Gene Pyramiding for Sustainable Crop Improvement against Biotic and Abiotic Stresses
Abstract
1. Introduction
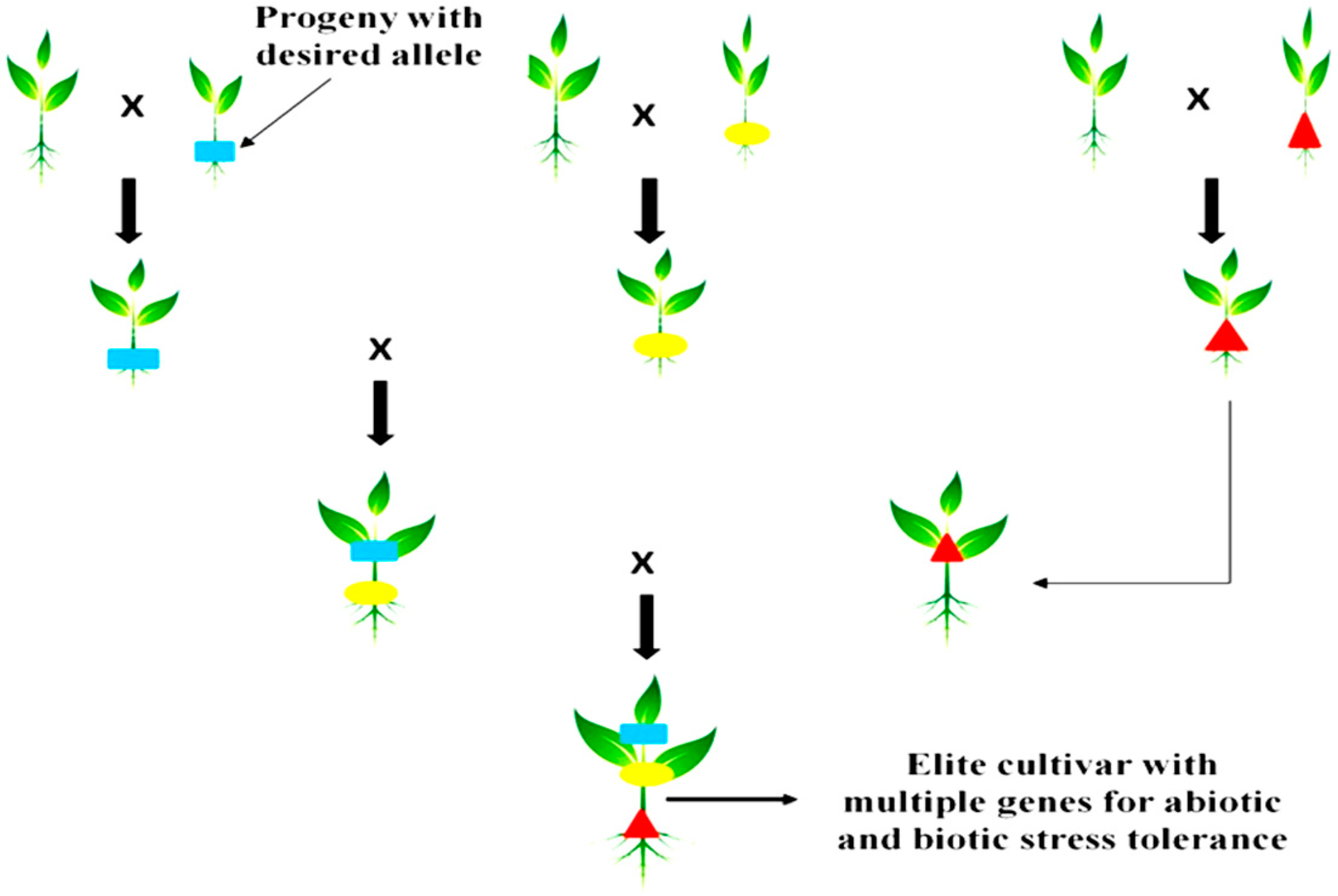
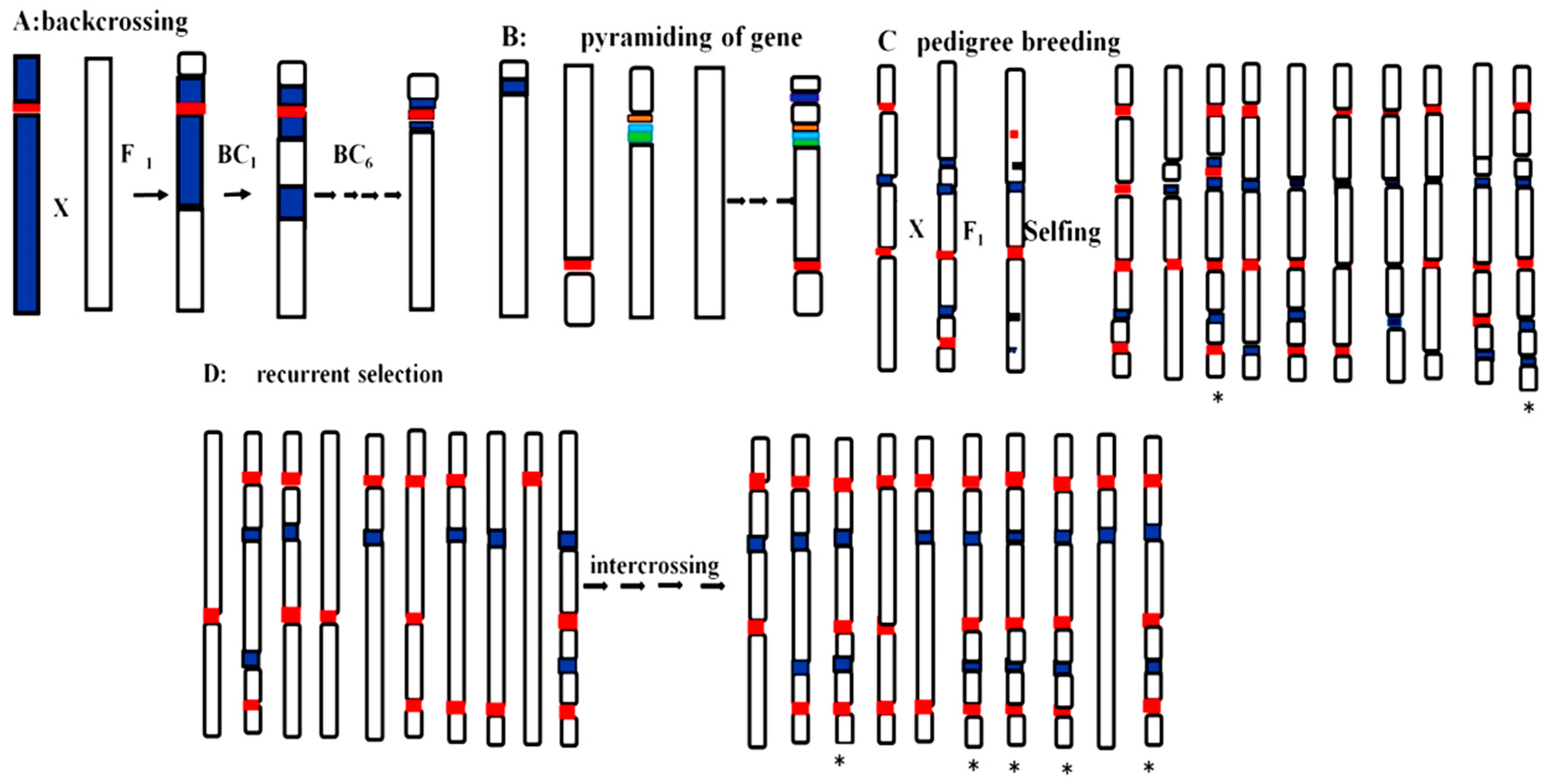
2. Types of Gene Pyramiding in Plant Breeding
3. Molecular Techniques in Breeding Programs
3.1. Molecular Marker-Assisted Selection
3.2. Marker-Assisted Backcrossing
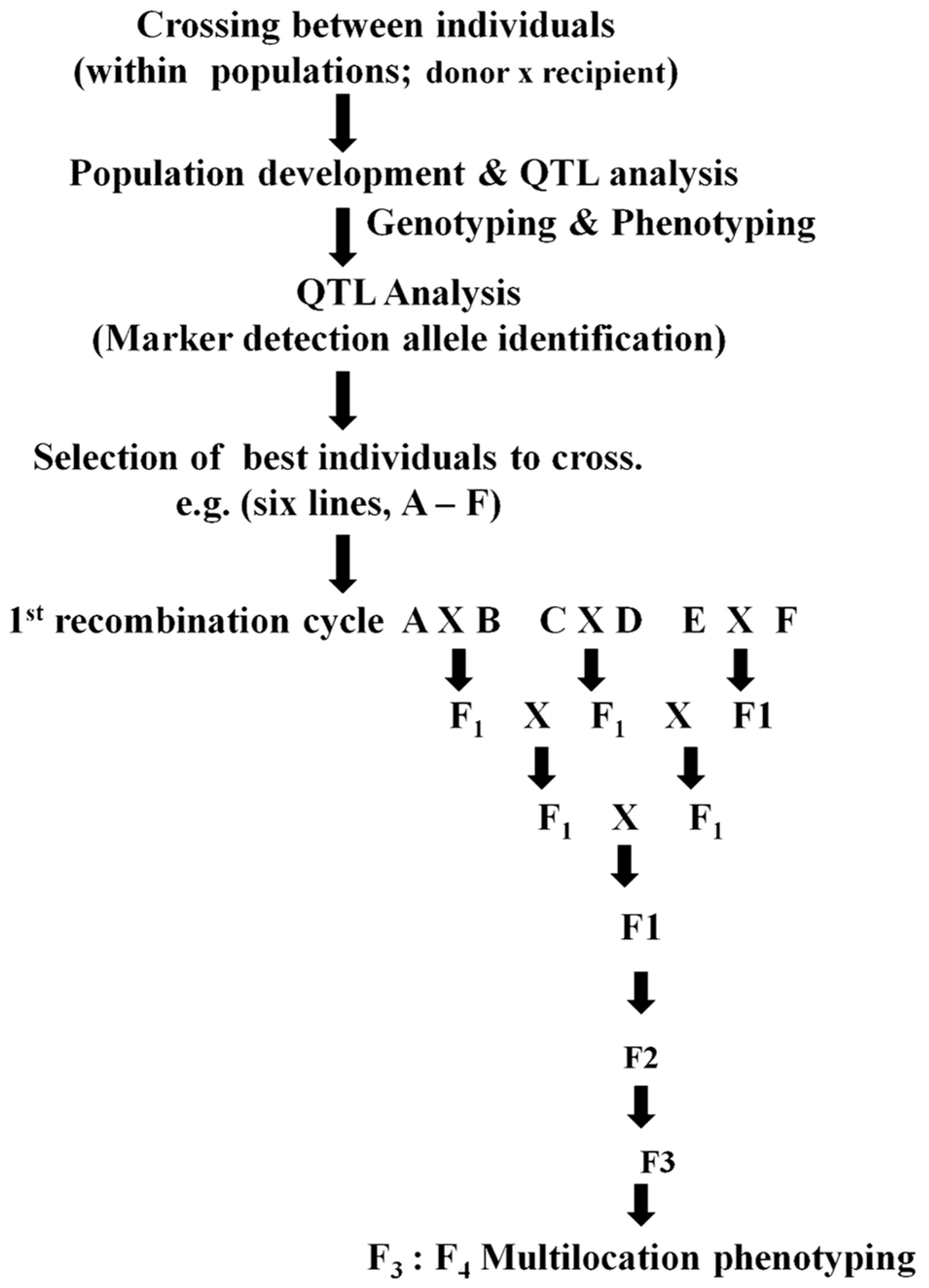
3.3. Marker-Assisted Recurrent Selection
3.4. Omics Techniques for Crop Improvement
3.5. Marker-Assisted Gene Pyramiding in Developing Resistant Crop Varieties
3.6. Gene Pyramiding Involving Polygenic Applications
4. Challenges in Molecular Markers Utilization in Plant Breeding
5. Contribution of the Gene Pyramiding Technique to Agricultural Sustainability
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Davis, K.F.; Chhatre, A.; Rao, N.D.; Singh, D.; Ghosh-Jerath, S.; Mridul, A.; Poblete-Cazenave, M.; Pradhan, N.; DeFries, R. Assessing the sustainability of post-Green Revolution cereals in India. Proc. Natl. Acad. Sci. USA 2019, 116, 25034–25041. [Google Scholar] [CrossRef] [PubMed]
- Barrett, C.B. Measuring Food Insecurity. Science 2010, 327, 825–828. [Google Scholar] [CrossRef] [PubMed]
- Atique, R.; Farooq, M.; Rashid, A.; Nadeem, F.; Stuerz, S.; Asch, F.; Bell, R.W.; Siddique, K.H.M. Boron nutrition of rice in different production systems: A review. Agron. Sustain. Dev. 2018, 38, 25. [Google Scholar] [CrossRef]
- Mwobobia, E.G.; Sichangi, A.W.; Thiong’o, K.B. Characterization of wheat production using earth-based observations: A case study of Meru County, Kenya. Model. Earth Syst. Environ. 2020, 6, 13–25. [Google Scholar] [CrossRef]
- Kage, U.; Kumar, A.; Dhokane, D.; Karre, S.; Kushalappa, A.C. Functional molecular markers for crop improvement. Crit. Rev. Biotechnol. 2016, 36, 917–930. [Google Scholar] [CrossRef]
- Pandey, P.; Irulappan, V.; Bagavathiannan, M.V.; Senthil-Kumar, M. Impact of Combined Abiotic and Biotic Stresses on Plant Growth and Avenues for Crop Improvement by Exploiting Physio-morphological Traits. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Waddington, S.R.; Li, X.; Dixon, J.; Hyman, G.; de Vicente, M.C. Getting the focus right: Production constraints for six major food crops in Asian and African farming systems. Food Secur. 2010, 2, 27–48. [Google Scholar] [CrossRef]
- Bowman, M.S.; Zilberman, D. Economic Factors Affecting Diversified Farming Systems. Ecol. Soc. 2013, 18. [Google Scholar] [CrossRef]
- Belsky, J.; Joshi, N.K. Impact of Biotic and Abiotic Stressors on Managed and Feral Bees. Insects 2019, 10, 233. [Google Scholar] [CrossRef]
- Choudhary, K.; Choudhary, O.P.; Shekhawat, N.S. Marker assisted selection: A novel approach for crop improvement. Am. Eurasian J. Agron. 2008, 1, 26–30. [Google Scholar]
- Womack, E.D.; Williams, W.P.; Smith, J.S.; Warburton, M.L.; Bhattramakki, D. Mapping Quantitative Trait Loci for Resistance to Fall Armyworm (Lepidoptera: Noctuidae) Leaf-Feeding Damage in Maize Inbred Mp705. J. Econ. Entomol. 2020, 113, 956–963. [Google Scholar] [CrossRef]
- Kansiime, M.K.; Mugambi, I.; Rwomushana, I.; Nunda, W.; Lamontagne-Godwin, J.; Rware, H.; Phiri, N.A.; Chipabika, G.; Ndlovu, M.; Day, R. Farmer perception of fall armyworm (Spodoptera frugiderda J.E. Smith) and farm-level management practices in Zambia. Pest Manag. Sci. 2019, 75, 2840–2850. [Google Scholar] [CrossRef]
- Assefa, F. Status of Fall Armyworm (Spodoptera frugiperda), Biology and Control Measures on Maize Crop in Ethiopia: A Review. Int. J. Entomol. Res. 2018, 6. [Google Scholar] [CrossRef]
- Nagoshi, R.N.; Goergen, G.; Tounou, K.A.; Agboka, K.; Koffi, D.; Meagher, R.L. Analysis of strain distribution, migratory potential, and invasion history of fall armyworm populations in northern Sub-Saharan Africa. Sci. Rep. 2018, 8, 3710. [Google Scholar] [CrossRef]
- Sharifzadeh, M.S.; Abdollahzadeh, G.; Damalas, C.A.; Rezaei, R. Farmers’ Criteria for Pesticide Selection and Use in the Pest Control Process. Agriculture 2018, 8, 24. [Google Scholar] [CrossRef]
- Vincelli, P. Genetic Engineering and Sustainable Crop Disease Management: Opportunities for Case-by-Case Decision-Making. Sustainability 2016, 8, 495. [Google Scholar] [CrossRef]
- Federico, A.C.; Darci, A.G.; Patrick, J.T. Empirical investigation of mutation rate for herbicide resistance. Weed Sci. 2019, 67, 361–368. [Google Scholar]
- Pandeya, D.; López-Arredondo, D.L.; Janga, M.R.; Campbell, L.M.; Estrella, H.P.; Bagavathiannan, M.V.; Herrera-Estrella, L.; Rathore, K.S. Selective fertilization with phosphite allows unhindered growth of cotton plants expressing the ptxD gene while suppressing weeds. Proc. Natl. Acad. Sci. USA 2018, 115, E6946–E6955. [Google Scholar] [CrossRef]
- Hanson, P.; Lu, S.-F.; Wang, J.-F.; Chen, W.; Kenyon, L.; Tan, C.-W.; Tee, K.L.; Wang, Y.-Y.; Hsu, Y.-C.; Schafleitner, R.; et al. Conventional and molecular marker-assisted selection and pyramiding of genes for multiple disease resistance in tomato. Sci. Hortic. 2016, 201, 346–354. [Google Scholar] [CrossRef]
- Mundt, C.C. Durable resistance: A key to sustainable management of pathogens and pests. Infect. Genet. Evol. 2014, 27, 446–455. [Google Scholar] [CrossRef]
- Johnson, R. A Critical Analysis of Durable Resistance. Annu. Rev. Phytopathol. 1984, 22, 309–330. [Google Scholar] [CrossRef]
- Kottapalli, K.R.; Lakshmi, N.M.; Jena, K.K. Effective strategy for pyramiding three bacterial blight resistance genes into fine grain rice cultivar, Samba Mahsuri, using sequence tagged site markers. Biotechnol. Lett. 2010, 32, 989–996. [Google Scholar] [CrossRef]
- Steiner, B.; Michel, S.; Maccaferri, M.; Lemmens, M.; Tuberosa, R.; Buerstmayr, H. Exploring and exploiting the genetic variation of Fusarium head blight resistance for genomic-assisted breeding in the elite durum wheat gene pool. Theor. Appl. Genet. 2019, 132, 969–988. [Google Scholar] [CrossRef]
- Suresh, S.; Malathi, D. Gene Pyramiding for Biotic Stress Tolerance In Crop plants. Wkly. Sci. Res. J. 2013, 1, 1–14. [Google Scholar]
