A Trp574Leu Target-Site Mutation Confers Imazamox Resistance in Multiple Herbicide-Resistant Wild Poinsettia Populations from Brazil
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Whole-Plant Imazamox Dose-Response
2.3. ALS Enzyme Assay
2.4. ALS Sequencing
2.5. Homology Modeling of S- and R-ALS from Wild Poinsettia
2.6. Docking of herbicides to S- and R-ALS
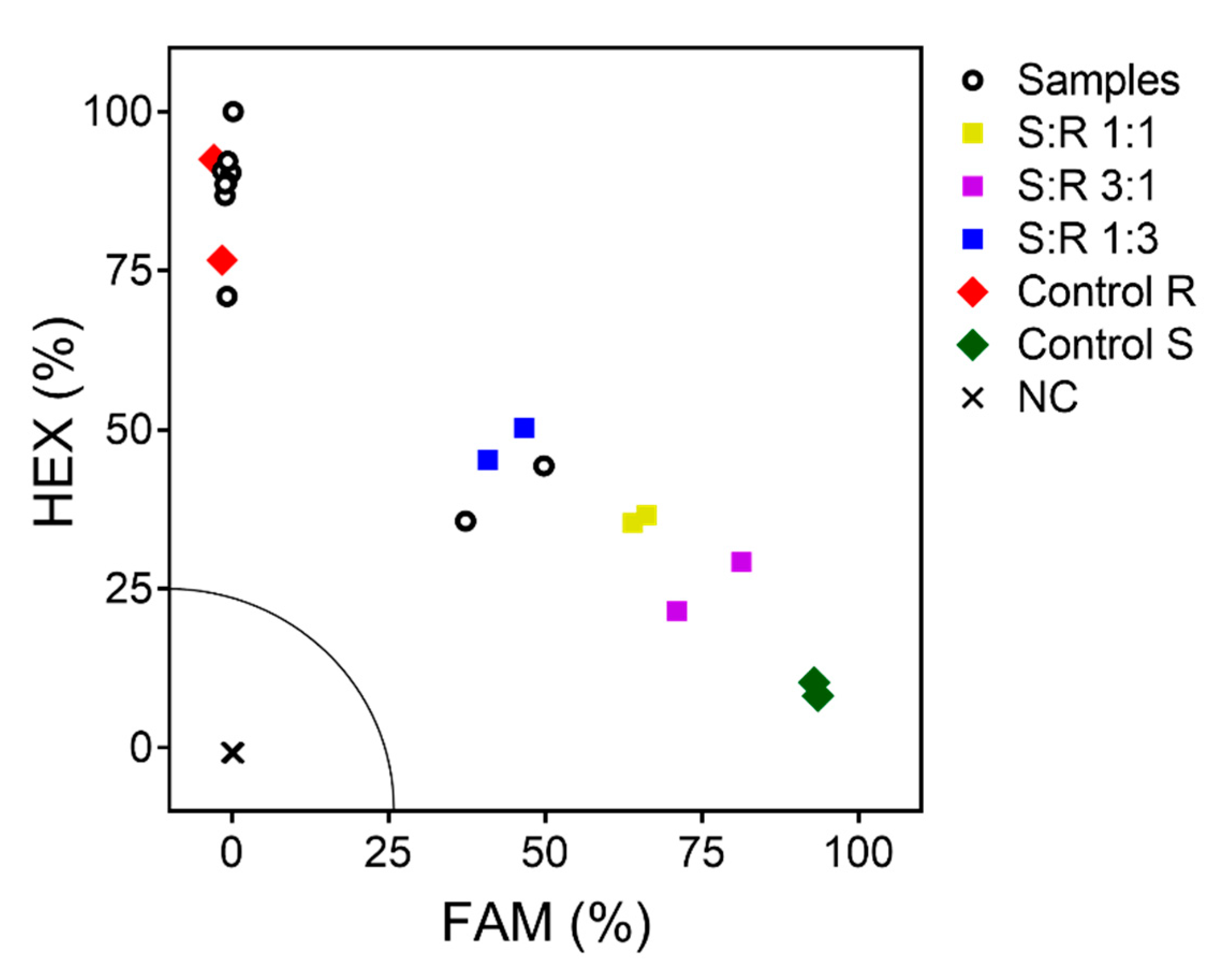
2.7. Genotyping Assay for Trp574Leu
2.8. Data Analysis
3. Results and Discussion
3.1. Imazamox Dose-Response
3.2. ALS Enzyme Assay
3.3. ALS Sequencing
3.4. Biding of Imazamox to ALS of R and S Wild Poinsettia
3.5. Genotyping Assay for Trp574Leu
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Dressler, R.L. A synopsis of poinsettia (Euphorbiaceae). Ann. Mo. Bot. Gard. 1962, 48, 329–341. [Google Scholar] [CrossRef]
- Brecke, B.J. Wild poinsettia (Euphorbia heterophylla) germination and emergence. Weed Sci. 1995, 43, 103–106. [Google Scholar] [CrossRef]
- Brecke, B.J.; Tobola, P. Growth and development of wild poinsettia (Euphorbia heterophylla) selections in peanut (Arachis hypogaea). Weed Sci. 1996, 44, 575–578. [Google Scholar] [CrossRef]
- Bridges, D.C.; Brecke, B.J.; Barbour, J.C. Wild poinsettia (Euphorbia heterophylla) interference by peanut (Arachis hypogaea). Weed Sci. 1996, 40, 37–42. [Google Scholar] [CrossRef]
- Duarte, D.J.; Bianco, S.; Leonardo, B.C.; Panosso, A.R. Estimativa da área foliar de Euphorbia heterophylla. Planta Daninha 2009, 27, 527–531. [Google Scholar] [CrossRef]
- Adelusi, A.A.; Odufeko, G.T.; Makinde, A.M. Interference of Euphorbia heterophylla Linn on the growth and reproductive yield of soybean (Glycine max (Linn) Merill). Res. J. Bot. 2006, 1, 85–94. [Google Scholar]
- Meschede, D.K.; Oliveira, R.S., Jr.; Constantin, J.; Scapim, C.A. Critical period of interference of Euphorbia heterophylla in soybean crop under low seeding rate. Planta Daninha 2002, 20, 381–387. [Google Scholar] [CrossRef]
- Machado, A.B.; Trezzi, M.M.I.; Patel, F.; Cieslik, L.F.; Debastiani, F. Grain yield of beans and weed economic threshold in two periods of competition with Euphorbia heterophylla. Planta Daninha 2015, 33, 41–48. [Google Scholar] [CrossRef][Green Version]
