traits
4. Discussion
4.1. Potential Use of the Spike Length QTLs in Breeding
4.2. Comparison of QSL-1, QSL-2, and QSL-3 with Previously Reported Spike Length QTLs
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Morgournov, A.; Sonder, K.; Abugalieva, A.; Bhadauria, V.; Cuthbert, R.D.; Shamanin, V.; Zeleskiy, Y.; DePauw, R.M. Effect of climate change on spring wheat yields in North America and Eurasia in 1981-2015 and implications for breeding. PLoS ONE 2018, 13, e0204932. [Google Scholar]
- Ray, D.K.; Mueller, N.D.; West, P.C.; Foley, J.A. Yield trends are insufficient to double global crop production by 2050. PLoS ONE 2013, 8, e66428. [Google Scholar] [CrossRef] [PubMed]
- Nadolska-Orczyk, A.; Rajchel, I.K.; Orczyk, W.; Gasparis, S. Major genes determining yield-related traits in wheat and barley. Theor. Appl. Genet. 2017, 130, 1081–1098. [Google Scholar] [CrossRef] [PubMed]
- Valluru, R.; Reynolds, M.P.; Salse, J. Genetic and molecular bases of yield-associated traits: A translational biology approach between rice and wheat. Theor. Appl. Genet. 2014, 127, 1463–1489. [Google Scholar] [CrossRef] [PubMed]
- Bennett, D.; Izanloo, A.; Reynolds, M.; Kuchel, H.; Langridge, P.; Schnurbusch, T. Genetic dissection of grain yield and physical grain quality in bread wheat (Triticum aestivum L.) under water-limited environments. Theor. Appl. Genet. 2012, 125, 255–271. [Google Scholar] [CrossRef] [PubMed]
- Edae, E.A.; Byrne, P.F.; Haley, S.D.; Lopes, M.S.; Reynolds, M.P. Genome-wide association mapping of yield and yield components of spring wheat under contrasting moisture regimes. Theor. Appl. Genet. 2014, 127, 791–807. [Google Scholar] [CrossRef]
- Reynolds, M.P.; Pellegrineschi, A.; Skovmand, B. Sink limitation to yield and biomass: A summary of investigations in spring wheat. Ann. Appl. Biol. 2005, 146, 39–49. [Google Scholar] [CrossRef]
- Reynolds, M.P.; Foulkes, M.J.; Furbank, R.; Griffiths, S.; King, J.; Murchie, M.; Parry, M.; Slafer, G. Achieving yield gains in wheat. Plant Cell Enviorn. 2012, 35, 1799–1823. [Google Scholar] [CrossRef]
- Fischer, R.A. Understanding the physiological basis of yield potential in wheat. J. Agric. Sci. 2007, 145, 99–113. [Google Scholar] [CrossRef]
- Bernardo, R. Molecular markers and selection for complex traits in plants: Learning from the last 20 years. Crop Sci. 2008, 48, 1649–1664. [Google Scholar] [CrossRef]
- Foulkes, M.J.; Slafer, G.A.; Davies, W.J.; Berry, P.M.; Sylvester-Bradley, R.; Martre, P.; Calderini, D.F.; Griffiths, S.; Reynolds, M.P. Raising yield potential of wheat. III. Optimizing partitioning to grain while maintaining lodging resistance. J. Exp. Bot. 2011, 62, 469–486. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, S.; Schnurbusch, T. Of floral fortune: Tinkering with the grain yield potential of cereal crops. New Phytol. 2020, 225, 1873–1882. [Google Scholar] [CrossRef] [PubMed]
- Calderini, D.F.; Drecer, M.F.; Slafer, G.A. Genetic improvement in wheat yield and associated traits. A re-examination of previous results and the latest trends. Plant Breed. 1995, 114, 108–112. [Google Scholar]
- Wurschum, T.; Leiser, W.L.; Langer, S.M.; Tucker, M.R.; Longin, C.F. Phenotypic and genetic analysis of spike and kernel characteristics in wheat reveals long-term genetic trends of grain yield components. Theor. Appl. Genet. 2018, 131, 2071–2084. [Google Scholar] [CrossRef] [PubMed]