- Muthurajan, R.; Balasubramanian, P. Pyramiding Genes for Enhancing Tolerance to Abiotic and Biotic Stresses. In Molecular Techniques in Crop Improvement; Jain, S., Brar, D., Eds.; Springer: Dordrecht, The Netherlands, 2010. [Google Scholar]
- Pancaldi, F.; Trindade, L.M. Marginal Lands to Grow Novel Bio-Based Crops: A Plant Breeding Perspective. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Rana, M.; Sood, A.; Hussain, W.; Kaldate, R.; Sharma, T.R.; Gill, R.K.; Kumar, S.; Singh, S. Chapter 6—Gene Pyramiding and Multiple Character Breeding. In Lentils; Singh, M., Ed.; Academic Press: Cambridge, MA, USA, 2019; pp. 83–124. [Google Scholar]
- Bai, Y.; Kissoudis, C.; Yan, Z.; Visser, R.G.F.; van der Linden, G. Plant behaviour under combined stress: Tomato responses to combined salinity and pathogen stress. Plant J. 2018, 93, 781–793. [Google Scholar] [CrossRef]
- Ruengphayak, S.; Chaichumpoo, E.; Phromphan, S.; Kamolsukyunyong, W.; Sukhaket, W.; Phuvanartnarubal, E.; Korinsak, S.; Korinsak, S.; Vanavichit, A. Pseudo-backcrossing design for rapidly pyramiding multiple traits into a preferential rice variety. Rice 2015, 8, 7. [Google Scholar] [CrossRef]
- Zhu, S.; Li, Y.; Vossen, J.H.; Visser, R.G.F.; Jacobsen, E. Functional stacking of three resistance genes against Phytophthora infestans in potato. Transgenic Res. 2012, 21, 89–99. [Google Scholar] [CrossRef]
- Piquerez, S.J.M.; Harvey, S.E.; Beynon, J.L.; Ntoukakis, V. Improving crop disease resistance: Lessons from research on Arabidopsis and tomato. Front. Plant Sci. 2014, 5, 671. [Google Scholar] [CrossRef]
- Alconi, C.; Stevanato, P.; Motto, M.; Biancardi, E. Breeding for Biotic Stress Resistance/Tolerance in Plants. In Crop Production for Agricultural Improvement; Ashraf, M., Öztürk, M., Ahmad, M., Aksoy, A., Eds.; Springer: Dordrecht, The Netherlands, 2012. [Google Scholar]
- Llorens, E.; González-Hernández, A.I.; Scalschi, L.; Fernández-Crespo, E.; Camañes, G.; Vicedo, B.; García-Agustín, P. Chapter 1—Priming mediated stress and cross-stress tolerance in plants: Concepts and opportunities. In Priming-Mediated Stress and Cross-Stress Tolerance in Crop Plants; Academic Press: Cambridge, MA, USA, 2020; pp. 1–20. [Google Scholar]
- Yu, Y.; Yang, Z. The isolation and characterization of a putative dehydrin gene in Triticum aestivum L. Biochem. Syst. Ecol. 2016, 66, 173–180. [Google Scholar] [CrossRef]
- Waqas, M.; Shahid, L.; Shoukat, K.; Aslam, U.; Azeem, F.; Atif, R.M. Chapter 1—Role of DNA-binding with one finger (Dof) transcription factors for abiotic stress tolerance in plants. In Transcription Factors for Abiotic Stress Tolerance in Plants; Academic Press: Cambridge, MA, USA, 2020; pp. 1–14. [Google Scholar]
- Loo, Y.Y.; Billa, L.; Singh, A. Effect of climate change on seasonal monsoon in Asia and its impact on the variability of monsoon rainfall in Southeast Asia. Geosci. Front. 2015, 6, 817–823. [Google Scholar] [CrossRef]
- Thitisaksakul, M.; Tananuwong, K.; Shoemaker, C.F.; Chun, A.; Tanadul, O.-U.-M.; Labavitch, J.M.; Beckles, D.M. Effects of Timing and Severity of Salinity Stress on Rice (Oryza sativa L.) Yield, Grain Composition, and Starch Functionality. J. Agric. Food Chem. 2015, 63, 2296–2304. [Google Scholar] [CrossRef]
- Tiwari, S.; Sl, K.; Kumar, V.; Singh, B.; Rao, A.R.; Mithra, S.A.; Rai, V.; Singh, A.K.; Singh, N.K. Mapping QTLs for Salt Tolerance in Rice (Oryza sativa L.) by Bulked Segregant Analysis of Recombinant Inbred Lines Using 50K SNP Chip. PLoS ONE 2016, 11, e0153610. [Google Scholar] [CrossRef]
- Bimpong, I.K.; Manneh, B.; Sock, M.; Diaw, F.; Amoah, N.K.A.; Ismail, A.M.; Gregorio, G.; Singh, R.K.; Wopereis, M. Improving salt tolerance of lowland rice cultivar ‘Rassi’ through marker-aided backcross breeding in West Africa. Plant Sci. 2016, 242, 288–299. [Google Scholar] [CrossRef]
- Ren, Z.-H.; Gao, J.-P.; Li, L.-G.; Cai, X.-L.; Huang, W.; Chao, D.-Y.; Zhu, M.-Z.; Wang, Z.-Y.; Luan, S.; Lin, H.-X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 2005, 37, 1141–1146. [Google Scholar] [CrossRef]
- Das, P.; Nutan, K.K.; Singla-Pareek, S.L.; Pareek, A. Understanding salinity responses and adopting ‘omics-based’ approaches to generate salinity tolerant cultivars of rice. Front. Plant Sci. 2015, 6, 712. [Google Scholar] [CrossRef]
- Oladosu, Y.; Rafii, M.Y.; Samuel, C.; Fatai, A.; Magaji, U.; Kareem, I.; Kamarudin, Z.S.; Muhammad, I.; Kolapo, K. Drought Resistance in Rice from Conventional to Molecular Breeding: A Review. Int. J. Mol. Sci. 2019, 20, 3519. [Google Scholar] [CrossRef]
- Sandhu, N.; Dixit, S.; Swamy, B.P.M.; Raman, A.; Kumar, S.; Singh, S.P.; Yadaw, R.B.; Singh, O.N.; Reddy, J.N.; Anandan, A.; et al. Marker Assisted Breeding to Develop Multiple Stress Tolerant Varieties for Flood and Drought Prone Areas. Rice 2019, 12, 8. [Google Scholar] [CrossRef]
- Rai, N.; Bellundagi, A.; Kumar, P.K.C.; Kalasapura, T.R.; Rani, S.; Sinha, N.; krishna, H.; Jain, N.; Singh, G.P.; Singh, P.K.; et al. Marker-assisted backcross breeding for improvement of drought tolerance in bread wheat (Triticum aestivum L. em Thell). Plant Breed. 2018, 137, 514–526. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, J.; Wei, P.; Zhang, B.; Gou, F.; Feng, Z.; Mao, Y.; Yang, L.; Zhang, H.; Xu, N.; et al. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 2014, 12, 797–807. [Google Scholar] [CrossRef]
- Shinada, H.; Iwata, N.; Sato, T.; Fujino, K. QTL pyramiding for improving of cold tolerance at fertilization stage in rice. Breed. Sci. 2014, 63, 483–488. [Google Scholar] [CrossRef][Green Version]
- Almeida, D.M.; Almadanim, M.C.; Lourenço, T.; Abreu, I.A.; Saibo, N.J.; Oliveira, M.M. Screening for Abiotic Stress Tolerance in Rice: Salt, Cold, and Drought. Methods Mol. Biol. 2016, 1398, 155–182. [Google Scholar] [CrossRef]
- Hasan, M.M.; Rafii, M.Y.; Ismail, M.R.; Mahmood, M.; Rahim, H.A.; Alam, M.A.; Ashkani, S.; Malek, M.A.; Latif, M.A. Marker-assisted backcrossing: A useful method for rice improvement. Biotechnol. Biotechnol. Equip. 2015, 29, 237–254. [Google Scholar] [CrossRef]
