Transcriptome Analysis of ‘Haegeum’ Gold Kiwifruit Following Ethylene Treatment to Improve Postharvest Ripening Quality
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Ethylene Treatment
2.2. RNA Extraction and Sequencing Using Illumina Truseq Stranded mRNA Library Prep Kit
2.3. Mapping Reads on a Reference Genome and Calculating Expression between Samples
2.4. Identification of DEGs and Functional Enrichment Analysis
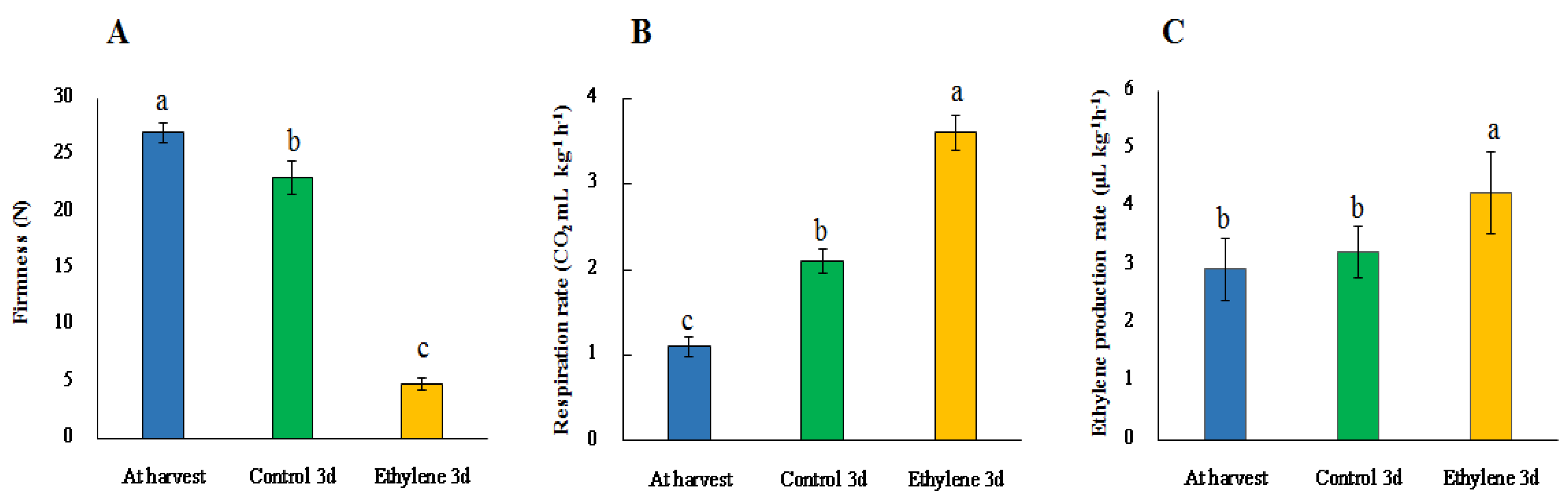