- Dayan, F.E.; Barker, A.; Takano, H.; Bough, R.; Ortiz, M.; Duke, S.O. Herbicide mechanism of action and resistance. In Comprehensive Biotechnology, 3rd ed.; Moo-Young, M., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; Volume 4, p. 4826. [Google Scholar]
- Garcia, M.D.; Nouwens, A.; Lonhienne, T.G.; Guddat, L.W. Comprehensive understanding of acetohydroxyacid synthase inhibition by different herbicide families. Proc. Nat. Acad. Sci. USA 2017, 114, 10911100. [Google Scholar] [CrossRef]
- Zhou, Q.; Liu, W.; Zhang, Y.; Liu, K.K. Action mechanism of acetolactate synthase-inhibiting herbicides. Pestic. Biochem. Physiol. 2007, 89, 89–96. [Google Scholar] [CrossRef]
- Duke, S.O. The history and current status of glyphosate. Pest Manag. Sci. 2018, 74, 1027–1034. [Google Scholar] [CrossRef] [PubMed]
- Beckie, H.J.; Tardif, F.J. Herbicide cross resistance in weeds. Crop. Prot. 2012, 35, 15–28. [Google Scholar] [CrossRef]
- Heap, I. International Herbicide Resistance Weed Database. Available online: www.weedscience.org (accessed on 30 May 2020).
- Gelmini, G.A.; Victória Filho, R.; Novo, M.C.S.S.; Adoryan, M.L. Resistance of Euphorbia heterophylla L. biotypes to ALS enzyme inhibitor herbicides in soybean crop. Bragantia 2001, 60, 93–99. [Google Scholar] [CrossRef]
- Prigol, A.; Galon, L.; Forte, C.T.; Kujawisky, R.; Concenço, G.; Trezzi, M.M.; Trevisol, R.; Radunz, A.L.; Perim, G.F. Evaluation of wild poinsettia biotypes with suspicion of resistant to ALS and Protox inhibitors herbicides. Rev. Bras. Herb. 2014, 13, 214–224. [Google Scholar]
- Trezzi, M.M.; Fellipi, C.L.; Mattei, D.; Silva, H.L.; Nunes, A.L.; Debastiani, C.; Vidal, R.A.; Marques, A. Multiple resistance of acetolactate synthase and protoporphyrinogen oxidase inhibitors in Euphorbia heterophylla biotypes. J. Environ. Sci. Health Part B 2005, 40, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Adegas, F.S.; Gazziero, D.L.P.; Oliveira, R.S., Jr.; Mendes, R.R.; Rodrigues, L.J. Euphorbia heterophylla: Um Novo Caso de Resistência ao Glifosato no Brasil. Available online: https://www.infoteca.cnptia.embrapa.br/infoteca/bitstream/doc/1121045/1/ComTec98.pdf (accessed on 30 May 2020).