- Sanghera, G.S.; Wani, S.H.; Hussain, W.; Singh, N.B. Engineering cold stress tolerance in crop plants. Curr. Genom. 2011, 12, 30–43. [Google Scholar] [CrossRef]
- Lv, H.; Fang, Z.; Yang, L.; Zhang, Y.; Wang, Y. An update on the arsenal: Mining resistance genes for disease management of Brassica crops in the genomic era. Hortic. Res. 2020, 7, 34. [Google Scholar] [CrossRef]
- Van Esse, H.P.; Reuber, T.L.; van der Does, D. Genetic modification to improve disease resistance in crops. New Phytol. 2020, 225, 70–86. [Google Scholar] [CrossRef]
- Borrelli, V.M.G.; Brambilla, V.; Rogowsky, P.; Marocco, A.; Lanubile, A. The Enhancement of Plant Disease Resistance Using CRISPR/Cas9 Technology. Front. Plant Sci. 2018, 9, 1245. [Google Scholar] [CrossRef]
- Sedeek, K.E.M.; Mahas, A.; Mahfouz, M. Plant Genome Engineering for Targeted Improvement of Crop Traits. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Kah, M.; Tufenkji, N.; White, J.C. Nano-enabled strategies to enhance crop nutrition and protection. Nat. Nanotechnol. 2019, 14, 532–540. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Parker, J.E.; Ainsworth, A.; Oldroyd, G.E.D.; Schroeder, J.I. Genetic strategies for improving crop yields. Nature 2019, 575, 109–118. [Google Scholar] [CrossRef]
- Fuchs, M. Pyramiding resistance-conferring gene sequences in crops. Curr. Opin. Virol. 2017, 26, 36–42. [Google Scholar] [CrossRef]
- Liu, R.; Lu, J.; Zhou, M.; Zheng, S.; Liu, Z.; Zhang, C.; Du, M.; Wang, M.; Li, Y.; Wu, Y.; et al. Developing stripe rust resistant wheat (Triticum aestivum L.) lines with gene pyramiding strategy and marker-assisted selection. Genet. Resour. Crop Evol. 2020, 67, 381–391. [Google Scholar] [CrossRef]
- Ashkani, S.; Rafii, M.Y.; Shabanimofrad, M.; Miah, G.; Sahebi, M.; Azizi, P.; Tanweer, F.A.; Akhtar, M.S.; Nasehi, A. Molecular Breeding Strategy and Challenges Towards Improvement of Blast Disease Resistance in Rice Crop. Front. Plant Sci. 2015, 6, 886. [Google Scholar] [CrossRef]
- Srivastava, V.; Thomson, J. Gene stacking by recombinases. Plant Biotechnol. J. 2016, 14, 471–482. [Google Scholar] [CrossRef]
- Gupta, S.K.; Pandey, M.K. Chapter 19—Breeding for Disease and Insect-Pest Resistance. In Integrated Pest Management; Abrol, D.P., Ed.; Academic Press: San Diego, CA, USA, 2014; pp. 401–417. [Google Scholar]
- Mago, R.; Lawrence, G.J.; Ellis, J.G. The application of DNA marker and doubled-haploid technology for stacking multiple stem rust resistance genes in wheat. Mol. Breed. 2011, 27, 329–335. [Google Scholar] [CrossRef]
- Liu, J.; Liu, D.; Tao, W.; Li, W.; Wang, S.; Chen, P.; Cheng, S.; Gao, D. Molecular marker-facilitated pyramiding of different genes for powdery mildew resistance in wheat. Plant Breed. 2000, 119, 21–24. [Google Scholar] [CrossRef]
- Nelson, R.; Wiesner-Hanks, T.; Wisser, R.; Balint-Kurti, P. Navigating complexity to breed disease-resistant crops. Nat. Rev. Genet. 2018, 19, 21–33. [Google Scholar] [CrossRef]
- Pathania, A.; Rialch, N.; Sharma, P.N. Marker-Assisted Selection in Disease Resistance Breeding: A Boon to Enhance Agriculture Production. In Current Developments in Biotechnology and Bioengineering; Dubey, S.K., Pandey, A., Sangwan, R.S., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 187–213. [Google Scholar]
- Dixit, S.; Singh, U.M.; Singh, A.K.; Alam, S.; Venkateshwarlu, C.; Nachimuthu, V.V.; Yadav, S.; Abbai, R.; Selvaraj, R.; Devi, M.N.; et al. Marker Assisted Forward Breeding to Combine Multiple Biotic-Abiotic Stress Resistance/Tolerance in Rice. Rice 2020, 13, 29. [Google Scholar] [CrossRef]
- Floros, J.D.; Newsome, R.; Fisher, W.; Barbosa-Cánovas, G.V.; Chen, H.; Dunne, C.P.; German, J.B.; Hall, R.L.; Heldman, D.R.; Karwe, M.V.; et al. Feeding the World Today and Tomorrow: The Importance of Food Science and Technology. Compr. Rev. Food Sci. Food Saf. 2010, 9, 572–599. [Google Scholar] [CrossRef]
- Su, J.; Jiang, J.; Zhang, F.; Liu, Y.; Ding, L.; Chen, S.; Chen, F. Current achievements and future prospects in the genetic breeding of chrysanthemum: A review. Hortic. Res. 2019, 6, 109. [Google Scholar] [CrossRef]
- Zhang, Y.; Massel, K.; Godwin, I.D.; Gao, C. Applications and potential of genome editing in crop improvement. Genome Biol. 2018, 19, 210. [Google Scholar] [CrossRef] [PubMed]
- Malav, A.K.; Chandrawat, K.S.I. Gene pyramiding: An overview. Int. J. Curr. Res. Biosci. Plant Biol. 2016, 3, 22–28. [Google Scholar] [CrossRef]
- Hall, L.M.; Booker, H.; Siloto, R.M.P.; Jhala, A.J.; Weselake, R.J. Chapter 6—Flax (Linum usitatissimum L.). In Industrial Oil Crops; McKeon, T.A., Hayes, D.G., Hildebrand, D.F., Weselake, R.J., Eds.; AOCS Press: Urbana, IL, USA, 2016; pp. 157–194. [Google Scholar]
- Cui, Y.; Patel, J.; Zou, J.; Keller, W.A. Oilseed Brassicas. In Compendium Transgenic Crop Plants; Kole, C., Hall, T.C., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2009. [Google Scholar]
- Moose, S.P.; Mumm, R.H. Molecular Plant Breeding as the Foundation for 21st Century Crop Improvement. Plant Physiol. 2008, 147, 969–977. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.K.; Nayak, S. Gene pyramiding-A broad spectrum technique for developing durable stress resistance in crops. Biotechnol. Mol. Biol. Rev. 2010, 5, 51–60. [Google Scholar]
- Miah, G.; Rafii, M.Y.; Ismail, M.R.; Puteh, A.B.; Rahim, H.A.; Islam, K.N.; Latif, M.A. A review of microsatellite markers and their applications in rice breeding programs to improve blast disease resistance. Int. J. Mol. Sci. 2013, 14, 22499–22528. [Google Scholar] [CrossRef]
- Das, G.; Rao, G.J.N. Molecular marker assisted gene stacking for biotic and abiotic stress resistance genes in an elite rice cultivar. Front. Plant Sci. 2015, 6, 698. [Google Scholar] [CrossRef]
- Hayashi, K.; Hashimoto, N.; Daigen, M.; Ashikawa, I. Development of PCR-based SNP markers for rice blast resistance genes at the Piz locus. Theor. Appl. Genet. 2004, 108, 1212–1220. [Google Scholar] [CrossRef]