2.5. Measurement of Firmness, Respiration Rate and Ethylene Production Rate
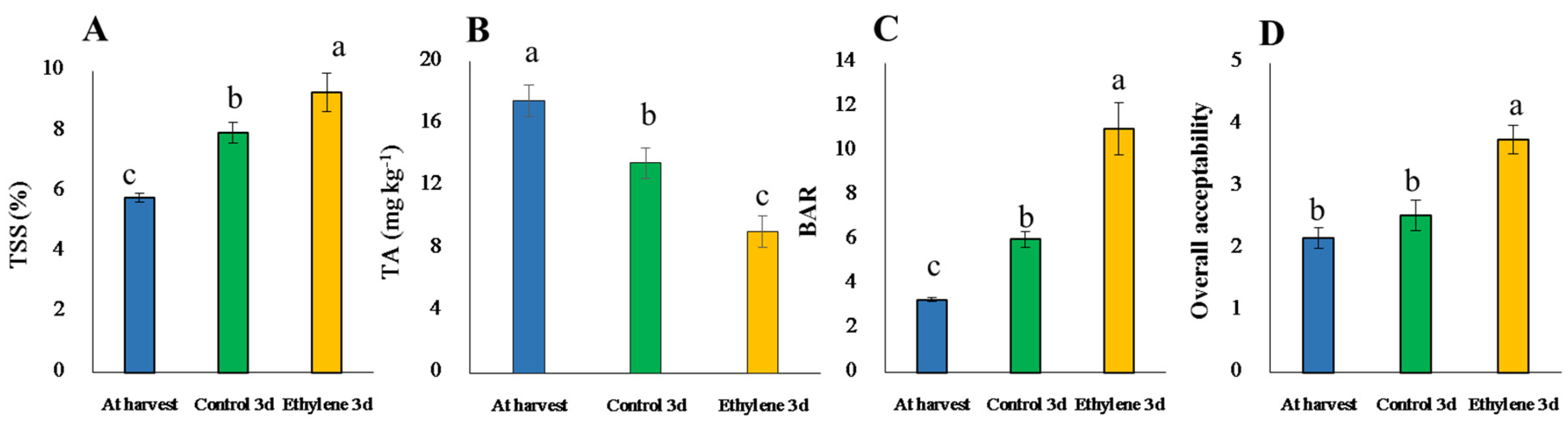
2.6. Measurement of Total Soluble Solids (TSS), Titratable Acidity (TA), Brix Acid Ratio (BAR) and Overall Acceptability
2.7. Statistical Analysis of Quality Parameters
3. Results and Discussion
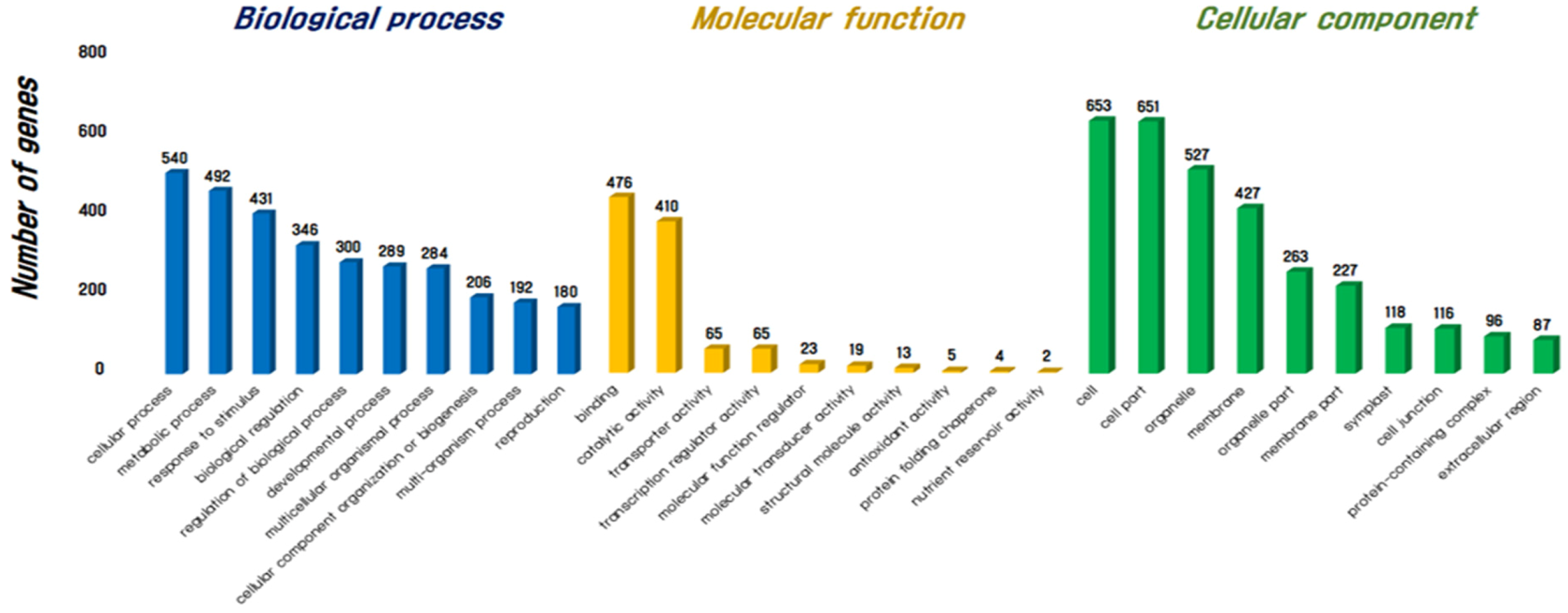
3.1. Assembly and Annotation
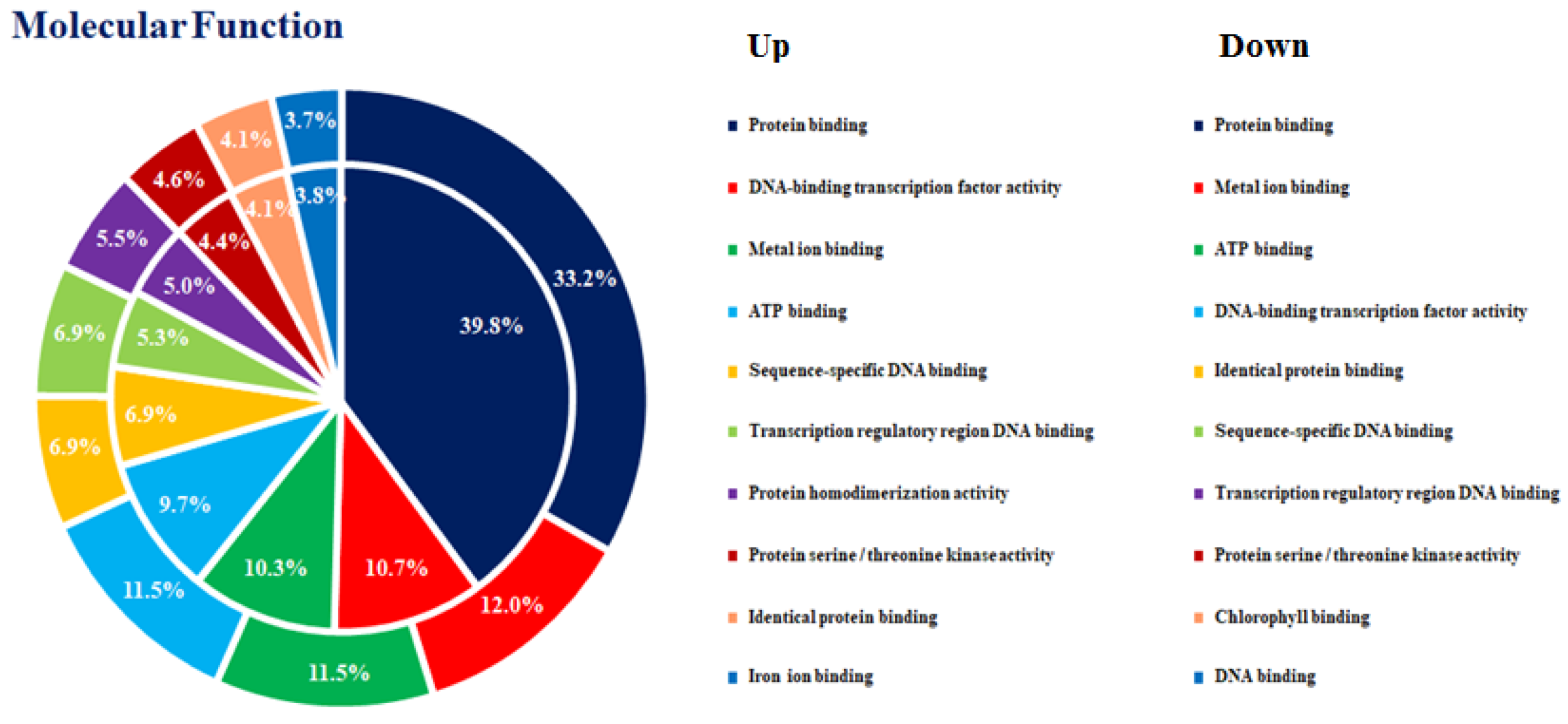
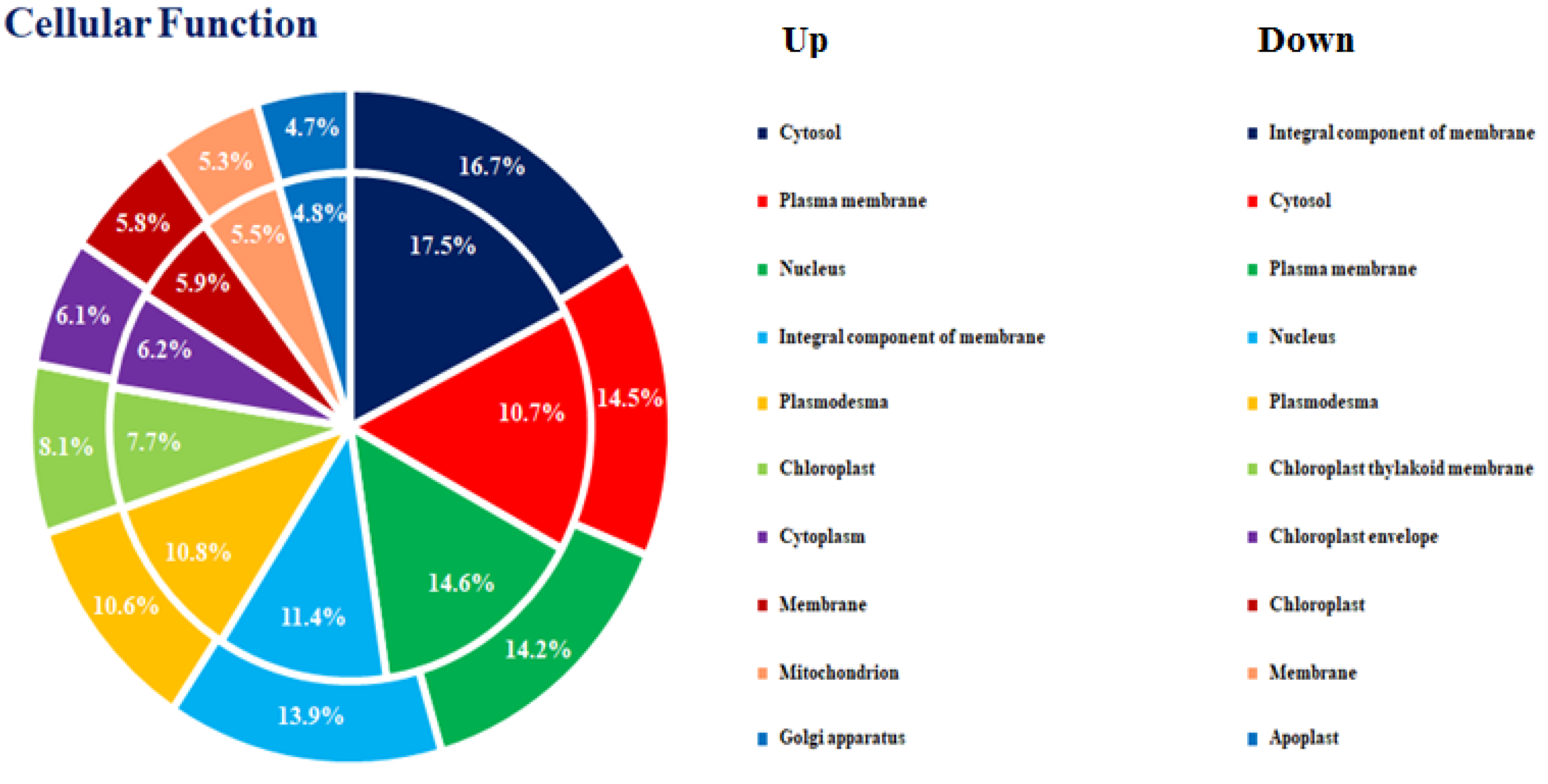