- Oliveira, R.S., Jr.; Constantin, J.; Toledo, R.; Kajihara, L.H.; Stasieviski, A.; Pagliari, P.H.; Arantes, J.G.Z.; Cavalieri, S.D.; Alonso, D.G.; Roso, A.C. Aplicações sequenciais de flumiclorac-pentyl para o controle de Euphorbia heterophylla na cultura da soja. Acta Sci. Agron. 2006, 28, 115–122. [Google Scholar]
- Mendes, R.R.; Takano, H.K.; Adegas, F.S.; Oliveira, R.S., Jr.; Gaines, T.A.; Dayan, F.E. R128L Target site mutation in PPO2 evolves in wild poinsettia (Euphorbia heterophylla L.) with cross-resistance to PPO-Inhibiting herbicides. Weed Sci. 2020, 1–31. (in press). [CrossRef]
- Giacomini, D. Sequence Database. Available online: http://www.weedscience.org/Pages/sequence.aspx (accessed on 30 May 2020).
- Tranel, P.J.; Wright, T.R.; Heap, I.M. Mutations in Herbicide-Resistant Weeds to ALS Inhibitors. Available online: http://www.weedscience.org/Pages/MutationDisplayAll.aspx (accessed on 15 May 2020).
- McCourt, J.A.; Pang, S.S.; King-Scott, J.; Guddat, L.W.; Duggleby, R.G. Herbicide-binding sites revealed in the structure of plant acetohydroxyacid synthase. Proc. Natl. Acad. Sci. USA 2006, 130, 569–573. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Protein Structure Modeling with MODELLER. In Functional Genomics: Methods in Molecular Biology; Kaufmann, M., Klinger, C., Savelsbergh, A., Eds.; Humana Press: New York, NY, USA, 2017; pp. 39–54. [Google Scholar]
- Brunharo, C.A.C.G.; Takano, H.K.; Mallory-Smith, C.A.; Dayan, F.E.; Hanson, B.D. Role of glutamine synthetase isogenes and herbicide metabolism in the mechanism of resistance to glufosinate in Lolium perenne L. spp. multiflorum biotypes from Oregon. J. Agric. Food Chem. 2019, 67, 8431–8440. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Van Zundert, G.C.P.; Rodrigues, J.P.G.L.M.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.J.; van Dijk, M.; de Vries, S.J.; Bonvin, A.M.J.J. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Molec. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef] [PubMed]
- Forli, S.; Huey, R.; Pique, M.E.; Sanner, M.; Goodsell, D.S.; Olson, A.J. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsl. Prot. Cryst. 2002, 40, 82–92. [Google Scholar]
- Streibig, J.C. Herbicide bioassay. Weed Res. 1988, 28, 479–484. [Google Scholar] [CrossRef]
- Ritz, C.; Baty, F.; Streibig, J.C.; Gerhard, D. Dose-response analysis using R. PLoS ONE 2015, 10, e0146021. [Google Scholar] [CrossRef]
- Rodrigues, B.N.; Almeida, F.S. Guia de Herbicidas, 7th ed.; Authors: Londrina, Brazil, 2018. [Google Scholar]
- Zheng, D.; Patzoldt, W.L.; Tranel, P.J. Association of the W574L ALS substitution with resistance to cloransulam and imazamox in common ragweed (Ambrosia artemisiifolia). Weed Sci. 2005, 53, 424–430. [Google Scholar] [CrossRef]
- Kumar, V.; Jha, P. First report of Ser653Arn mutation endowing high level of resistance to imazamox in downy brome (Bromus tectorum L.). Pest Manag. Sci. 2017, 73, 2585–2591. [Google Scholar] [CrossRef]
- Ntoanidou, S.; Madesis, P.; Diamantidis, G.; Eleftherohorinos, I. Trp574 substitution in the acetolactate synthase of Sinapis arvensis confers cross-resistance to tribenuron and imazamox. Pestci. Biochem. Physiol. 2017, 142, 9–14. [Google Scholar] [CrossRef]
- Gazziero, D.L.P.; Brighenti, A.M.; Maciel, C.D.G.; Christoffoleti, P.J.; Adegas, F.S.; Voll, E. Resistance of the weed wild poinsettia to ALS inhibitors herbicides. Planta Daninha 1998, 16, 117–125. [Google Scholar] [CrossRef]