- Liu, W.; Maurer, H.P.; Li, G.; Tucker, M.R.; Gowda, M.; Weissmann, E.A.; Hahn, V.; Würschum, T. Genetic architecture of winter hardiness and frost tolerance in triticale. PLoS ONE 2014, 9, e99848. [Google Scholar] [CrossRef]
- Tullu, A.; Tar’an, B.; Warkentin, T.; Vandenberg, A. Construction of an Intraspecific Linkage Map and QTL Analysis for Earliness and Plant Height in Lentil. Crop Sci. 2008, 48, 2254–2264. [Google Scholar] [CrossRef]
- Taylor, P.W.J.; Ades, P.K.; Ford, R. QTL mapping of resistance in lentil (Lens culinaris ssp. culinaris) to ascochyta blight (Ascochyta lentis). Plant Breed. 2006, 125, 506–512. [Google Scholar] [CrossRef]
- Fedoruk, M.J.; Vandenberg, A.; Bett, K.E. Quantitative Trait Loci Analysis of Seed Quality Characteristics in Lentil using Single Nucleotide Polymorphism Markers. Plant Genome 2013, 6. [Google Scholar] [CrossRef]
- Van Eeuwijk, F.A.; Bustos-Korts, D.; Millet, E.J.; Boer, M.P.; Kruijer, W.; Thompson, A.; Malosetti, M.; Iwata, H.; Quiroz, R.; Kuppe, C.; et al. Modelling strategies for assessing and increasing the effectiveness of new phenotyping techniques in plant breeding. Plant Sci. 2019, 282, 23–39. [Google Scholar] [CrossRef]
- Ahmed, M.M.; Huang, C.; Shen, C.; Khan, A.Q.; Lin, Z. Map-based cloning of qBWT-c12 discovered brassinosteroid-mediated control of organ size in cotton. Plant Sci. 2020, 291, 110315. [Google Scholar] [CrossRef] [PubMed]
- Ribaut, J.M.; de Vicente, M.C.; Delannay, X. Molecular breeding in developing countries: Challenges and perspectives. Curr. Opin. Plant Biol. 2010, 13, 213–218. [Google Scholar] [CrossRef]
- Collard, B.C.Y.; Mackill, D.J. Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008, 363, 557–572. [Google Scholar] [CrossRef] [PubMed]
- Chukwu, S.C.; Rafii, M.Y.; Ramlee, S.I.; Ismail, S.I.; Hasan, M.M.; Oladosu, Y.A.; Magaji, U.G.; Akos, I.; Olalekan, K.K. Bacterial leaf blight resistance in rice: A review of conventional breeding to molecular approach. Mol. Biol. Rep. 2019, 46, 1519–1532. [Google Scholar] [CrossRef] [PubMed]
- Oladosu, Y.; Rafii, M.Y.; Magaji, U.; Abdullah, N.; Miah, G.; Chukwu, S.C.; Hussin, G.; Ramli, A.; Kareem, I. Genotypic and Phenotypic Relationship among Yield Components in Rice under Tropical Conditions. Biomed Res. Int. 2018, 2018, 8936767. [Google Scholar] [CrossRef]
- Quibod, I.L.; Perez-Quintero, A.; Booher, N.J.; Dossa, G.S.; Grande, G.; Szurek, B.; Vera Cruz, C.; Bogdanove, A.J.; Oliva, R. Effector Diversification Contributes to Xanthomonas oryzae pv. oryzae Phenotypic Adaptation in a Semi-Isolated Environment. Sci. Rep. 2016, 6, 34137. [Google Scholar] [CrossRef]
- Das, G.; Patra, J.K.; Baek, K.-H. Insight into MAS: A Molecular Tool for Development of Stress Resistant and Quality of Rice through Gene Stacking. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Poczai, P.; Varga, I.; Laos, M.; Cseh, A.; Bell, N.; Valkonen, J.P.T.; Hyvönen, J. Advances in plant gene-targeted and functional markers: A review. Plant Methods 2013, 9, 6. [Google Scholar] [CrossRef]
- Wang, Z.; Baulcombe, D.C. Transposon age and non-CG methylation. Nat. Commun. 2020, 11, 1221. [Google Scholar] [CrossRef] [PubMed]
- Shirasawa, K.; Hirakawa, H. DNA marker applications to molecular genetics and genomics in tomato. Breed. Sci. 2013, 63, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Bohle, H.M.; Gabaldón, T. Selection of marker genes using whole-genome DNA polymorphism analysis. Evol. Bioinform. Online 2012, 8, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Nadeem, M.A.; Nawaz, M.A.; Shahid, M.Q.; Doğan, Y.; Comertpay, G.; Yıldız, M.; Hatipoğlu, R.; Ahmad, F.; Alsaleh, A.; Labhane, N.; et al. DNA molecular markers in plant breeding: Current status and recent advancements in genomic selection and genome editing. Biotechnol. Biotechnol. Equip. 2018, 32, 261–285. [Google Scholar] [CrossRef]
- Tong, H.; Küken, A.; Nikoloski, Z. Integrating molecular markers into metabolic models improves genomic selection for Arabidopsis growth. Nat. Commun. 2020, 11, 2410. [Google Scholar] [CrossRef]
- Pilet-Nayel, M.-L.; Moury, B.; Caffier, V.; Montarry, J.; Kerlan, M.-C.; Fournet, S.; Durel, C.-E.; Delourme, R. Quantitative Resistance to Plant Pathogens in Pyramiding Strategies for Durable Crop Protection. Front. Plant Sci. 2017, 8, 1838. [Google Scholar] [CrossRef]
- Ye, Y.; Cai, M.; Ju, Y.; Jiao, Y.; Feng, L.; Pan, H.; Cheng, T.; Zhang, Q. Identification and Validation of SNP Markers Linked to Dwarf Traits Using SLAF-Seq Technology in Lagerstroemia. PLoS ONE 2016, 11, e0158970. [Google Scholar] [CrossRef]
- Kushwah, A.; Gupta, S.; Bindra, S.; Johal, N.; Singh, I.; Bharadwaj, C.; Dixit, G.P.; Gaur, P.M.; Nayyar, H.; Singh, S. Chapter 6—Gene pyramiding and multiple character breeding. In Chickpea: Crop Wild Relatives for Enhancing Genetic Gains; Academic Press: Cambridge, MA, USA, 2020; pp. 131–165. [Google Scholar]
- Pazhamala, L.; Saxena, R.K.; Singh, V.K.; Sameerkumar, C.V.; Kumar, V.; Sinha, P.; Patel, K.; Obala, J.; Kaoneka, S.R.; Tongoona, P.; et al. Genomics-assisted breeding for boosting crop improvement in pigeonpea (Cajanus cajan). Front. Plant Sci. 2015, 6, 50. [Google Scholar] [CrossRef]
- Hu, J.; Xiao, C.; He, Y. Recent progress on the genetics and molecular breeding of brown planthopper resistance in rice. Rice 2016, 9, 30. [Google Scholar] [CrossRef]
- Jiang, G.L. Molecular Marker-Assisted Breeding: A Plant Breeder’s Review. In Advances in Plant Breeding Strategies: Breeding, Biotechnology and Molecular Tools; Al-Khayri, J., Jain, S., Johns, D., Eds.; Springer: Cham, Switzerland, 2015. [Google Scholar]
- Panigrahi, J.; Mishra, R.R.; Sahu, A.R.; Rath, S.C.; Kole, C.R. Marker-Assisted Breeding for Stress Resistance in Crop Plants. In Advances in Plant Breeding Strategies: Breeding, Biotechnology and Molecular Tools; Al-Khayri, J., Jain, S., Johnson, D., Eds.; Springer: Cham, Switzerland, 2013. [Google Scholar]
- Rezene, Y.; Tesfaye, K.; Mukankusi, C.; Gepts, P. Marker-Assisted Pyramiding Resistance Genes Against Angular Leaf Spot and Common Bacterial Blight Disease into Preferred Common Bean Cultivar “REDWOLAITA”. Adv. Crop Sci. Technol. 2019, 7, 416. [Google Scholar] [CrossRef]