3.2. DEGs in the Comparison of Ethylene-Treated vs. Control Kiwifruit
3.3. Firmness and Genes related to Softening
3.4. Ethylene Production and Respiration Rates and Related Genes
3.5. TSS, TA, BAR and Sensory Evaluation and the Related Genes
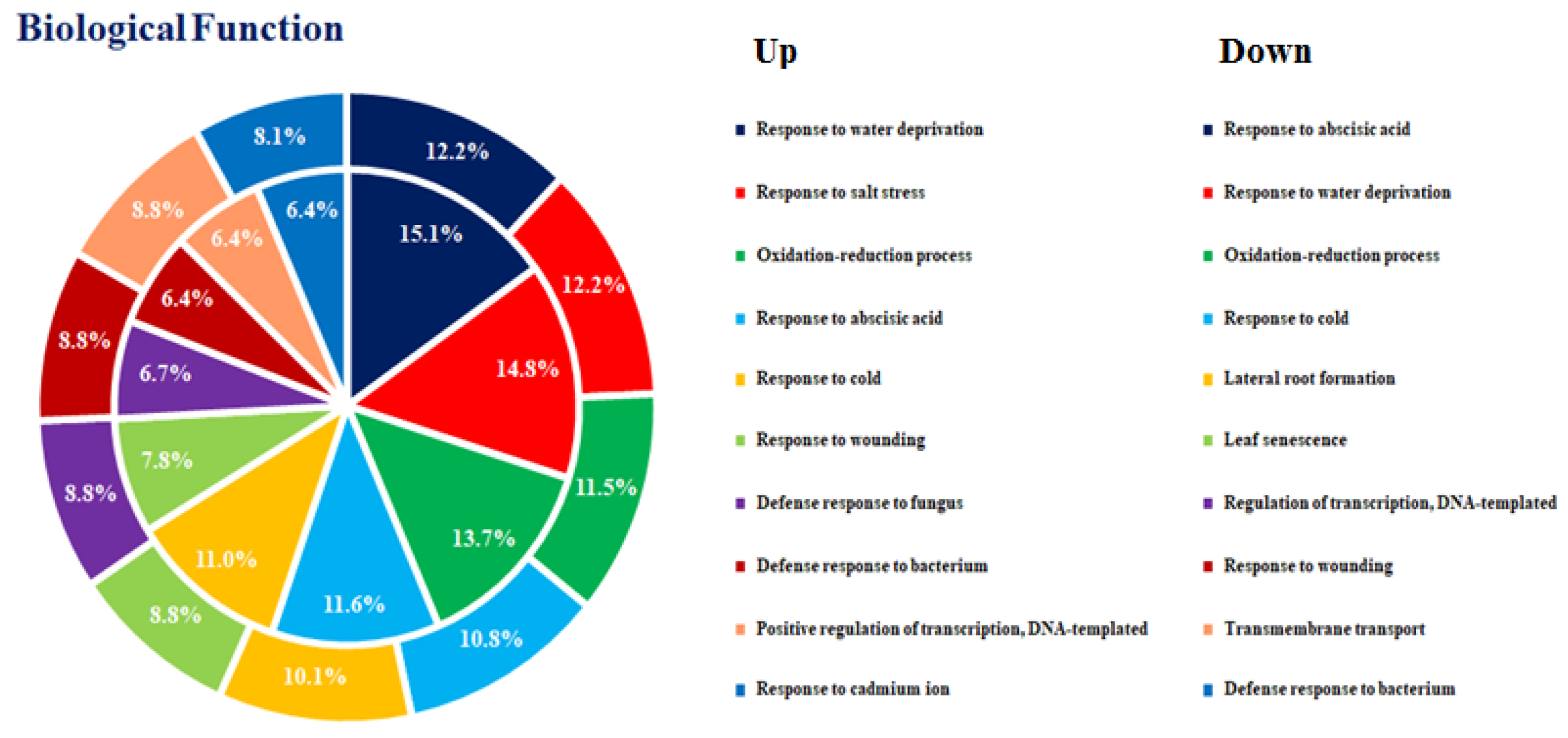
3.6. Stress-Related Genes Due to Ethylene Treatment
3.7. Color and Other Changes and the Related Genes
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kwack, Y.-B.; Kim, H.-L.; Choi, Y.-H.; Lee, J.-H.; Kim, J.G.; Lee, Y.-B. Fruit Quality and Fruit Locule Air Hole of Kiwifruit (Actinidia deliciosa cv. Hayward) Affected by Early Defoliation. Korean J. Environ. Agric. 2012, 31, 229–234. [Google Scholar] [CrossRef] [Green Version]
- FAOSTAT. Food and Agriculture Organization of the United Nations Cropping Database. 2017. Available online: http://faostat3.fao.org/home/index.html (accessed on 29 December 2018).