- Vidal, R.A.; Winkler, L.M.; Hernandez, G.C.; Fleck, N.G.; Merotto, A., Jr.; Trezzi, M.M. A field survey of crop management practices and distribution of ALS resistant Euphorbia heterophylla in two states in southern Brazil. Planta Daninha 2004, 22, 403–410. [Google Scholar] [CrossRef]
- Rojano-Delgado, A.M.; Portugal, J.M.; Pauma-Bautista, C.; Alcantara-de-la-Cruz, R.; Torra, J.; Alcántara, E.; Prado, R. Target site as the main mechanism of resistance to imazamox in a Euphorbia heterophylla population. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Shanner, D.L.; Nadler-Hassar, T.; Henry, W.B.; Koger, C.H. A rapid in vivo shikimate accumulation assay with excised leaf discs. Weed Sci. 2005, 53, 769–774. [Google Scholar] [CrossRef]
- Dayan, F.E.; Owens, D.K.; Corniani, N.; Silva, F.M.L.; Watson, S.B.; Howell, J.L.; Shaner, D.L. Biochemical markers and enzyme assays for herbicide mode of action and resistance studies. Weed Sci. 2015, 63, 23–63. [Google Scholar] [CrossRef]
- Wang, M.; Liu, B.; Li, Y.; Luo, X.; Li, L. Non–target site based resistance to the ALS-inhibiting herbicide mesosulfuron-methyl in American sloughgrass (Beckmannia syzigachne). Weed Sci. 2019, 67, 527–533. [Google Scholar] [CrossRef]
- Ntoanidou, S.; Madesis, P.; Eleftherohorinos, I. Resistance of Rapistrum rugosum to tribenuron and imazamox due to Trp574 or Pro197 substitution in the acetolactate synthase. Pestic. Biochem. Physiol. 2019, 154, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Wang, J.X.; Zhang, H.J.; Liu, J.L.; Bian, S.N. Study on mutations in ALS for resistance to tribenuron-methyl in Galium aparine L. Agric. Sci. China 2011, 10, 86–91. [Google Scholar] [CrossRef]