- Jiang, W.; Zhou, H.; Bi, H.; Fromm, M.; Yang, B.; Weeks, D.P. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013, 41, e188. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Wang, S.; Su, Z.; Li, T.; Zhang, X.; Bai, G. Meta-analysis of QTL for Fusarium head blight resistance in Chinese wheat landraces. Crop J. 2019, 7, 784–798. [Google Scholar] [CrossRef]
- Almeida, G.D.; Makumbi, D.; Magorokosho, C.; Nair, S.; Borém, A.; Ribaut, J.-M.; Bänziger, M.; Prasanna, B.M.; Crossa, J.; Babu, R. QTL mapping in three tropical maize populations reveals a set of constitutive and adaptive genomic regions for drought tolerance. Theor. Appl. Genet. 2013, 126, 583–600. [Google Scholar] [CrossRef] [PubMed]
- Crosbie, T.M.; Eathington, S.R.; Johnson, G.R.; Edwards, M.; Reiter, R.; Stark, S.; Mohanty, R.G.; Oyervides, M.; Buehler, R.E.; Walker, A.K.; et al. Plant Breeding: Past, Present, and Future. In Plant Breeding: The Arnel R. Hallauer International Symposium; Lamkey, K.R., Lee, M., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2006. [Google Scholar]
- Gu, R.; Chen, F.; Long, L.; Cai, H.; Liu, Z.; Yang, J.; Wang, L.; Li, H.; Li, J.; Liu, W.; et al. Enhancing phosphorus uptake efficiency through QTL-based selection for root system architecture in maize. J. Genet. Genom. 2016, 43, 663–672. [Google Scholar] [CrossRef]
- Huang, X.; Feng, Q.; Qian, Q.; Zhao, Q.; Wang, L.; Wang, A.; Guan, J.; Fan, D.; Weng, Q.; Huang, T.; et al. High-throughput genotyping by whole-genome resequencing. Genome Res. 2009, 19, 1068–1076. [Google Scholar] [CrossRef]
- Xie, W.; Feng, Q.; Yu, H.; Huang, X.; Zhao, Q.; Xing, Y.; Yu, S.; Han, B.; Zhang, Q. Parent-independent genotyping for constructing an ultrahigh-density linkage map based on population sequencing. Proc. Natl. Acad. Sci. USA 2010, 107, 10578–10583. [Google Scholar] [CrossRef]
- Burbano, H.A.; Hodges, E.; Green, R.E.; Briggs, A.W.; Krause, J.; Meyer, M.; Good, J.M.; Maricic, T.; Johnson, P.L.F.; Xuan, Z.; et al. Targeted Investigation of the Neandertal Genome by Array-Based Sequence Capture. Science 2010, 328, 723–725. [Google Scholar] [CrossRef]
- Potokina, E.; Druka, A.; Luo, Z.; Wise, R.; Waugh, R.; Kearsey, M. Gene expression quantitative trait locus analysis of 16,000 barley genes reveals a complex pattern of genome-wide transcriptional regulation. Plant J. 2008, 53, 90–101. [Google Scholar] [CrossRef]
- Druka, A.; Druka, I.; Centeno, A.G.; Li, H.; Sun, Z.; Thomas, W.T.B.; Bonar, N.; Steffenson, B.J.; Ullrich, S.E.; Kleinhofs, A.; et al. Towards systems genetic analyses in barley: Integration of phenotypic, expression and genotype data into GeneNetwork. BMC Genet. 2008, 9, 73. [Google Scholar] [CrossRef]
- Marino, R.; Ponnaiah, M.; Krajewski, P.; Frova, C.; Gianfranceschi, L.; Pè, M.E.; Sari-Gorla, M. Addressing drought tolerance in maize by transcriptional profiling and mapping. Mol. Genet. Genom. 2009, 281, 163–179. [Google Scholar] [CrossRef]
- Ribaut, J.-M.; Ragot, M. Marker-assisted selection to improve drought adaptation in maize: The backcross approach, perspectives, limitations, and alternatives. J. Exp. Bot. 2006, 58, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Bankole, F.; Menkir, A.; Olaoye, G.; Crossa, J.; Hearne, S.; Unachukwu, N.; Gedil, M. Genetic Gains in Yield and Yield Related Traits under Drought Stress and Favorable Environments in a Maize Population Improved Using Marker Assisted Recurrent Selection. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Zhang, L.; DeLacy, I.; Arief, V.; Dieters, M.; Pfeiffer, W.H.; Wang, J.; Li, H. Modeling and simulation of recurrent phenotypic and genomic selections in plant breeding under the presence of epistasis. Crop J. 2020. [Google Scholar] [CrossRef]
- EFSA(European-Food-sqafety-Authority); Aguilera, J.; Aguilera-Gomez, M.; Barrucci, F.; Cocconcelli, P.S.; Davies, H.; Denslow, N.; Dorne, J.L.; Grohmann, L. EFSA Scientific Colloquium 24-‘omics in risk assessment: State of the art and next steps. EFSA Support. Publ. 2018, 15, 1512. [Google Scholar] [CrossRef]
- Varshney, R.K.; Graner, A.; Sorrells, M.E. Genomics-assisted breeding for crop improvement. Trends Plant Sci. 2005, 10, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.K.; Varshney, R.K. Cereal Genomics: An Overview. In Cereal Genomics; Gupta, P.K., Varshney, R.K., Eds.; Springer: Dordrecht, The Netherlands, 2004. [Google Scholar]
- Jacobs, D.I.; van der Heijden, R.; Verpoorte, R. Proteomics in plant biotechnology and secondary metabolism research. Phytochem. Anal. 2000, 11, 277–287. [Google Scholar] [CrossRef]
- Hirano, H.; Islam, N.; Kawasaki, H. Technical aspects of functional proteomics in plants. Phytochemistry 2004, 65, 1487–1498. [Google Scholar] [CrossRef]
- Bhushan, D.; Pandey, A.; Choudhary, M.K.; Datta, A.; Chakraborty, S.; Chakraborty, N. Comparative proteomics analysis of differentially expressed proteins in chickpea extracellular matrix during dehydration stress. Mol. Cell. Proteom. 2007, 6, 1868–1884. [Google Scholar] [CrossRef]
- Pandey, A.; Chakraborty, S.; Datta, A.; Chakraborty, N. Proteomics approach to identify dehydration responsive nuclear proteins from chickpea (Cicer arietinum L.). Mol. Cell. Proteom. 2008, 7, 88–107. [Google Scholar] [CrossRef]
- Razzaq, A.; Sadia, B.; Raza, A.; Khalid, H.M.; Saleem, F. Metabolomics: A Way Forward for Crop Improvement. Metabolites 2019, 9, 303. [Google Scholar] [CrossRef]
- Piasecka, A.; Kachlicki, P.; Stobiecki, M. Analytical Methods for Detection of Plant Metabolomes Changes in Response to Biotic and Abiotic Stresses. Int. J. Mol. Sci. 2019, 20, 379. [Google Scholar] [CrossRef]
- Sung, J.; Lee, S.; Lee, Y.; Ha, S.; Song, B.; Kim, T.; Waters, B.M.; Krishnan, H.B. Metabolomic profiling from leaves and roots of tomato (Solanum lycopersicum L.) plants grown under nitrogen, phosphorus or potassium-deficient condition. Plant Sci. 2015, 241, 55–64. [Google Scholar] [CrossRef]
