Effect of Plant Growth Promoting Bacteria on the Growth of Wheat Seedlings Subjected to Phosphate Starvation
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Bacterial Inocula
2.2. Identification of 12A and 25A
2.3. Preparation of Seeds
2.4. Greenhouse Experiment
2.5. Plant and Root Sampling
2.6. Total RNA Extraction and cDNA Synthesis
2.7. Semi-Quantitative Reverse Transcriptase (RT)-PCR
2.8. Statistical Analysis
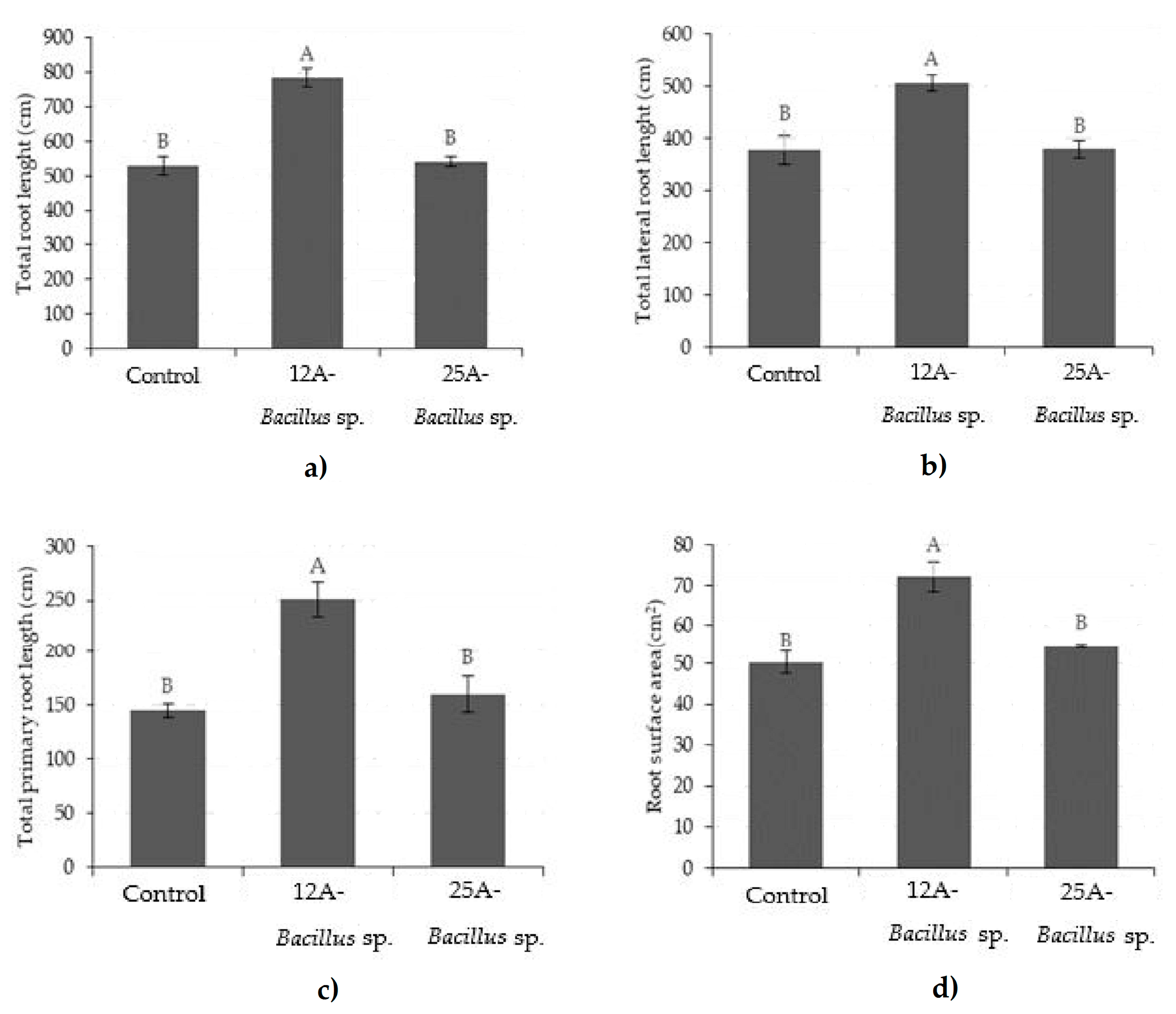
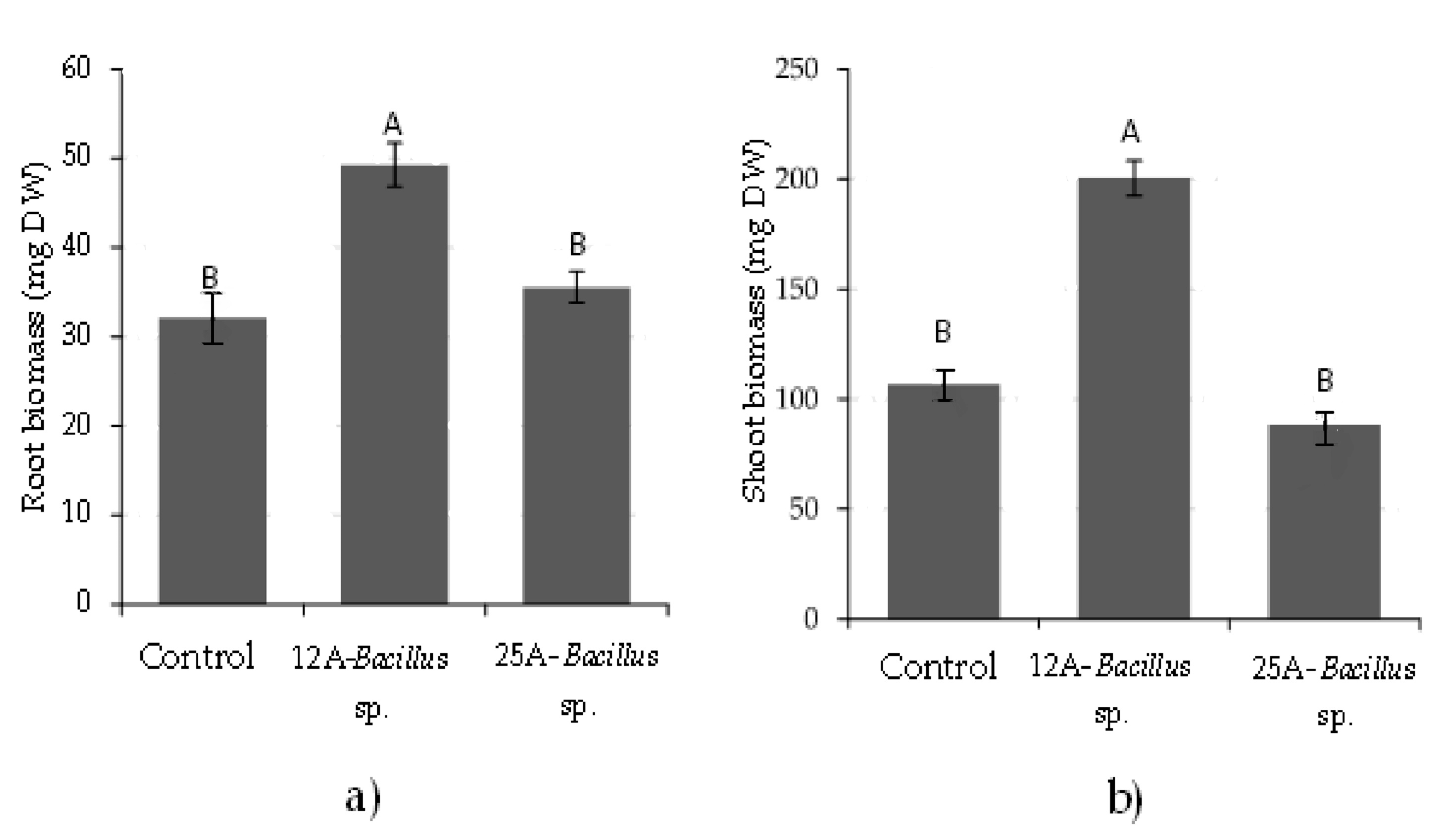
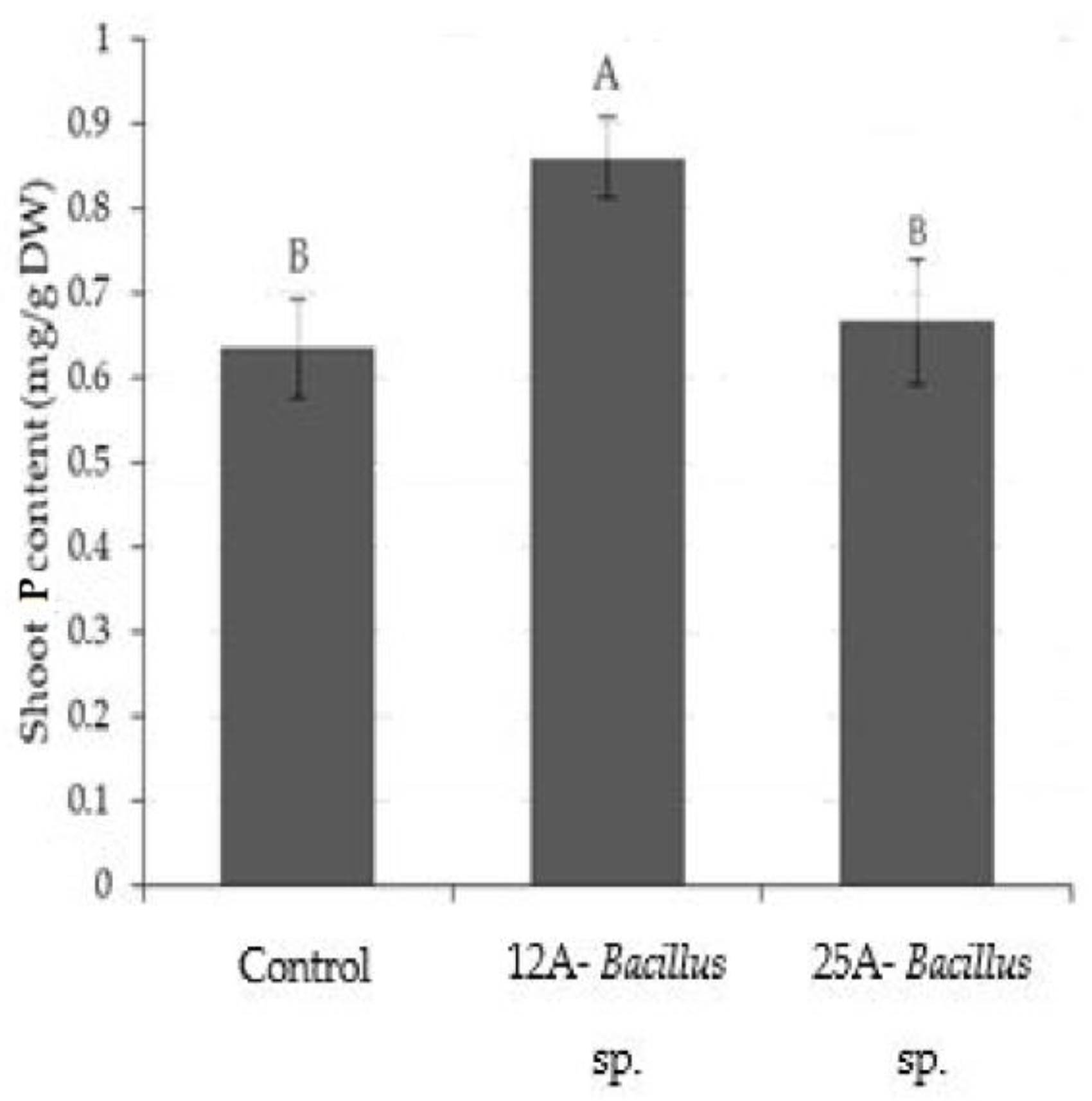
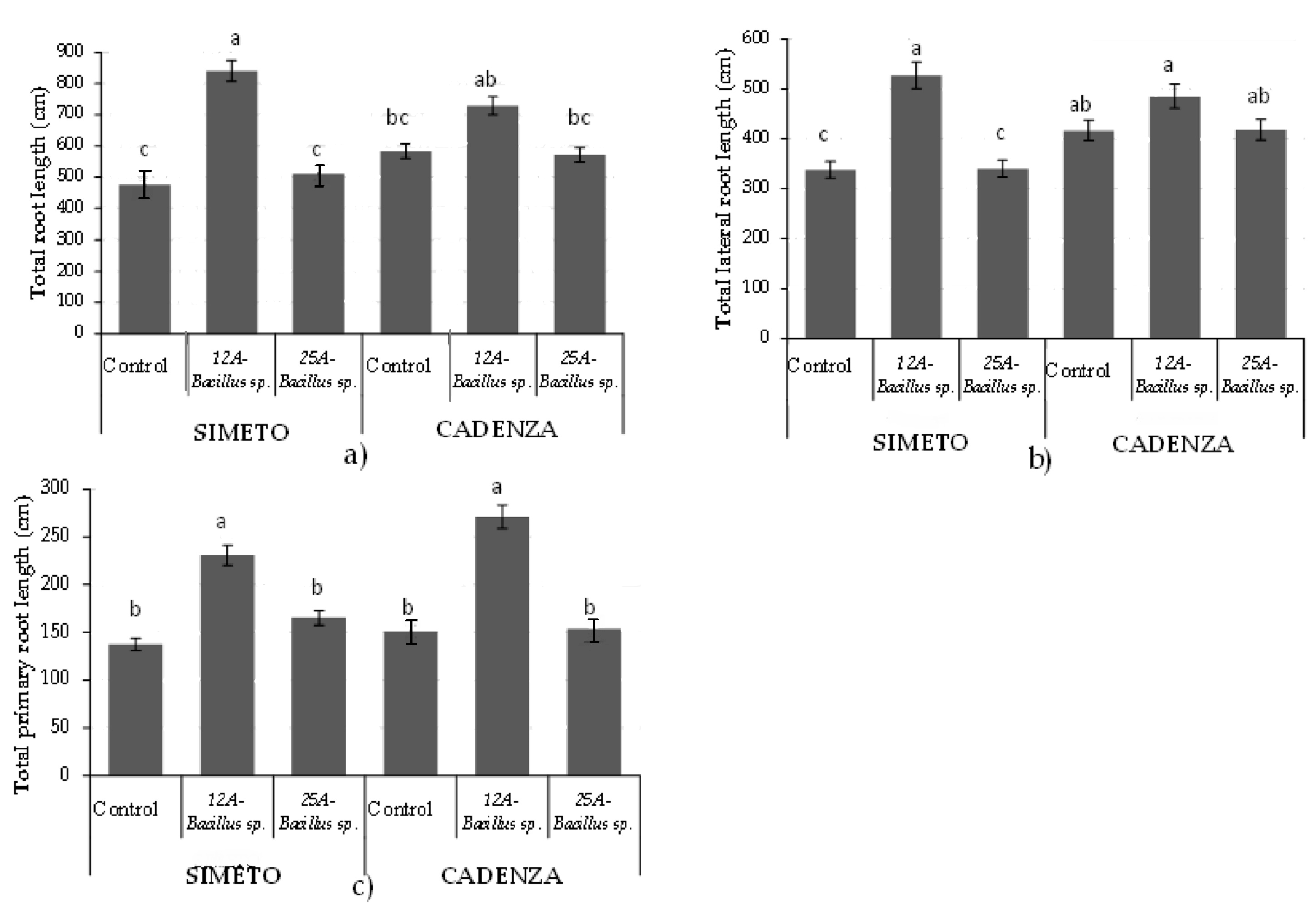
3. Results
3.1. Root Imaging and Plant Biomass
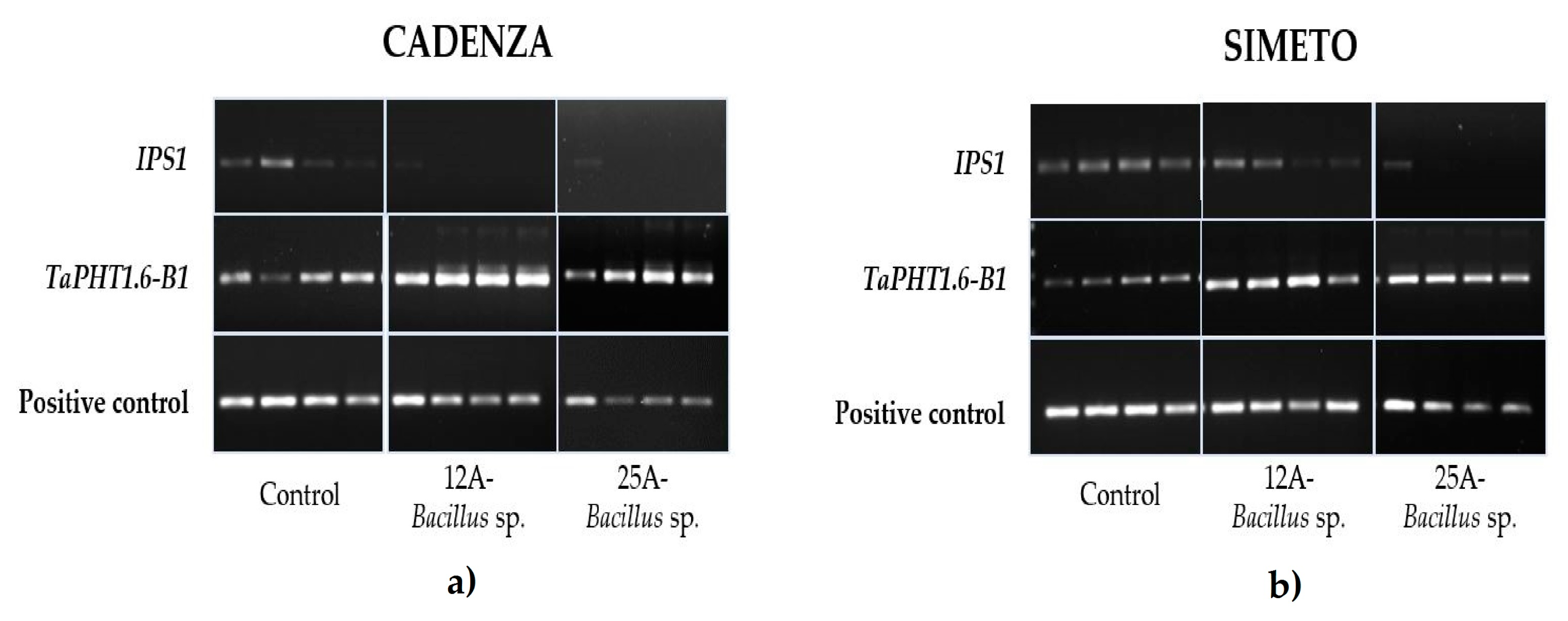
3.2. Expression Analysis of Representative Pi-Starvation Induced Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- de Souza, R.; Ambrosini, A.; Passaglia, L.M. Plant