- Milani, A.; Scarabel, L.; Sattin, M. A family affair: Resistance mechanism and alternative control of three Amaranthus species resistant to acetolactate synthase inhibitors in Italy. Pest Manag. Sci. 2020, 76, 1205–1213. [Google Scholar] [CrossRef]
- Li, J.; Li, M.; Gao, X.; Fang, F. A novel amino acid substitution Trp574Arg in acetolactate synthase (ALS) confers broad resistance to ALS-inhibiting herbicides in crabgrass (Digitaria sanguinalis). Pest Manag. Sci. 2017, 73, 2538–2543. [Google Scholar] [CrossRef]
- Küpper, A.; Borgato, E.A.; Patterson, E.L.; Netto, A.G.; Nicolai, M.; Carvalho, S.J.; Nissen, S.; Gaines, T.A.; Christoffoleti, P.J. Multiple resistance to glyphosate and acetolactate synthase inhibitors in Palmer amaranth (Amaranthus palmeri) identified in Brazil. Weed Sci. 2017, 65, 317–326. [Google Scholar] [CrossRef]
- Patterson, E.L.; Fleming, M.B.; Kessler, K.C.; Nissen, S.J.; Gaines, T.A. A KASP genotyping method to identify Northern watermilfoil, Eurasian watermilfoil, and their interspecific hybrids. Front. Plant Sci. 2017, 8, 725. [Google Scholar] [CrossRef]
- Song, D.; Wu, G.; Vrinten, P.; Qiu, X. Development of imidazolinone herbicide tolerant borage (Borago officinalis L.). Plant Sci. 2017, 262, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Takano, H.K.; Fernandes, V.N.; Adegas, F.S.; Oliveira, R.S., Jr.; Westra, P.; Gaines, T.A.; Dayan, F.E. A novel TIPT double mutation in EPSPS conferring glyphosate resistance in tetraploid Bidens subalternans. Pest Manag. Sci. 2020, 76, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Burgos, N.R.; Tranel, P.J.; Streibig, J.C.; Davis, V.M.; Shaner, D.; Norsworthy, J.K.; Ritz, C. Confirmation of resistance to herbicides and evaluation of resistance levels. Weed Sci. 2013, 61, 4–20. [Google Scholar] [CrossRef]