- Dawid, C.; Hille, K. Functional Metabolomics—A useful tool to characterize stress-induced metabolome alterations opening new avenues towards tailoring food crop quality. Agronomy 2018, 8, 138. [Google Scholar] [CrossRef]
- Fernie, A.R.; Schauer, N. Metabolomics-assisted breeding: A viable option for crop improvement? Trends Genet. 2009, 25, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Sauer, U.G.; Deferme, L.; Gribaldo, L.; Hackermüller, J.; Tralau, T.; van Ravenzwaay, B.; Yauk, C.; Poole, A.; Tong, W.; Gant, T.W. The challenge of the application of ‘omics technologies in chemicals risk assessment: Background and outlook. Regul. Toxicol. Pharmacol. 2017, 91, S14–S26. [Google Scholar] [CrossRef] [PubMed]
- Horgan, R.P.; Kenny, L.C. ‘Omic’ technologies: Genomics, transcriptomics, proteomics and metabolomics. Obstet. Gynaecol. 2011, 13, 189–195. [Google Scholar] [CrossRef]
- Rakwal, R.; Hayashi, G.; Shibato, J.; Deepak, S.A.; Gundimeda, S.; Simha, U.; Padmanaban, A.; Gupta, R.; Han, S.-I.; Kim, S.T.; et al. Progress Toward Rice Seed OMICS in Low-Level Gamma Radiation Environment in Iitate Village, Fukushima. J. Hered. 2017, 109, 206–211. [Google Scholar] [CrossRef]
- Heinig, U.; Gutensohn, M.; Dudareva, N.; Aharoni, A. The challenges of cellular compartmentalization in plant metabolic engineering. Curr. Opin. Biotechnol. 2013, 24, 239–246. [Google Scholar] [CrossRef]
- Saito, K.; Matsuda, F. Metabolomics for functional genomics, systems biology, and biotechnology. Annu. Rev. Plant Biol. 2010, 61, 463–489. [Google Scholar] [CrossRef]
- Tugizimana, F.; Mhlongo, M.I.; Piater, L.A.; Dubery, I.A. Metabolomics in Plant Priming Research: The Way Forward? Int. J. Mol. Sci. 2018, 19, 1759. [Google Scholar] [CrossRef]
- Agarrwal, R.; Nair, S. Chapter 16—Metabolomics-assisted crop improvement. In Advancement in Crop Improvement Techniques; Tuteja, N., Tuteja, R., Passricha, N., Saifi, S.K., Eds.; Woodhead Publishing: Cambridge, UK, 2020; pp. 263–274. [Google Scholar]
- Perumalsamy, S.; Bharani, M.; Sudha, M.; Nagarajan, P.; Arul, L.; Saraswathi, R.; Balasubramanian, P.; Ramalingam, J. Functional marker-assisted selection for bacterial leaf blight resistance genes in rice (Oryza sativa L.). Plant Breed. 2010, 129, 400–406. [Google Scholar] [CrossRef]
- Arunakumari, K.; Durgarani, C.V.; Satturu, V.; Sarikonda, K.R.; Chittoor, P.D.R.; Vutukuri, B.; Laha, G.S.; Nelli, A.P.K.; Gattu, S.; Jamal, M.; et al. Marker-Assisted Pyramiding of Genes Conferring Resistance Against Bacterial Blight and Blast Diseases into Indian Rice Variety MTU1010. Rice Sci. 2016, 23, 306–316. [Google Scholar] [CrossRef]
- Van Ooijen, J.W. JoinMap®4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations; Kyazma BV: Wageningen, The Netherlands, 2006. [Google Scholar]
- Richards, E.J. Inherited epigenetic variation—Revisiting soft inheritance. Nat. Rev. Genet. 2006, 7, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Sandhu, N.; Dixit, S.; Yadav, S.; Swamy, B.P.M.; Shamsudin, N.A.A. Marker-assisted selection strategy to pyramid two or more QTLs for quantitative trait-grain yield under drought. Rice 2018, 11, 35. [Google Scholar] [CrossRef] [PubMed]
- Angeles-Shim, R.B.; Reyes, V.P.; del Valle, M.M.; Lapis, R.S.; Shim, J.; Sunohara, H.; Jena, K.K.; Ashikari, M.; Doi, K. Marker-Assisted Introgression of Quantitative Resistance Gene pi21 Confers Broad Spectrum Resistance to Rice Blast. Rice Sci. 2020, 27, 113–123. [Google Scholar] [CrossRef]
- Zorrilla-Fontanesi, Y.; Cabeza, A.; Domínguez, P.; Medina, J.J.; Valpuesta, V.; Denoyes-Rothan, B.; Sánchez-Sevilla, J.F.; Amaya, I. Quantitative trait loci and underlying candidate genes controlling agronomical and fruit quality traits in octoploid strawberry (Fragaria × ananassa). Theor. Appl. Genet. 2011, 123, 755–778. [Google Scholar] [CrossRef]
- Verma, S.; Zurn, J.D.; Salinas, N.; Mathey, M.M.; Denoyes, B.; Hancock, J.F.; Finn, C.E.; Bassil, N.V.; Whitaker, V.M. Clarifying sub-genomic positions of QTLs for flowering habit and fruit quality in U.S. strawberry (Fragaria × ananassa) breeding populations using pedigree-based QTL analysis. Hortic. Res. 2017, 4, 17062. [Google Scholar] [CrossRef]
- Xin, F.; Zhu, T.; Wei, S.; Han, Y.; Zhao, Y.; Zhang, D.; Ma, L.; Ding, Q. QTL Mapping of Kernel Traits and Validation of a Major QTL for Kernel Length-Width Ratio Using SNP and Bulked Segregant Analysis in Wheat. Sci. Rep. 2020, 10, 25. [Google Scholar] [CrossRef]
- Yousef, G.G.; Juvik, J.A. Comparison of Phenotypic and Marker-Assisted Selection for Quantitative Traits in Sweet Corn. Crop Sci. 2001, 41, 645–655. [Google Scholar] [CrossRef]
- Robert, V.J.M.; West, M.A.L.; Inai, S.; Caines, A.; Arntzen, L.; Smith, J.K.; Clair, D.A. Marker-assisted introgression of blackmold resistance QTL alleles from wild Lycopersicon cheesmanii to cultivated tomato (L. esculentum) and evaluation of QTL phenotypic effects. Mol. Breed. 2001, 8, 217–233. [Google Scholar] [CrossRef]
- Śliwka, J.; Jakuczun, H.; Chmielarz, M.; Hara, S.A.; Tomczyńska, I.; Kilian, A.; Zimnoch-Guzowska, E. Late blight resistance gene from Solanum ruiz-ceballosii is located on potato chromosome X and linked to violet flower colour. BMC Genet. 2012, 13, 11. [Google Scholar] [CrossRef]
- Maruthasalam, S.; Kalpana, K.; Kumar, K.K.; Loganathan, M.; Poovannan, K.; Raja, J.A.J.; Kokiladevi, E.; Samiyappan, R.; Sudhakar, D.; Balasubramanian, P. Pyramiding transgenic resistance in elite indica rice cultivars against the sheath blight and bacterial blight. Plant Cell Rep. 2007, 26, 791–804. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.E.; Bradley, J.R.J.; Van Duyn, J.W. Performance of Feral and Cry1Ac-Selected Helicoverpa zea (Lepidoptera: Noctuidae) Strains on Transgenic Cottons Expressing One or Two Bacillus thuringiensis ssp. kurstaki Proteins Under Greenhouse Conditions. J. Entomol. Sci. 2004, 39, 46–55. [Google Scholar] [CrossRef]
- Puspito, A.N.; Rao, A.Q.; Hafeez, M.N.; Iqbal, M.S.; Bajwa, K.S.; Ali, Q.; Rashid, B.; Abbas, M.A.; Latif, A.; Shahid, A.A.; et al. Transformation and Evaluation of Cry1Ac+Cry2A and GTGene in Gossypium hirsutum L. Front. Plant Sci. 2015, 6, 943. [Google Scholar] [CrossRef]