- Koh, Y.; Jung, J.; Hur, J. Current Status of Occurrence of Major Diseases on Kiwifruits and Their Control in KorEA. Acta Hortic. 2003, 610, 437–443. [Google Scholar] [CrossRef]
- Park, Y.; Jung, S.; Gorinstein, S. Ethylene treatment of ‘Hayward’ kiwifruits (Actinidia deliciosa) during ripening and its influence on ethylene biosynthesis and antioxidant activity. Sci. Hortic. 2006, 108, 22–28. [Google Scholar] [CrossRef]
- Kwack, Y.-B.; Kim, H.L.; Lee, J.H.; Chung, K.H.; Chae, W.B. ‘Goldone’, a Yellow—Fleshed Kiwifruit Cultivar with Large Fruit Size. Korean J. Hortic. Sci. 2017, 35, 142–146. [Google Scholar]
- Ferguson, A. Kiwifruit: Evolution of a crop. Acta Hortic. 2011, 913, 31–42. [Google Scholar] [CrossRef]
- Kim, G.H.; Jung, J.S.; Koh, Y.J. Occurrence and Epidemics of Bacterial Canker of Kiwifruit in Korea. Plant Pathol. J. 2017, 33, 351–361. [Google Scholar] [CrossRef] [Green Version]
- Choi, H.R.; Tilahun, S.; Park, D.S.; Lee, Y.M.; Choi, J.H.; Baek, M.W.; Jeong, C.S. Harvest time affects quality and storability of kiwifruit (Actinidia spp.). Sci. Hortic. 2019, 256, 108523. [Google Scholar] [CrossRef]
- Tilahun, S.; Choi, H.R.; Park, D.S.; Lee, Y.M.; Choi, J.H.; Baek, M.W.; Hyok, K.; Park, S.M.; Jeong, C.S. Ripening quality of kiwifruit cultivars is affected by harvest time. Sci. Hortic. 2020, 261, 108936. [Google Scholar] [CrossRef]
- Petkou, I.T.; Pritsa, T.S.; Sfakiotakis, E.M. Effects of polyamines on ethylene production, respiration and ripening of kiwifruit. J. Hortic. Sci. Biotechnol. 2004, 79, 977–980. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, K.; Zhang, S.; Ferguson, I. The role of salicylic acid in postharvest ripening of kiwifruit. Postharvest Boil. Technol. 2003, 28, 67–74. [Google Scholar] [CrossRef]
- Antunes, M.D.; Sfakiotakis, E. Effect of high temperature stress on ethylene biosynthesis, respiration and ripening of ‘Hayward’ kiwifruit. Postharvest Boil. Technol. 2000, 20, 251–259. [Google Scholar] [CrossRef]
- Richardson, A.; Boldingh, H.; A McAtee, P.; Gunaseelan, K.; Luo, Z.; Atkinson, R.G.; David, K.; Burdon, J.; Schaffer, R. Fruit development of the diploid kiwifruit, Actinidia chinensis ’Hort16A’. BMC Plant Boil. 2011, 11, 182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitalo, O.W.; Asiche, W.O.; Kasahara, Y.; Tosa, Y.; Owino, W.; Mworia, E.G.; Ushijima, K.; Nakano, R.; Kubo, Y. Characterization of Ripening-related Genes Involved in Ethylene-independent Low Temperature-modulated Ripening in ‘Rainbow Red’ Kiwifruit during Storage and On-vine. Hortic. J. 2018, 87, 421–429. [Google Scholar] [CrossRef] [Green Version]
- Fei, Z.; Tang, X.; Alba, R.M.; White, J.A.; Ronning, C.M.; Martin, G.B.; Tanksley, S.D.; Giovannoni, J.J. Comprehensive EST analysis of tomato and comparative genomics of fruit ripening. Plant J. 2004, 40, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Newcomb, R.; Crowhurst, R.; Gleave, A.; Rikkerink, E.H.; Allan, A.C.; Beuning, L.L.; Bowen, J.H.; Gera, E.; Jamieson, K.R.; Janssen, B.J.; et al. Analyses of Expressed Sequence Tags from Apple1. Plant Physiol. 2006, 141, 147–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asif, M.H.; Lakhwani, D.; Pathak, S.; Gupta, P.; Bag, S.; Nath, P.; Trivedi, P.K. Transcriptome analysis of ripe and unripe fruit tissue of banana identifies major metabolic networks involved in fruit ripening process. BMC Plant Boil. 2014, 14, 316. [Google Scholar] [CrossRef] [Green Version]
- Park, D.S.; Tilahun, S.; Park, K.C.; Choi, I.Y.; Jeong, C.S. Transcriptome analysis of astringent ‘Cheongdo-Bansi’ persimmon fruit treated with ethylene for removal of astringency. Postharvest Boil. Technol. 2019, 150, 52–59. [Google Scholar] [CrossRef]
- Li, W.; Liu, Y.; Zeng, S.; Xiao, G.; Wang, G.; Wang, Y.; Peng, M.; Huang, H. Gene expression profiling of development and anthocyanin accumulation in kiwifruit (Actinidia chinensis) based on transcriptome sequencing. PLoS One 2015, 10, e0138743. [Google Scholar]
- Park, D.S.; Tilahun, S.; Heo, J.Y.; Jeong, C.S. Quality and expression of ethylene response genes of ‘Daebong’ persimmon fruit during ripening at different temperatures. Postharvest Boil. Technol. 2017, 133, 57–63. [Google Scholar] [CrossRef]