growth-promoting bacteria as inoculants in agricultural soils. Genet. Mol. Biol. 2015, 38, 401–419. [Google Scholar] [CrossRef] [PubMed]
- Heuer, S.; Gaxiola, R.; Schilling, R.; Herrera-Estrella, L.; López-Arredondo, D.; Wissuwa, M.; Delhaize, E.; Rouached, H. Improving phosphorus use efficiency: A complex trait with emerging opportunities. Plant J. 2017, 90, 868–885. [Google Scholar] [CrossRef] [PubMed]
- López-Arredondo, D.L.; Leyva-Gonzalez, M.A.; Gonzalez-Morales, S.A.; Lopez-Bucio, J.; Herrera-Estrella, L. Phosphate nutrition: Improving low-phosphate tolerance in crops. Annu. Rev. Plant Biol. 2014, 65, 95–123. [Google Scholar] [CrossRef]
- Dhillon, J.; Torres, G.; Driver, E.; Figueiredo, B.; Raun, W.R. World phosphorus use efficiency in cereal crops. Agron. J. 2017, 109, 1–8. [Google Scholar] [CrossRef]
- Sharma, S.B.; Sayyed, R.Z.; Trivedi, M.H.; Gobi, T.A. Phosphate solubilizing microbes: Sustainable approach for managing phosphorus deficiency in agricultural soils. Springerplus 2013, 2, 587. [Google Scholar] [CrossRef]
- Oono, Y.; Kobayashi, F.; Kawahara, Y.; Yazawa, T.; Handa, H.; Itoh, T.; Matsumoto, T. Characterisation of the wheat (triticum aestivum L.) transcriptome by de novo assembly for the discovery of phosphate starvation-responsive genes: Gene expression in Pi-stressed wheat. BMC Genom. 2013, 14, 77. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, I.; Weigel, A.; Garcıa, D.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007, 39, 1033–1037. [Google Scholar] [CrossRef] [PubMed]
- Briat, J.F.; Rouached, H.; Tissot, N.; Gaymard, F.; Dubos, C. Integration of P, S, Fe, and Zn nutrition signals in Arabidopsis thaliana: Potential involvement of phosphate starvation response 1 (PHR1). Front. Plant Sci. 2015, 6, 290. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Verma, S.; Trivedi, P.K. Involvement of small RNAs in phosphorus and sulfur sensing, signaling and stress: Current update. Front. Plant Sci. 2017, 8, 285. [Google Scholar] [CrossRef]
- Aung, K.; Lin, S.I.; Wu, C.C.; Huang, Y.T.; Su, C.L.; Chiou, T.J. pho2, a phosphate over accumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol. 2006, 141, 1000–1011. [Google Scholar] [CrossRef]
- Bari, R.; Datt Pant, B.; Stitt, M.; Scheible, W.R. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 2006, 141, 988–999. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Ying, S.; Huang, H.; Li, K.; Wu, P.; Shou, H.X. Involvement of OsSPX1 in phosphate homeostasis in rice. Plant J. 2009, 57, 895–904. [Google Scholar] [CrossRef] [PubMed]
- Ai, P.; Sun, S.; Zhao, J.; Fan, X.; Xin, W.; Guo, Q.; Yu, L.; Shen, Q.; Wu, P.; Miller, A.J.; et al. Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J. 2009, 57, 798–809. [Google Scholar] [CrossRef] [PubMed]
- Jeong, K.; Mattes, N.; Catausan, S.; Chin, J.H.; Paszkowski, U.; Heuer, S. Genetic diversity for mycorrhizal symbiosis and phosphate transporters in rice. JIPB 2015, 57, 969–979. [Google Scholar] [CrossRef] [PubMed]
- Grün, A.; Buchner, P.; Broadley, M.R.; Hawkesford, M.J. Identification and expression profiling of Pht1 phosphate transporters in wheat in controlled environments and in the field. Plant Biol. 2017, 20, 374–389. [Google Scholar] [CrossRef]
- FAO. Available online: http://www.fao.org/news/story/en/item/273303/icode (accessed on 19 December 2014).