- Rajaram, S.; Reynolds, M.P. International Wheat breeding: Abstracts of XVIth Eucarpia Congress-Plant Breeding: Sustaining the Future, Proceedings of the 6th Eucarpia Congress, Edinburgh, UK, 10–14 September 2001; Edinburgh University Press: Edinburgh, UK, 2001. [Google Scholar]
- Gaju, O.; Reynolds, M.P.; Sparkes, D.L.; Foulkes, M.J. Relationships between large spike phenotype, grain number and yield potential in spring wheat. Crop Sci. 2009, 49, 961–973. [Google Scholar] [CrossRef]
- Gaju, O.; Reynolds, M.P.; Sparkes, D.L.; Mayes, S.; RibasVargas, G.; Crossa, J.; Foulkes, M.J. Relationships between physiological traits, grain number and yield potential in a wheat DH population of large spike phenotype. Field Crops Res. 2014, 164, 126–135. [Google Scholar] [CrossRef]
- Kang, C.S.; Seo, S.J.; Kim, K.H.; Jung, Y.G.; Kim, H.S.; Park, J.C.; Choi, I.D.; Kim, K.H.; Hyun, J.N.; Kim, K.J.; et al. A New Plant Type Wheat “Iksan 370” with Long Spikes, High Yield and Hard Wheat and Breeding Method. No. 10-2013-0120136, 8 October 2013. [Google Scholar]
- Kim, K.M.; Kim, K.H.; Cheong, Y.K.; Choi, C.H.; Kim, Y.K.; Park, J.H.; Kim, K.H.; Jang, J.; Choi, J.K.; Bae, J.S.; et al. Taejoong’ a Wheat Variety with Good Noodle Quality, Red Grain Wheat, Long Spike, and Moderate Resistance to Fusarium Head Blight. Korean J. Breed. Sci. 2019, 51, 445–452. [Google Scholar] [CrossRef]
- Song, T.H.; Kim, K.H.; Cheong, Y.K.; Son, J.H.; Park, J.C.; OH, Y.J.; Park, J.H.; Kim, Y.K.; Kim, K.H.; Park, T.I.; et al. Evaluation of feed value and fermentation quality of new wheat cultivar, “Taejoong’. Korean J. Breed. Sci. 2017, 37, 61–67. [Google Scholar] [CrossRef]
- Wang, J.; Li, H.; Zhang, L.; Meng, L. Users’ Manual of QTL IciMapping; The Quantitative Genetics Group, Institute of Crop Science, Chinese Academy of Agricultural Sciences (CAAS): Beijing, China; Genetic Resources Program, International Maize and Wheat Improvement Center (CIMMYT): Mexico City, Mexico, 2014. [Google Scholar]
- Fischer, R.A. Yield Potential in a Dwarf Spring Wheat and the Effect of Shading. Crop Sci. 1975, 5, 607–613. [Google Scholar] [CrossRef]
- Fischer, R.A.; Rees, D.; Sayre, K.D.; Lu, Z.M.; Condon, A.G.; Saavedra, A.L. Wheat Yield Progress Associated with Higher Stomatal Conductance and Photosynthetic Rate, and Cooler Canopies. Crop Sci. 1998, 38, 1467–1475. [Google Scholar] [CrossRef]
- Foulkes, M.J.; Snape, J.W.V.; Shearman, V.J.; Reynolds, M.P.; Gaju, O.; Sylvester-Bradley, R. Genetic progress in yield potential in wheat: Recent advances and future prospects. J. Agric. Sci. 2007, 145, 17–29. [Google Scholar] [CrossRef]
- Singh, R.P.; Singh, P.K.; Rutkoski, J.; Hodson, D.P.; He, X.; Jørgensen, L.N.; Hovmøller, M.S.; Huerta-Espino, J. Disease Impact on Wheat Yield Potential and Prospects of Genetic Control. Annu. Rev. Phytopathol. 2016, 54, 303–325. [Google Scholar] [CrossRef] [PubMed]