- Johnson, M.T.J.; Carpenter, E.J.; Tian, Z.; Bruskiewich, R.; Burris, J.N.; Carrigan, C.T.; Chase, M.W.; Clarke, N.D.; Covshoff, S.; Depamphilis, C.W.; et al. Evaluating Methods for Isolating Total RNA and Predicting the Success of Sequencing Phylogenetically Diverse Plant Transcriptomes. PLoS ONE 2012, 7, e50226. [Google Scholar] [CrossRef]
- Kim, S.H. Study on CEACAM1 Mediated Cell Death and Antitumor Effects of Metformin in 5-Fluorouracil Resistant Gastrointestinal Cancer Cells. Ph.D Thesis, Seoul National University, Seoul, Korea, 2017. [Google Scholar]
- Tilahun, S.; Seo, M.H.; Park, D.S.; Jeong, C.S. Effect of cultivar and growing medium on the fruit quality attributes and antioxidant properties of tomato (Solanum lycopersicum L.). Hortic. Environ. Biotechnol. 2018, 59, 215–223. [Google Scholar] [CrossRef]
- Belew, D.; Park, D.S.; Tilahun, S.; Jeong, C.S. The Effects of Treatment with Ethylene-Producing Tablets on the Quality and Storability of Banana (Musa sp.). Korean J. Hortic. Sci. 2016, 34, 746–754. [Google Scholar]
- Tilahun, S.; Park, D.S.; Taye, A.M.; Jeong, C.S. Effects of Storage Duration on Physicochemical and Antioxidant Properties of Tomato (Lycopersicon esculentum Mill.). Korean J. Hortic. Sci. 2017, 35, 88–97. [Google Scholar]
- Tilahun, S.; Park, D.S.; Taye, A.M.; Jeong, C.S. Effect of ripening conditions on the physicochemical and antioxidant properties of tomato (Lycopersicon esculentum Mill.). Food Sci. Biotechnol. 2017, 26, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.H.; Tilahun, S.; Park, D.S.; Melaku, A.; Jeong, C.S. Effect of Ripening Conditions on the Quality and Storability of Muskmelon (Cucumis melo L.) Fruits. Korean J. Hortic. Sci. 2018, 36, 741–755. [Google Scholar]
- Chang, Y.-Y.; Chu, Y.-W.; Chen, C.-W.; Leu, W.-M.; Hsu, H.-F.; Yang, C.-H. Characterization of Oncidium ‘Gower Ramsey’ Transcriptomes using 454 GS-FLX Pyrosequencing and Their Application to the Identification of Genes Associated with Flowering Time. Plant Cell Physiol. 2011, 52, 1532–1545. [Google Scholar] [CrossRef] [Green Version]
- Parchman, T.L.; Geist, K.; Grahnen, J.A.; Benkman, C.W.; Buerkle, C.A. Transcriptome sequencing in an ecologically important tree species: Assembly, annotation, and marker discovery. BMC Genom. 2010, 11, 180. [Google Scholar] [CrossRef] [Green Version]
- Sui, C.; Zhang, J.; Wei, J.-H.; Chen, S.; Li, Y.; Xu, J.; Jin, Y.; Xie, C.-X.; Gao, Z.; Chen, H.; et al. Transcriptome analysis of Bupleurum chinense focusing on genes involved in the biosynthesis of saikosaponins. BMC Genom. 2011, 12, 539. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.-M.; Zhao, L.; Larson-Rabin, Z.; Li, D.-Z.; Guo, Z.-H. De Novo Sequencing and Characterization of the Floral Transcriptome of Dendrocalamus latiflorus (Poaceae: Bambusoideae). PLoS ONE 2012, 7, e42082. [Google Scholar] [CrossRef]
- Goulao, L.; Oliveira, C. Cell wall modifications during fruit ripening: When a fruit is not the fruit. Trends Food Sci. Technol. 2008, 19, 4–25. [Google Scholar] [CrossRef] [Green Version]
- Bennett, A.B.; Labavitch, J.M. Ethylene and ripening-regulated expression and function of fruit cell wall modifying proteins. Plant Sci. 2008, 175, 130–136. [Google Scholar] [CrossRef]
- Glissant, D.; Dédaldéchamp, F.; Delrot, S. Transcriptomic analysis of grape berry softening during ripening. OENO ONE 2008, 42, 1. [Google Scholar] [CrossRef]
- Figueroa, C.; Pimentel, P.; Gaete, C.; Moya, M.; Herrera, R.; Caligari, P.D.; Moya-León, M.A. Softening rate of the Chilean strawberry (Fragaria chiloensis) fruit reflects the expression of polygalacturonase and pectate lyase genes. Postharvest Boil. Technol. 2008, 49, 210–220. [Google Scholar] [CrossRef]
- Tavarini, S.; Degl’Innocenti, E.; Remorini, D.; Massai, R.; Guidi, L. Polygalacturonase and β-galactosidase activities in Hayward kiwifruit as affected by light exposure, maturity stage and storage time. Sci. Hortic. 2009, 120, 342–347. [Google Scholar] [CrossRef]
- Schroder, R.; Atkinson, R.G. Kiwifruit cell walls: Towards an understanding of softening? NZ. J. For. Sci. 2006, 36, 112–129. [Google Scholar]