- Jiang, H.; Qi, P.; Wang, T.; Chen, M.; Chen, N.; Pan, L.; Chi, X. Isolation and characterization of halotolerant phosphate-solubilizing microorganisms from saline soils. 3Biotech 2018, 8, 461. [Google Scholar] [CrossRef]
- Tao, G.C.; Tian, S.J.; Cai, M.Y.; Xie, G.H. Phosphate-solubilizing and -mineralizing abilities of bacteria isolated from soils. Pedosphere 2008, 18, 515–523. [Google Scholar] [CrossRef]
- Singh, S.; Kapoor, K.K. Inoculation with phosphate-solubilizing microorganisms and a vesicular-arbuscular mycorrhizal fungus improves dry matter yield and nutrient uptake by wheat grown in a sandy soil. Biol. Fertil. Soils 1999, 28, 139–144. [Google Scholar] [CrossRef]
- Krey, T.; Vassilev, N.; Baum, C.; Eichler-Löbermann, B. Effects of long-term phosphorus application and plant-growth promoting rhizobacteria on maize phosphorus nutrition under field conditions. Eur. J. Soil Biol. 2013, 55, 124–130. [Google Scholar] [CrossRef]
- Rodríguez, H.; Fraga, R. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 1999, 17, 319–339. [Google Scholar] [CrossRef]
- Alori, E.T.; Glick, B.R.; Babalola, O.O. Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front. Microbiol. 2017, 8, 971. [Google Scholar] [CrossRef] [PubMed]
- Spaepen, S.; Vanderleyden, J.; Remans, R. Indole-3-acetic acid in microbial and microorganism-plant signalling. FEMS Microbiol. Rev. 2007, 4, 425–448. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Bauddh, K.; Barman, S.C.; Singh, R.P. Amendments of microbial biofertilizers and organic substances reduces requirement of urea and DAP with enhanced nutrient availability and productivity of wheat (Triticum aestivum L.). Ecol. Eng. 2014, 71, 432–437. [Google Scholar] [CrossRef]
- Saia, S.; Rappa, V.; Ruisi, P.; Abenavoli, M.R.; Sunseri, F.; Giambalvo, D.; Frenda, A.S.; Martinelli, F. Soil inoculation with symbiotic microorganisms promotes plant growth and nutrient transporter genes expression in durum wheat. Front. Plant Sci. 2015, 6, 815. [Google Scholar] [CrossRef] [PubMed]
- Ogut, M.; Er, F. Mineral composition of field grown winter wheat inoculated with phosphorus solubilizing bacteria at different plant growth stages. J. Plant Nutr. 2016, 39, 479–490. [Google Scholar] [CrossRef]
- Gupta, M.; Kiran, S.; Gulati, A.; Singh, B.; Tewari, R. Isolation and identification of phosphate solubilizing bacteria able to enhance the growth and aloin-A biosynthesis of Aloe barbadensis Miller. Microbiol. Res. 2012, 167, 358–363. [Google Scholar] [CrossRef]
- Sadiq, H.M.; Jahangir, G.Z.; Nasir, I.A.; Iqtidar, M.; Iqbal, M. Isolation and characterization of phosphate-solubilizing bacteria from rhizosphere soil. Biotechnol. Biotechnol. Equip. 2013, 27, 4248–4255. [Google Scholar] [CrossRef]
- Sarker, A.; Islam, T. Phosphate solubilizing bacteria promote growth and enhance nutrient uptake by wheat. Plant Sci. Today 2014, 1, 86–93. [Google Scholar] [CrossRef]
- Zhao, K.; Penttinen, P.; Zhang, X.; Ao, X.; Liu, M.; Yu, X.; Chen, Q. Maize rhizosphere in Sichuan, China, hosts plant growth promoting Burkholderia cepacia with phosphate solubilizing and antifungal abilities. Microbiol. Res. 2014, 169, 76–82. [Google Scholar] [CrossRef]
- Manzoor, M.; Abbasi, M.K.; Tariq, S. Isolation of phosphate solubilizing bacteria from maize rhizosphere and their potential for rock phosphate solubilization–mineralization and plant growth promotion. Geomicrobiol. J. 2017, 34, 81–95. [Google Scholar] [CrossRef]