- Rey-Caballero, J.; Menéndez, J.; Osuna, M.D.; Salas, M.; Torra, J. Target-site and non-target-site resistance mechanisms to ALS inhibiting herbicides in Papaver rhoeas. Pestic. Biochem. Physiol. 2017, 138, 57–65. [Google Scholar] [CrossRef]
- Bolaji, A.O.; Olojede, C.B.; Famurewa, A.A.; Faluyi, J.O. Morphological and cytological studies of Euphorbia hyssopifolia L. and Euphorbia heterophylla L. from Ile-Ife, Nigeria. Niger. J. Genet. 2014, 28, 15–18. [Google Scholar] [CrossRef]
- Frigo, M.J.; Mangolin, C.A.; Oliveira, R.S.; Maria de Fátima, P.S. Esterase polymorphism for analysis of genetic diversity and structure of wild poinsettia (Euphorbia heterophylla) populations. Weed Sci. 2009, 57, 54–60. [Google Scholar] [CrossRef]
- Cronquist, A. An Integrated System of Classification of Flowering Plants; Columbia University Press: New York, NY, USA, 1981; p. 1262. [Google Scholar]
- Loureiro, I.; Escorial, M.C.; Chueca, M.C. Pollen-mediated movement of herbicide resistance genes in Lolium rigidum. PLoS ONE 2016, 11, e0157892. [Google Scholar] [CrossRef][Green Version]
- Wu, C.; Davis, A.S.; Tranel, P.J. Limited fitness costs of herbicide-resistance traits in Amaranthus tuberculatus facilitate resistance evolution. Pest Manag. Sci. 2018, 74, 293–301. [Google Scholar] [CrossRef]
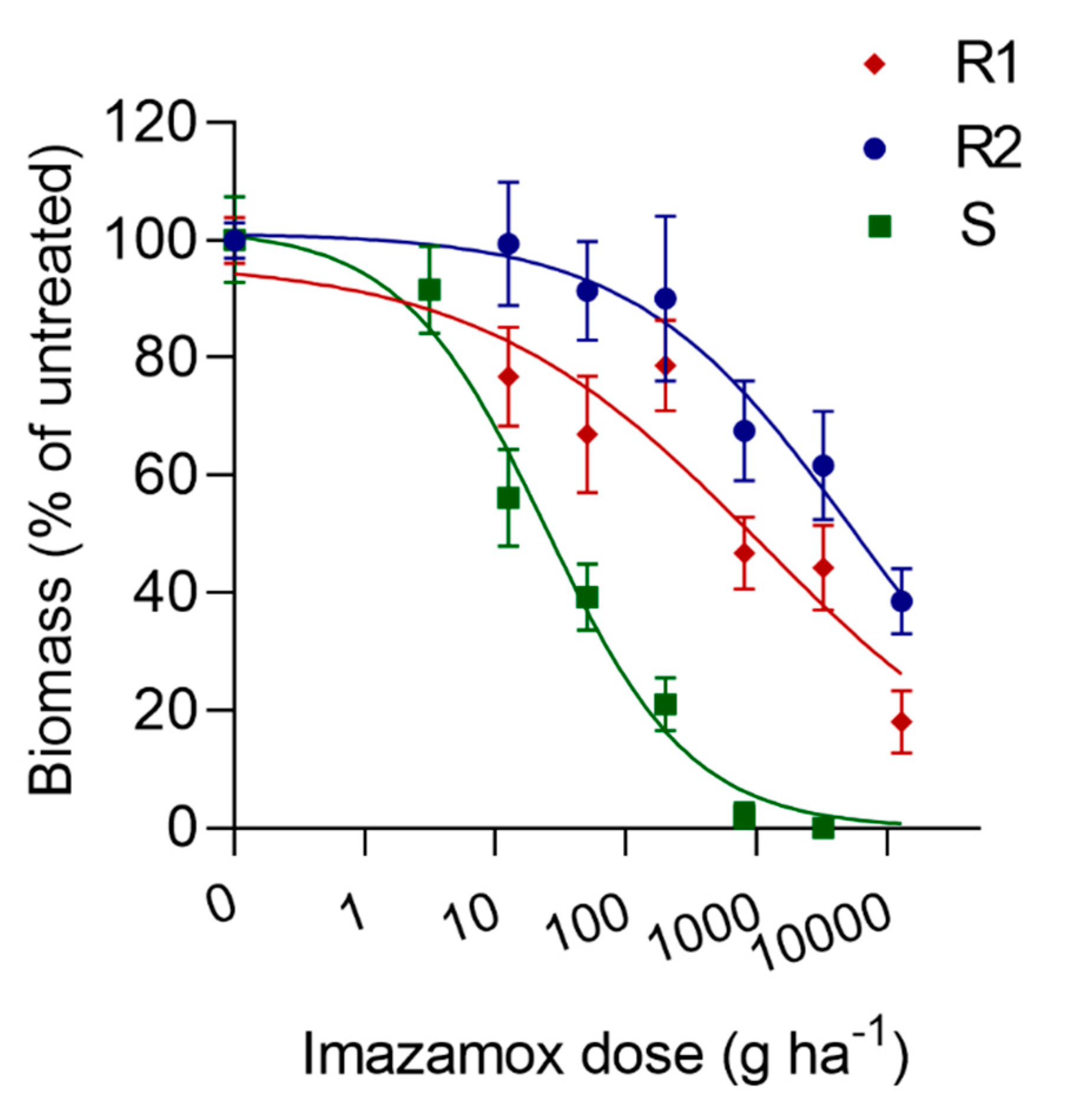
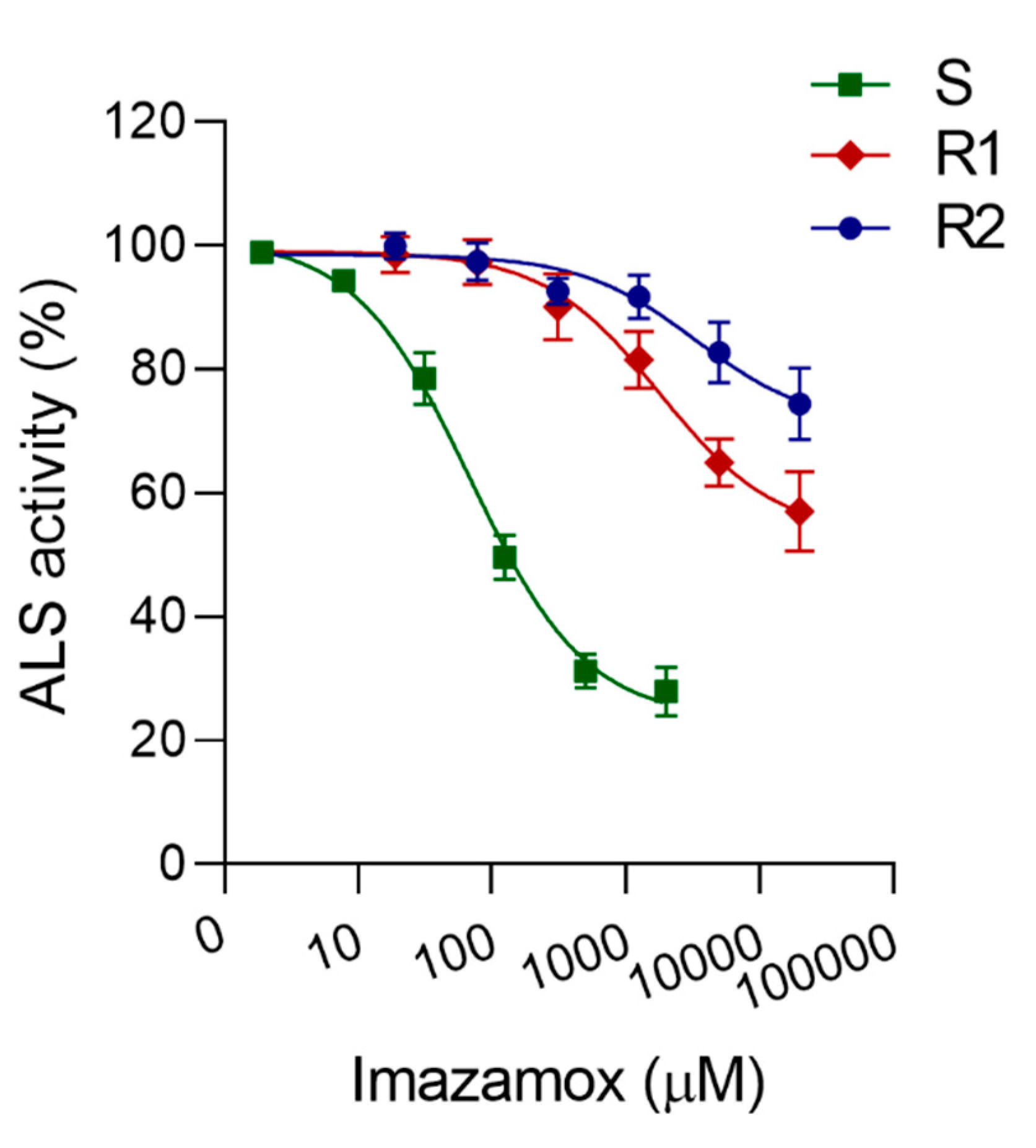
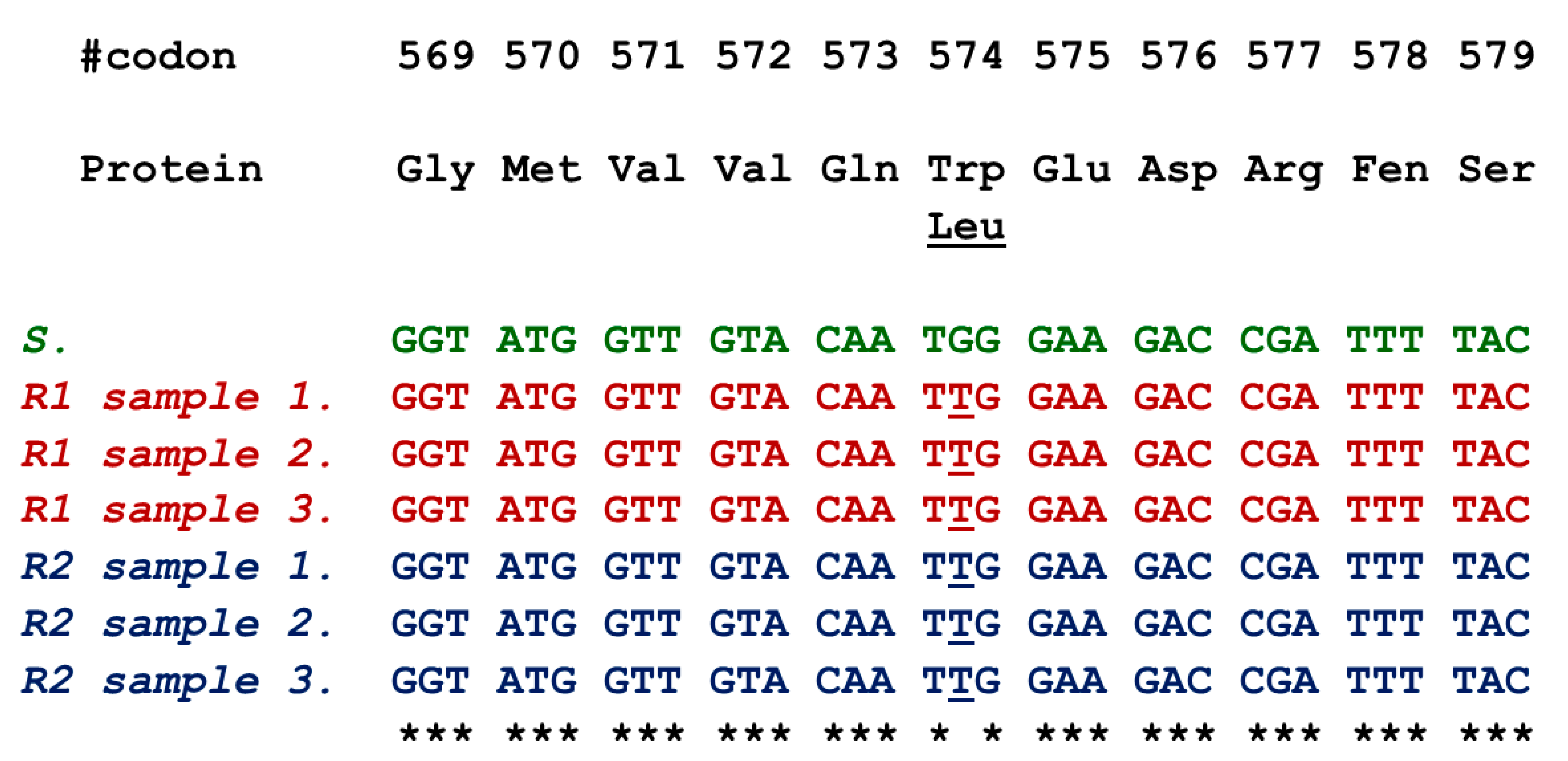

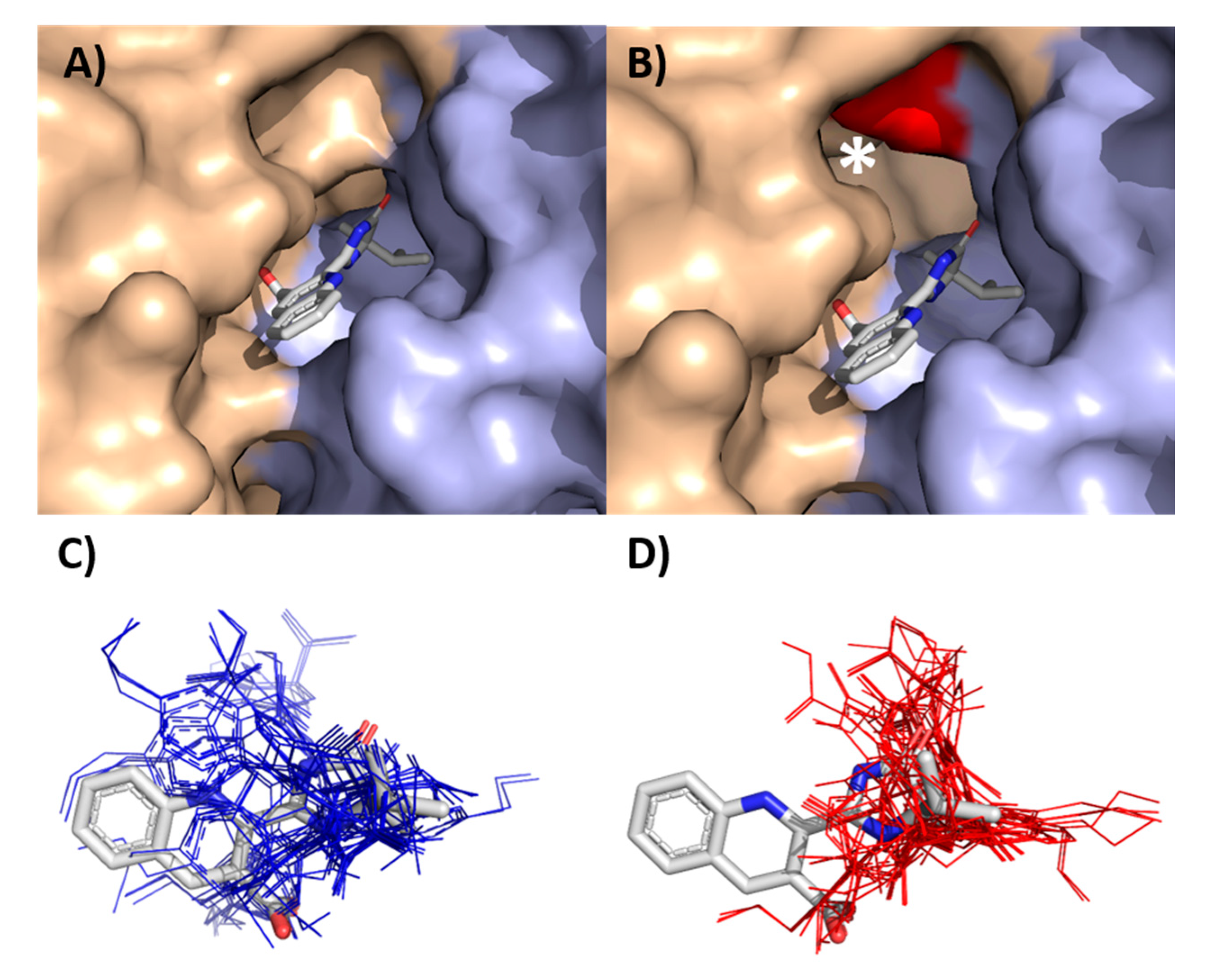