- Zhang, B.; Chi, D.; Hiebert, C.; Fetch, T.; McCallum, B.; Xue, A.; Cao, W.; Depauw, R.; Fedak, G. Pyramiding stem rust resistance genes to race TTKSK (Ug99) in wheat. Can. J. Plant Pathol. 2019, 41, 443–449. [Google Scholar] [CrossRef]
- Ali, M.A.; Shahzadi, M.; Zahoor, A.; Dababat, A.A.; Toktay, H.; Bakhsh, A.; Nawaz, M.A.; Li, H. Resistance to Cereal Cyst Nematodes in Wheat and Barley: An Emphasis on Classical and Modern Approaches. Int. J. Mol. Sci. 2019, 20, 432. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Santi, D.V. m5C RNA and m5C DNA methyl transferases use different cysteine residues as catalysts. Proc. Natl. Acad. Sci. USA 2000, 97, 8263–8265. [Google Scholar] [CrossRef]
- Joseph, M.; Gopalakrishnan, S.; Sharma, R.K.; Singh, V.P.; Singh, A.K.; Singh, N.K.; Mohapatra, T. Combining bacterial blight resistance and Basmati quality characteristics by phenotypic and molecular marker-assisted selection in rice. Mol. Breed. 2004, 13, 377–387. [Google Scholar] [CrossRef]
- Singh, S.; Sidhu, J.S.; Huang, N.; Vikal, Y.; Li, Z.; Brar, D.S.; Dhaliwal, H.S.; Khush, G.S. Pyramiding three bacterial blight resistance genes (xa5, xa13 and Xa21) using marker-assisted selection into indica rice cultivar PR106. Theor. Appl. Genet. 2001, 102, 1011–1015. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, L.; Liu, Y.; Dai, H.; He, J.; Kang, H.; Pan, G.; Huang, J.; Qiu, Z.; Wang, Q.; et al. Marker assisted pyramiding of two brown planthopper resistance genes, Bph3 and Bph27 (t), into elite rice Cultivars. Rice 2016, 9, 27. [Google Scholar] [CrossRef]
- Narayanan, N.N.; Baisakh, N.; Vera Cruz, C.M.; Gnanamanickam, S.S.; Datta, K.; Datta, S.K. Molecular Breeding for the Development of Blast and Bacterial Blight Resistance in Rice cv. IR50. Crop Sci. 2002, 42, 2072–2079. [Google Scholar] [CrossRef]
- Datta, K.; Baisakh, N.; Thet, K.M.; Tu, J.; Datta, S.K. Pyramiding transgenes for multiple resistance in rice against bacterial blight, yellow stem borer and sheath blight. Theor. Appl. Genet. 2002, 106, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Verlaan, M.G.; Hutton, S.F.; Ibrahem, R.M.; Kormelink, R.; Visser, R.G.F.; Scott, J.W.; Edwards, J.D.; Bai, Y. The Tomato Yellow Leaf Curl Virus resistance genes Ty-1 and Ty-3 are allelic and code for DFDGD-class RNA-dependent RNA polymerases. PLoS Genet. 2013, 9, e1003399. [Google Scholar] [CrossRef]
- Werner, K.; Friedt, W.; Ordon, F. Strategies for Pyramiding Resistance Genes Against the Barley Yellow Mosaic Virus Complex (BaMMV, BaYMV, BaYMV-2). Mol. Breed. 2005, 16, 45–55. [Google Scholar] [CrossRef]
- Castro, A.J.; Capettini, F.; Corey, A.E.; Filichkina, T.; Hayes, P.M.; Kleinhofs, A.; Kudrna, D.; Richardson, K.; Sandoval-Islas, S.; Rossi, C.; et al. Mapping and pyramiding of qualitative and quantitative resistance to stripe rust in barley. Theor. Appl. Genet. 2003, 107, 922–930. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Zhang, T.; Bai, S.; Wang, Z.; He, K. Evaluation of Bt Corn with Pyramided Genes on Efficacy and Insect Resistance Management for the Asian Corn Borer in China. PLoS ONE 2016, 11, e0168442. [Google Scholar] [CrossRef] [PubMed]
- Mehrotra, M.; Singh, A.K.; Sanyal, I.; Altosaar, I.; Amla, D.V. Pyramiding of modified cry1Ab and cry1Ac genes of Bacillus thuringiensis in transgenic chickpea (Cicer arietinum L.) for improved resistance to pod borer insect Helicoverpa armigera. Euphytica 2011, 182, 87. [Google Scholar] [CrossRef]
- Djian, C.; Palloix, A.; Fazari, A.; Marteu, N.; Barbary, A.; Abad, P.; Sage-Palloix, A.-M.; Mateille, T.; Risso, S.; Lanza, R.; et al. Pyramiding, alternating or mixing: Comparative performances of deployment strategies of nematode resistance genes to promote plant resistance efficiency and durability. BMC Plant Biol. 2014, 14, 53. [Google Scholar] [CrossRef]
- Yang, Z.; Huang, D.; Tang, W.; Zheng, Y.; Liang, K.; Cutler, A.J.; Wu, W. Mapping of quantitative trait loci underlying cold tolerance in rice seedlings via high-throughput sequencing of pooled extremes. PLoS ONE 2013, 8, e68433. [Google Scholar] [CrossRef]
- Fujino, K.; Hirayama, Y.; Kaji, R. Marker-assisted selection in rice breeding programs in Hokkaido. Breed. Sci. 2019, 69, 383–392. [Google Scholar] [CrossRef]
- Lin, R.; Zhao, W.; Meng, X.; Wang, M.; Peng, Y. Rice gene OsNAC19 encodes a novel NAC-domain transcription factor and responds to infection by Magnaporthe grisea. Plant Sci. 2007, 172, 120–130. [Google Scholar] [CrossRef]
- Kim, Y.S.; Kim, I.S.; Shin, S.Y.; Park, T.H.; Park, H.M.; Kim, Y.H.; Lee, G.S.; Kang, H.G.; Lee, S.H.; Yoon, H.S. Overexpression of Dehydroascorbate Reductase Confers Enhanced Tolerance to Salt Stress in Rice Plants (Oryza sativa L. japonica). J. Agron. Crop Sci. 2014, 200, 444–456. [Google Scholar] [CrossRef]
- Wang, A.; Yu, X.; Mao, Y.; Liu, Y.; Liu, G.; Liu, Y.; Niu, X. Overexpression of a small heat-shock-protein gene enhances tolerance to abiotic stresses in rice. Plant Breed. 2015, 134, 384–393. [Google Scholar] [CrossRef]
- Kurokawa, Y.; Noda, T.; Yamagata, Y.; Angeles-Shim, R.; Sunohara, H.; Uehara, K.; Furuta, T.; Nagai, K.; Jena, K.K.; Yasui, H.; et al. Construction of a versatile SNP array for pyramiding useful genes of rice. Plant Sci. 2016, 242, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Roorkiwal, M.; Kale, S.; Garg, V.; Yadala, R.; Varshney, R.K. InDel markers: An extended marker resource for molecular breeding in chickpea. PLoS ONE 2019, 14, e0213999. [Google Scholar] [CrossRef] [PubMed]
- Gelli, M.; Konda, A.R.; Liu, K.; Zhang, C.; Clemente, T.E.; Holding, D.R.; Dweikat, I.M. Validation of QTL mapping and transcriptome profiling for identification of candidate genes associated with nitrogen stress tolerance in sorghum. BMC Plant Biol. 2017, 17, 123. [Google Scholar] [CrossRef]
- Forster, B.P.; Ellis, R.P.; Thomas, W.T.B.; Newton, A.C.; Tuberosa, R.; This, D.; El-Enein, R.A.; Bahri, M.H.; Ben Salem, M. The development and application of molecular markers for abiotic stress tolerance in barley. J. Exp. Bot. 2000, 51, 19–27. [Google Scholar] [CrossRef]