- Di Benedetto, N.A.; Campaniello, D.; Bevilacqua, A.; Cataldi, M.P.; Sinigaglia, M.; Flagella, Z.; Corbo, M.A. A study on a multi-step characterization and selection of Plant Growth Promoting Bacteria from durum wheat rhizosphere to improve nutrient use efficiency. Microorganisms 2019, 7, 541. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wang, Z.; Ren, H.; Shen, C.; Li, Y.; Ling, H.Q.; Wu, C.; Lian, X.; Wu, P. OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice. Plant J. 2010, 62, 508–517. [Google Scholar] [CrossRef]
- Robinson, R.J.; Fraaije, B.A.; Clark, I.M.; Jackson, R.W.; Hirsch, P.R.; Mauchline, T.H. Wheat seed embryo excision enables the creation of axenic seedlings and Koch’s postulates testing of putative bacterial endophytes. Sci. Rep. 2016, 6, 25581. [Google Scholar] [CrossRef] [PubMed]
- Masters-Clark, E.; Shone, E.; Paradelo, M.; Hirsch, P.R.; Clark, I.M.; Otten, W.; Brennan, F.; Mauchline, T.H. Development of a defined compost system for the study of plant-microbe interactions. Sci. Rep. 2020, 10, 7521. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R. Plant Growth-Promoting Bacteria: Mechanisms and applications. Scientifica 2012, 2012, 963401. [Google Scholar] [CrossRef] [PubMed]
- Di Benedetto, N.A.; Corbo, M.R.; Campaniello, D.; Cataldi, M.P.; Bevilacqua, A.; Sinigaglia, M.; Flagella, Z. The role of Plant Growth Promoting Bacteria in improving nitrogen use efficiency for sustainable crop production: A focus on wheat. AIMS Microbiol. 2017, 3, 413–434. [Google Scholar] [CrossRef]
- Spaepen, S.; Dobbelaere, S.; Croonenborghs, A.; Vanderleyden, J. Effects of Azospirillum brasilense indole-3-acetic acid production on inoculated wheat plants. Plant Soil 2008, 312, 15–23. [Google Scholar] [CrossRef]
- Ali, B.; Sabri, A.N.; Ljung, K.; Hasnain, S. Auxin production by plant associated bacteria: Impact on endogenous IAA content and growth of Triticum aestivum L. Lett. Appl. Microbiol. 2009, 48, 542–547. [Google Scholar] [CrossRef]
- Majeed, A.; Abbasi, M.K.; Hameed, S.; Imran, A.; Rahim, N. Isolation and characterization of Plant Growth-Promoting Rhizobacteria from wheat rhizosphere and their effect on plant growth promotion. Front. Microbiol. 2015, 6, 198. [Google Scholar] [CrossRef]
- Patten, L.; Glick, B.R. Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl. Environ. Microbiol. 2002, 68, 3795–3801. [Google Scholar] [CrossRef]
- Ribeiro, V.P.; Marriel, I.E.; Sousa, S.M.; Lana, U.G.P.; Mattos, B.B.; Oliveira, C.A.; Gomes, E.A. Endophytic Bacillus strains enhance pearl millet growth and nutrient uptake under low-P. Microbiol. Braz. J. Microbiol. 2018, 1, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Dodd, I.C.; Zinovkina, N.Y.; Safronova, V.I.; Belimov, A.A. Rhizobacterial mediation of plant hormone status. Ann. Appl. Biol. 2010, 157, 361–379. [Google Scholar] [CrossRef]
- Pérez-de-Luque, A.; Tille, S.; Johnson, I.; Pascual-Pardo, D.; Jurriaan Ton, J.; Cameron, D.D. The interactive effects of arbuscular mycorrhiza and plant growth-promoting rhizobacteria synergistically enhance host plant defences against pathogen. Sci. Rep. 2017, 7, 16409. [Google Scholar] [CrossRef]