| Population | a | b | c (GR50, I50) | R2 | RF 1 |
|---|---|---|---|---|---|
| Dry biomass (c in g ha−1) | |||||
| S | 102 | 0.8 | 24.2 | 0.91 | - |
| R1 | 96.5 | 0.4 | 1084.3 | 0.75 | 45 |
| R2 | 101.3 | 0.5 | 5434.0 | 0.65 | 224.5 |
| Enzyme activity (c in µM) | |||||
| S | 104 | 0.6 | 163 | 0.92 | - |
| R1 | NA | NA | >20,000 | NA | >123 |
| R2 | NA | NA | >20,000 | NA | >123 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mendes, R.R.; Takano, H.K.; Oliveira, R.S., Jr.; Adegas, F.S.; Gaines, T.A.; Dayan, F.E. A Trp574Leu Target-Site Mutation Confers Imazamox Resistance in Multiple Herbicide-Resistant Wild Poinsettia Populations from Brazil. Agronomy 2020, 10, 1057. https://doi.org/10.3390/agronomy10081057
Mendes RR, Takano HK, Oliveira RS Jr., Adegas FS, Gaines TA, Dayan FE. A Trp574Leu Target-Site Mutation Confers Imazamox Resistance in Multiple Herbicide-Resistant Wild Poinsettia Populations from Brazil. Agronomy. 2020; 10(8):1057. https://doi.org/10.3390/agronomy10081057
Chicago/Turabian StyleMendes, Rafael R., Hudson K. Takano, Rubem S. Oliveira, Jr., Fernando S. Adegas, Todd A. Gaines, and Franck E. Dayan. 2020. "A Trp574Leu Target-Site Mutation Confers Imazamox Resistance in Multiple Herbicide-Resistant Wild Poinsettia Populations from Brazil" Agronomy 10, no. 8: 1057. https://doi.org/10.3390/agronomy10081057
APA StyleMendes, R. R., Takano, H. K., Oliveira, R. S., Jr., Adegas, F. S., Gaines, T. A., & Dayan, F. E. (2020). A Trp574Leu Target-Site Mutation Confers Imazamox Resistance in Multiple Herbicide-Resistant Wild Poinsettia Populations from Brazil. Agronomy, 10(8), 1057. https://doi.org/10.3390/agronomy10081057