- Lubkowitz, M. The Oligopeptide Transporters: A Small Gene Family with a Diverse Group of Substrates and Functions? Mol. Plant 2011, 4, 407–415. [Google Scholar] [CrossRef] [Green Version]
- Sharma, R.; Tan, F.; Jung, K.-H.; Sharma, M.K.; Peng, Z.; Ronald, P.C. Transcriptional dynamics during cell wall removal and regeneration reveals key genes involved in cell wall development in rice. Plant Mol. Boil. 2011, 77, 391–406. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Suo, J.; Han, Y.; Liang, C.; Jin, M.; Zhang, Z.; Rao, J. The effect of 1-methylcyclopropene, methyl jasmonate and methyl salicylate on lignin accumulation and gene expression in postharvest ‘Xuxiang’ kiwifruit during cold storage. Postharvest Boil. Technol. 2017, 124, 107–118. [Google Scholar] [CrossRef]
- Li, X.-J.; Nakagawa, N.; Sakurai, N. Purification and Biochemical Characterization of Cell Wall-bound Trehalase from Pericarp Tissues of Actinidia deliciosa. J. Jpn. Soc. Hortic. Sci. 2008, 77, 229–235. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.-W.; Hsu, Y.-K.; Cheng, Y.-H.; Yen, H.-C.; Wu, Y.-P.; Wang, C.-S.; Lai, C. Proteomic analysis of salt-responsive ubiquitin-related proteins in rice roots. Rapid Commun. Mass Spectrom. 2012, 26, 1649–1660. [Google Scholar] [CrossRef]
- Zenoni, S.; Fasoli, M.; Tornielli, G.B.; Santo, S.D.; Sanson, A.; De Groot, P.; Sordo, S.; Citterio, S.; Monti, F.; Pezzotti, M. Overexpression of PhEXPA1 increases cell size, modifies cell wall polymer composition and affects the timing of axillary meristem development in Petunia hybrida. New Phytol. 2011, 191, 662–677. [Google Scholar] [CrossRef]
- Schröder, R.; Atkinson, R.G.; Redgwell, R.J. Re-interpreting the role of endo-β-mannanases as mannan endotransglycosylase/hydrolases in the plant cell wall. Ann. Bot. 2009, 104, 197–204. [Google Scholar] [CrossRef] [Green Version]
- Saleme, M.D.L.S.; Cesarino, I.; Vargas, L.; Kim, H.; Vanholme, R.; Goeminne, G.; Van Acker, R.; Fonseca, F.; Pallidis, A.; Voorend, W.; et al. Silencing Caffeoyl Shikimate Esterase Affects Lignification and Improves Saccharification in Poplar. Plant Physiol. 2017, 175, 1040–1057. [Google Scholar] [CrossRef] [Green Version]
- Choat, B.; Gambetta, G.; Shackel, K.A.; Matthews, M.A. Vascular Function in Grape Berries across Development and Its Relevance to Apparent Hydraulic Isolation. Plant Physiol. 2009, 151, 1677–1687. [Google Scholar] [CrossRef] [Green Version]
- Gao, F.; Niu, Y.-D.; Hao, J.-F.; Bade, R.; Zhang, L.-Q.; Hasi, A. Identification of Differentially Expressed Genes During Ethylene Climacteric of Melon Fruit by Suppression Subtractive Hybridization. J. Integr. Agric. 2013, 12, 1431–1440. [Google Scholar] [CrossRef]
- Xu, S.; Rahman, A.; Baskin, T.I.; Kieber, J.J. Two Leucine-Rich Repeat Receptor Kinases Mediate Signaling, Linking Cell Wall Biosynthesis and ACC Synthase in Arabidopsis. Plant Cell 2008, 20, 3065–3079. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, H.; Takebayashi, S.I.; Sakamoto, A.; Igata, T.; Nakatsu, Y.; Saitoh, N.; Hino, S.; Nakao, M. The SETD8/PR-Set7 methyltransferase functions as a barrier to prevent senescence-associated metabolic remodeling. Cell Rep. 2017, 18, 2148–2161. [Google Scholar] [CrossRef] [Green Version]
- Weng, L.; Zhao, F.; Li, R.; Xu, C.; Chen, K.; Xiao, H. The Zinc Finger Transcription Factor SlZFP2 Negatively Regulates Abscisic Acid Biosynthesis and Fruit Ripening in Tomato1. Plant Physiol. 2015, 167, 931–949. [Google Scholar] [CrossRef] [Green Version]
- Roy, M.; Wu, R. Overexpression of S-adenosylmethionine decarboxylase gene in rice increases polyamine level and enhances sodium chloride-stress tolerance. Plant Sci. 2002, 163, 987–992. [Google Scholar] [CrossRef]
- Apelbaum, A.; Burgoon, A.C.; Anderson, J.D.; Lieberman, M.; Ben-Arie, R.; Mattoo, A.K. Polyamines Inhibit Biosynthesis of Ethylene in Higher Plant Tissue and Fruit Protoplasts. Plant Physiol. 1981, 68, 453–456. [Google Scholar] [CrossRef] [Green Version]
- Fujimoto, S.Y.; Ohta, M.; Usui, A.; Shinshi, H.; Ohme-Takagi, M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 2000, 12, 393–404. [Google Scholar] [PubMed] [Green Version]