- Collard, B.C.Y.; Jahufer, M.Z.Z.; Brouwer, J.B.; Pang, E.C.K. An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: The basic concepts. Euphytica 2005, 142, 169–196. [Google Scholar] [CrossRef]
- Nevame, A.Y.M.; Xia, L.; Nchongboh, C.G.; Hasan, M.M.; Alam, M.A.; Yongbo, L.; Wenting, Z.; Yafei, H.; Emon, R.M.; Ismail, M.R.; et al. Development of a New Molecular Marker for the Resistance to Tomato Yellow Leaf Curl Virus. Biomed Res. Int. 2018, 2018, 8120281. [Google Scholar] [CrossRef]
- Beukeboom, L.W.; Niehuis, O.; Pannebakker, B.A.; Koevoets, T.; Gibson, J.D.; Shuker, D.M.; van de Zande, L.; Gadau, J. A comparison of recombination frequencies in intraspecific versus interspecific mapping populations of Nasonia. Heredity 2010, 104, 302–309. [Google Scholar] [CrossRef]
- Mageto, E.K.; Lee, M.; Dhliwayo, T.; Palacios-Rojas, N.; San Vicente, F.; Burgueño, J.; Hallauer, A.R. An Evaluation of Kernel Zinc in Hybrids of Elite Quality Protein Maize (QPM) and Non-QPM Inbred Lines Adapted to the Tropics Based on a Mating Design. Agronomy 2020, 10, 695. [Google Scholar] [CrossRef]
- Wang, X.; Xu, Y.; Hu, Z.; Xu, C. Genomic selection methods for crop improvement: Current status and prospects. Crop J. 2018, 6, 330–340. [Google Scholar] [CrossRef]
- Bhatia, D. Advanced Quantitative Genetics Technologies for Accelerating Plant Breeding. In Accelerated Plant Breeding; Gosal, S., Wani, S., Eds.; Springer: Cham, Switzerland, 2020; Volume 1. [Google Scholar]
- Weiwei, W.; Haijun, L.; Zhou, Y.; Min, J.; Ning, Y.; Dong, L.; Jie, L.; Yingjie, X.; Qingchun, P.; Takayuki, T.; et al. Combining Quantitative Genetics Approaches with Regulatory Network Analysis to Dissect the Complex Metabolism of the Maize Kernel. Plant Physiol. 2016, 170, 136–146. [Google Scholar]
- Watson, A.; Ghosh, S.; Williams, M.J.; Cuddy, W.; Simmonds, J.; Rey, M.-D.; Asyraf Hatta, M.; Hinchliffe, A.; Steed, A.; Reynolds, D.; et al. Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants 2018, 4, 23–29. [Google Scholar] [CrossRef]
- Gosal, S.S.; Pathak, D.; Wani, S.H.; Vij, S.; Pathak, M. Accelerated Breeding of Plants: Methods and Applications. In Accelerated Plant Breeding; Gosal, S., Wani, S., Eds.; Springer: Cham, Switzerland, 2020; Volume 1. [Google Scholar]
- Gao, H.; Christensen, O.F.; Madsen, P.; Nielsen, U.S.; Zhang, Y.; Lund, M.S.; Su, G. Comparison on genomic predictions using three GBLUP methods and two single-step blending methods in the Nordic Holstein population. Genet. Sel. Evol. 2012, 44, 8. [Google Scholar] [CrossRef] [PubMed]
- Libbrecht, M.W.; Noble, W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015, 16, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Wen, X. Bayesian model selection in complex linear systems, as illustrated in genetic association studies. Biometrics 2014, 70, 73–83. [Google Scholar] [CrossRef]
- Devi, E.L.; Devi, C.P.; Kumar, S.; Sharma, S.K.; Beemrote, A.; Chongtham, S.K.; Singh, C.H.; Tania, C.; Singh, T.B.; Ningombam, A.; et al. Marker assisted selection (MAS) towards generating stress tolerant crop plants. Plant Gene 2017, 11, 205–218. [Google Scholar] [CrossRef]

| Crop | Traits | Pyramided Genes | References |
|---|---|---|---|
| Biotic stress tolerance | |||
| Potato | Late blight resistance | Rpi-phu 1, Rpi-rzc | [147] |
| Cotton | Bacterial blight/sheath resistance | Chi11, t1p, Xa21 | [148] |
| Bollworm resistance | Cry1Ac, Cry2Ab | [149] | |
| Weed and pathogen resistance | ptxD/Phi | [6] | |
| Insect pest resistance | Cry1Ac, Cry2Ac | [150] | |
| Wheat | Leaf and stem rust resistance | SrCad, Sr33, Lr34, Fhb | [151] |
| Cereal cyst nematode resistance | CreX, CreY, CRISPR-Cas9 | [152] | |
| Aphid resistance | Gn2, Gn4 | [153] | |
| Rice | Gall midge resistance | Gm1, Gm2, Gm4 | [75] |
| Blast resistance | Pi(2)t, Pi25, Pi(t)a, Xa4, Xa5, Xa13, Xa21 | [154,155] | |
| BPH resistance | Bph1, Bph2 | [156] | |
| Blight resistance | Xa5, Xa13, Xa21 | [157] | |
| Bacterial, sheath blight, stem borer | Xa12, Rc7, Cry1AB1, Cry14c | [158] | |
| Soybean | Mosaic virus resistance | Rsv1, Rsv3, Rsv4 | [30] |
| Tomato | Leaf curl/spotted virus | Ty-1, Ty-3, Sw-5 | [159] |
| Barley | Mosaic virus resistance | rym4, rym5, rym9, rym11 | [160] |
| Strip rust resistance | 3 QTL | [161] | |
| Com | Com borer resistance | Cry1le, Cry1Ac | [162] |
| Chickpea | Lepidopteran resistance | Cry1Ac, Cry1Ab | [163] |
| Pepper | Root-knot nematode resistance | Me1, Me2 | [164] |
| Abiotic stress tolerance | |||
| Rice | Cold tolerance | 9PssT-3, 9PssT-7, 9PssT9, | [165] |
| Cold tolerance | 9SCT1a, 9SCT2 | [166] | |
| Drought tolerance | Soltol | [75] | |
| Drought tolerance | QTLs | [167] | |
| Cold tolerance | qPSST-3, qPSST-7, qPSST-9, qSCT1a, TSF4-1 | [168] | |
| Heat, drought, salt, and cold resistance | OsHSP18.6 | [169] | |
| Quantitative and qualitative traits | |||
| Cereal | High yield | Gn1a/OsCKX2, APO1, WFP/OsSPL 14 | [170] |
| Seed shape | GW2, GS 3, 9SW5 | [170] | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dormatey, R.; Sun, C.; Ali, K.; Coulter, J.A.; Bi, Z.; Bai, J. Gene Pyramiding for Sustainable Crop Improvement against Biotic and Abiotic Stresses. Agronomy 2020, 10, 1255. https://doi.org/10.3390/agronomy10091255
Dormatey R, Sun C, Ali K, Coulter JA, Bi Z, Bai J. Gene Pyramiding for Sustainable Crop Improvement against Biotic and Abiotic Stresses. Agronomy. 2020; 10(9):1255. https://doi.org/10.3390/agronomy10091255
Chicago/Turabian StyleDormatey, Richard, Chao Sun, Kazim Ali, Jeffrey A. Coulter, Zhenzhen Bi, and Jiangping Bai. 2020. "Gene Pyramiding for Sustainable Crop Improvement against Biotic and Abiotic Stresses" Agronomy 10, no. 9: 1255. https://doi.org/10.3390/agronomy10091255
APA StyleDormatey, R., Sun, C., Ali, K., Coulter, J. A., Bi, Z., & Bai, J. (2020). Gene Pyramiding for Sustainable Crop Improvement against Biotic and Abiotic Stresses. Agronomy, 10(9), 1255. https://doi.org/10.3390/agronomy10091255