- Denćić, S. Designing a d wheat ideotype with increased sink capacity. Plant Breed. 1994, 112, 311–317. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, Y.; Shi, H.; Wu, F. Identification and validation of novel low-tiller number QTL in common wheat. Theor. Appl. Genet. 2016, 129, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Bastos, L.M.; Carciochi, W.; Lollato, R.P.; Jaenisch, B.R.; Renzende, C.R.; Schwalbert, R.; Prasad, P.V.V.; Zhang, G.; Frotz, A.K.; Foster, C.; et al. Winter wheat yield response to plant density as a function of yield environment and tillering potential: A review and field studies. Front. Plant Sci. 2020, 11, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Ojha, R.; Sarkar, A.; Aryal, A.; Rahul, K.C.; Tiwari, S.; Poudel, M.; Pant, K.R.; Shrestha, J. Correlation and path coefficient analysis of wheat (Triticum aestivum L.) genotypes. Fmg. Mngmt. 2018, 3, 136–141. [Google Scholar]
- Guo, Z.; Zhao, Y.; Röder, M.; Reif, J.C.; Ganal, M.W.; Chen, D.; Schnurbusch, T. Manipulation and prediction of spike morphology traits for the improvement of grain yield in wheat. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- International Wheat Genome Sequencing Consortium (IWGSC). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Wen, W.; Liu, J.; Rasheed, A.; Yin, G.; Xia, X.; Wu, X.; He, Z. Genome-wide linkage mapping of QTL for yield components, plant height and yield-related physiological traits in the Chinese wheat cross Zhou 8425B/Chinese Spring. Front. Plant Sci. 2015, 6, 1099. [Google Scholar] [CrossRef] [PubMed]
- Zhai, H.; Feng, Z.; Li, J.; Liu, X.; Xiao, S.; Ni, Z.; Sun, Q. QTL Analysis of spike morphological traits and plant height in winter wheat (Triticum aestivum L.) using a high-density SNP and SSR-based linkage map. Front. Plant Sci. 2016, 7, 1617. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Cui, F.; Ji, J.; Zhang, W.; Zhao, X.; Liu, J.; Meng, D.; Tong, Y.; Wang, T.; Li, J. Dissection of pleiotropic QTL regions controlling wheat spike characteristics under different nitrogen treatments using traditional and conditional QTL mapping. Front. Plant Sci. 2019, 10, 187. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wang, X.; Zhang, G.; Jiang, P.; Chen, W.; Hao, Y.; Ma, X.; Xu, S.; Jia, J.; Kong, L.; et al. QTL mapping for yield-related traits in wheat based on four RIL populations. Theor. Appl. Genet. 2020, 133, 917–933. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Jia, L.; Lu, L.; Qin, D.; Zhang, J.; Guan, P.; Ni, Z.; Yao, Y.; Sun, Q.; Peng, H. Mapping QTLs of yield-related traits using RIL population derived from common wheat and Tibetan semi-wild wheat. Theor. Appl. Genet. 2014, 127, 2415–2432. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Conway, B.; Miller, D.; Marshall, D.; Cooper, A.; Murphy, P.; Chao, S.; Brown-Guedira, G.; Costa, J. Quantitative trait loci mapping for spike characteristics in hexaploid wheat. Plant Genome 2017, 10, 1–15. [Google Scholar] [CrossRef]

| QTL a | Year | Chromosome | Position (cM) | Left Marker | Right Marker | Interval (cM) b | LOD c | PVE (%) d | Add e |
|---|---|---|---|---|---|---|---|---|---|
| QDHD-1 | 2017 | 7B | 172.0 | AX-94684729 | AX-94474044 | 5.0 | 3.90 | 11.8 | 1.37 |
| 2019 | 4.24 | 13.7 | 1.30 | ||||||
| QSL-1 | 2017 | 1D | 268.0 | AX-94390275 | AX-94918964 | 5.0 | 3.63 | 11.2 | 0.59 |
| 2018 | 5.37 | 15.6 | 0.72 | ||||||
| 2019 | 4.04 | 15.1 | 0.62 | ||||||
| QSL-2 | 2018 | 5A | 208.0 | AX-94831718 | AX-94846616 | 16.0 | 3.82 | 12.2 | 0.62 |