- Hou, X.L.; Wu, P.; Jiao, F.C.; Jia, Q.J.; Chen, H.M.; Yu, J.; Song, X.W.; Yi, K.K. Regulation of the expression of OsIPS1 and OsIPS2 in rice via systemic and local Pi signaling and hormones. Plant Cell Environ. 2005, 28, 353–364. [Google Scholar] [CrossRef]
- Shin, H.; Shin, H.S.; Chen, R.; Harrison, M.J. Loss of At4 function impacts phosphate distribution between the roots and the shoots during phosphate starvation. Plant J. 2006, 45, 712–726. [Google Scholar] [CrossRef] [PubMed]
- Hammond, J.P.; White, P.J. Sucrose transport in the phloem: Integrating root responses to phosphorus starvation. J. Exp. Bot. 2008, 59, 93–109. [Google Scholar] [CrossRef] [PubMed]
- Doerner, P. Phosphate starvation signaling: A threesome controls systemic Pi homeostasis. Curr. Opin. Plant Biol. 2008, 11, 536–540. [Google Scholar] [CrossRef]
- Huang, C.Y.; Shirley, N.; Genc, Y.; Shi, B.; Langridge, P. Phosphate Utilization Efficiency Correlates with Expression of Low-Affinity Phosphate Transporters and Noncoding RNA, IPS1, in Barley. Plant Physiol. 2011, 156, 1217–1229. [Google Scholar] [CrossRef]
| Assay | 12A | 25A |
|---|---|---|
| catalase | 1 | 1 |
| oxidase | 1 | 1 |
| urease | nd | nd |
| motility | nd | nd |
| siderophores production | nd | 1 |
| ammonium production | 1 | 1 |
| phosphate-solubilization Ca3(PO4)2 (HALO mm) | 2 | 3 |
| phosphate-solubilization AlPO4 (HALO mm) | 6 | 2 |
| phosphate-solubilization Fe(PO4)3 (HALO mm) | 3 | 2 |
| P-mineralization (μM) 1 | 8.01 ± 0.33 | 3.96 ± 0.20 |
| IAA (μg/mL) 2 | 5.82 ± 0.13 | 0 |
| Name | Sequences | Gene Names and Accession Numbers |
|---|---|---|
| IPS1 F1 | 5′ CGCACACCATGTAGGCCACAA 3′ | TaIPS1;1: EU753150_1 |
| IPS1 R3 | 5′ CTTCATATCAATTTGGTATAGCTAGCTA 3′ | TaIPS1;2: EU753151_1 |
| TaIPS1;3: EU753152_1 | ||
| PHT1.6-B1 F1 | 5′ AATTAACCTGGACAACTCGACC 3′ | TraesCS5D02G472000.1 |
| PHTI.6-B1 R3 | 5′ CTGGCGCAGAACAAGGACC 3‘ | TraesCS5B02G470100.1 |
| TaCyc F | 5‘ CAAGCCGCTGCACTACAAGG 3‘ | TraesCS6A02G068900 |
| TaCyc R | 5‘ AGGGGACGGTGCAGATGAA 3‘ | TraesCS6D02G066700 |
| TraesCS6B02G093100 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cataldi, M.P.; Heuer, S.; Mauchline, T.H.; Wilkinson, M.D.; Masters-Clark, E.; Di Benedetto, N.A.; Corbo, M.R.; Flagella, Z. Effect of Plant Growth Promoting Bacteria on the Growth of Wheat Seedlings Subjected to Phosphate Starvation. Agronomy 2020, 10, 978. https://doi.org/10.3390/agronomy10070978
Cataldi MP, Heuer S, Mauchline TH, Wilkinson MD, Masters-Clark E, Di Benedetto NA, Corbo MR, Flagella Z. Effect of Plant Growth Promoting Bacteria on the Growth of Wheat Seedlings Subjected to Phosphate Starvation. Agronomy. 2020; 10(7):978. https://doi.org/10.3390/agronomy10070978
Chicago/Turabian StyleCataldi, Mariagrazia P., Sigrid Heuer, Tim H. Mauchline, Mark D. Wilkinson, Emily Masters-Clark, Nilde A. Di Benedetto, Maria Rosaria Corbo, and Zina Flagella. 2020. "Effect of Plant Growth Promoting Bacteria on the Growth of Wheat Seedlings Subjected to Phosphate Starvation" Agronomy 10, no. 7: 978. https://doi.org/10.3390/agronomy10070978
APA StyleCataldi, M. P., Heuer, S., Mauchline, T. H., Wilkinson, M. D., Masters-Clark, E., Di Benedetto, N. A., Corbo, M. R., & Flagella, Z. (2020). Effect of Plant Growth Promoting Bacteria on the Growth of Wheat Seedlings Subjected to Phosphate Starvation. Agronomy, 10(7), 978. https://doi.org/10.3390/agronomy10070978