| 2019 | 3.55 | 15.1 | 0.61 | ||||||
| QSL-3 | 2017 | 6A | 175.0 | AX-94411823 | AX-94602621 | 13.5 | 7.59 | 26.0 | 0.88 |
| 2018 | 7.25 | 28.8 | 0.95 | ||||||
| 2019 | 5.88 | 28.8 | 0.84 | ||||||
| QTN-1 | 2017 | 5B | 269.0 | AX-95176502 | AX-94429067 | 18.0 | 4.17 | 16.3 | −0.75 |
| 2018 | 4.10 | 16.4 | −0.74 | ||||||
| 2019 | 4.15 | 16.4 | −0.75 | ||||||
| QLCR-1 | 2017 | 1B | 330.0 | AX-94414200 | AX-94757158 | 3.0 | 8.67 | 19.1 | −0.05 |
| 2018 | 3.62 | 8.7 | −0.03 | ||||||
| 2019 | 3.75 | 8.7 | −0.04 | ||||||
| QLCR-2 | 2017 | 5B | 419.0 | AX-94904781 | AX-95257493 | 7.0 | 9.14 | 22.2 | 0.06 |
| 2018 | 10.82 | 34.5 | 0.07 | ||||||
| 2019 | 9.73 | 30.3 | 0.07 | ||||||
| QLCR-3 | 2018 | 6A | 171.0 | AX-94411823 | AX-94602621 | 17.5 | 5.54 | 17.4 | 0.05 |
| 2019 | 6.77 | 22.7 | 0.05 |
| Trait a | Allele b | Main Effect c | Interaction d | |||||
|---|---|---|---|---|---|---|---|---|
| QSL-1 | QSL-2 | QSL-3 | Q1 × Q2 | Q1 × Q3 | Q2 × Q3 | Q1 × Q2 × Q3 | ||
| SL (cm) | T | 12.2 | 12.2 | 12.5 | ||||
| K | 11.0 | 11.0 | 10.7 | |||||
| P-value | **** | **** | **** | ns | ns | ns | ns | |
| PVE (%) | 20.2 | 15.2 | 18.6 | |||||
| DHD (no.) | T | 185.5 | 185.4 | 185.1 | ||||
| K | 184.5 | 184.6 | 184.9 | |||||
| P-value | **** | ** | ns | ns | ns | ns | *** | |
| PVE (%) | 5.7 | 2.6 | 4.7 | |||||
| CL (cm) | T | 84.4 | 84.5 | 87.4 | ||||
| K | 86.5 | 86.4 | 83.5 | |||||
| P-value | ns | ns | **** | ns | ns | ** | ** | |
| PVE (%) | 6.5 | 3.0 | 2.3 | |||||
| COM (no./cm) | T | 4.4 | 4.3 | 4.3 | ||||
| K | 4.6 | 4.7 | 4.7 | |||||
| P-value | **** | **** | **** | ns | ns | ns | ns | |
| PVE (%) | 5.4 | 7.5 | 5.9 | |||||
| KS (no.) | T | 51.7 | 50.6 | 51.7 | ||||
| K | 48.9 | 50.0 | 48.9 | |||||
| P-value | **** | ns | ** | ns | ns | ns | ns | |
| PVE (%) | 6.0 | 3.6 | ||||||
| TN (no.) | T | 5.7 | 5.8 | 6.0 | ||||
| K | 6.4 | 6.3 | 6.1 | |||||
| P-value | **** | ** | * | *** | ns | * | ns | |
| PVE (%) | 8.3 | 2.6 | 1.6 | 3.6 | 1.6 | |||
| LCR (cm) | T | 0.71 | 0.73 | 0.73 | ||||
| K | 0.66 | 0.64 | 0.64 | |||||
| P-value | **** | **** | **** | ** | ** | ns | ns | |
| PVE (%) | 9.9 | 21.6 | 11.9 | 2.0 | 1.4 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, C.-S.; Mo, Y.-J.; Kim, K.-M.; Kim, K.-H.; Chun, J.-B.; Park, C.-S.; Cho, S.-W. Mapping Quantitative Trait Loci for Yield Potential Traits in Wheat Recombinant Inbred Lines. Agronomy 2021, 11, 22. https://doi.org/10.3390/agronomy11010022
Kang C-S, Mo Y-J, Kim K-M, Kim K-H, Chun J-B, Park C-S, Cho S-W. Mapping Quantitative Trait Loci for Yield Potential Traits in Wheat Recombinant Inbred Lines. Agronomy. 2021; 11(1):22. https://doi.org/10.3390/agronomy11010022
Chicago/Turabian StyleKang, Chon-Sik, Young-Jun Mo, Kyeong-Min Kim, Kyeong-Hoon Kim, Jae-Buhm Chun, Chul-Soo Park, and Seong-Woo Cho. 2021. "Mapping Quantitative Trait Loci for Yield Potential Traits in Wheat Recombinant Inbred Lines" Agronomy 11, no. 1: 22. https://doi.org/10.3390/agronomy11010022
APA StyleKang, C.-S., Mo, Y.-J., Kim, K.-M., Kim, K.-H., Chun, J.-B., Park, C.-S., & Cho, S.-W. (2021). Mapping Quantitative Trait Loci for Yield Potential Traits in Wheat Recombinant Inbred Lines. Agronomy, 11(1), 22. https://doi.org/10.3390/agronomy11010022