- Shi, Y.; Jiang, L.; Zhang, L.; Kang, R.; Yu, Z. Dynamic changes in proteins during apple (Malus x domestica) fruit ripening and storage. Hortic. Res. 2014, 1, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moing, A.; Rothan, C.; Svanella, L.; Just, D.; Diakou, P.; Raymond, P.; Gaudillère, J.P.; Monet, R. Role of phosphoenolpyruvate carboxylase in organic acid accumulation during peach fruit development. Physiol. Plant. 2000, 108, 1–10. [Google Scholar] [CrossRef]
- Bumee, S.; Ingkasuwan, P.; Kalapanulak, S.; Meechai, A.; Cheevadhanarak, S.; Saithong, T. Transcriptional Regulatory Network of Arabidopsis Starch Metabolism under Extensive Light Condition: A Potential Model of Transcription-modulated Starch Metabolism in Roots of Starchy Crops. Procedia Comput. Sci. 2013, 23, 113–121. [Google Scholar] [CrossRef] [Green Version]
- Duque, P.; Barreiro, M.G.; Arrabaça, J.D. Respiratory metabolism during cold storage of apple fruit. I. Sucrose metabolism and glycolysis. Physiol. Plant. 1999, 107, 14–23. [Google Scholar] [CrossRef]
- Wang, J.; De Luca, V. The biosynthesis and regulation of biosynthesis of Concord grape fruit esters, including ‘foxy’ methylanthranilate. Plant J. 2005, 44, 606–619. [Google Scholar] [CrossRef]
- Rippert, P.; Matringe, M. Purification and kinetic analysis of the two recombinant arogenate dehydrogenase isoforms of Arabidopsis thaliana. JBIC J. Boil. Inorg. Chem. 2002, 269, 4753–4761. [Google Scholar] [CrossRef]
- Zhang, B.; Yin, X.-R.; Li, X.; Yang, S.-L.; Ferguson, I.B.; Chen, K.-S. Lipoxygenase Gene Expression in Ripening Kiwifruit in Relation to Ethylene and Aroma Production. J. Agric. Food Chem. 2009, 57, 2875–2881. [Google Scholar] [CrossRef]
- Yuan, Y.; Yu, S.; Yu, J.; Zhan, Z.; Li, M.; Liu, G.; Wang, X.; Huang, L. Predicting the Function of 4-Coumarate:CoA Ligase (LJ4CL1) in Lonicera japonica. Int. J. Mol. Sci. 2014, 15, 2386–2399. [Google Scholar] [CrossRef] [Green Version]
- Gershater, M.C.; Edwards, R. Regulating biological activity in plants with carboxylesterases. Encycl. Appl. Plant Sci. 2007, 173, 579–588. [Google Scholar] [CrossRef]
- Yauk, Y.-K.; Ged, C.; Wang, M.Y.; Matich, A.J.; Tessarotto, L.; Cooney, J.; Chervin, C.; Atkinson, R.G. Manipulation of flavour and aroma compound sequestration and release using a glycosyltransferase with specificity for terpene alcohols. Plant J. 2014, 80, 317–330. [Google Scholar] [CrossRef]
- Kumar, V.; Prakash, O.; Kumar, S.; Narwal, S. α-glucosidase inhibitors from plants: A natural approach to treat diabetes. Pharmacogn. Rev. 2011, 5, 19–29. [Google Scholar] [CrossRef] [Green Version]
- Nishiyama, I.; Yamashita, Y.; Yamanaka, M.; Shimohashi, A.; Fukuda, T.; Oota, T. Varietal Difference in Vitamin C Content in the Fruit of Kiwifruit and OtherActinidiaSpecies. J. Agric. Food Chem. 2004, 52, 5472–5475. [Google Scholar] [CrossRef]
- Wilson, J.X. The physiological role of dehydroascorbic acid. FEBS Lett. 2002, 527, 5–9. [Google Scholar] [CrossRef] [Green Version]
- Vissers, M.C.M.; Carr, A.C.; Pullar, J.M.; Bozonet, S.M. The Bioavailability of Vitamin C from Kiwifruit. Adv. Food Nutr. Res. 2013, 68, 125–147. [Google Scholar]
- Shin, N.; Moon, S.-J.; Han, S.; Kim, B.-G.; Park, S.R.; Lee, S.-K.; Yoon, H.-J.; Lee, H.E.; Kwon, H.-B.; Baek, N.; et al. Expression of StMYB1R-1, a Novel Potato Single MYB-Like Domain Transcription Factor, Increases Drought Tolerance. Plant Physiol. 2010, 155, 421–432. [Google Scholar] [CrossRef] [Green Version]
- Lopez, A.P.; Portales, R.B.; Muñoz-Blanco, J.; Rodríguez-Franco, A.; López-Ráez, J.A.; Medina-Escobar, N. Characterization of a strawberry late-expressed and fruit-specific peptide methionine sulphoxide reductase. Physiol. Plant. 2006, 126, 129–139. [Google Scholar] [CrossRef]
- Tkaczuk, K.L.; Shumilin, I.A.; Chruszcz, M.; Evdokimova, E.; Savchenko, A.; Minor, W. Structural and functional insight into the universal stress protein family. Evol. Appl. 2013, 6, 434–449. [Google Scholar] [CrossRef]
- Bansal, N.; Kanwar, S.S. Peroxidase(s) in Environment Protection. Sci. World J. 2013, 2013, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Palafox-Carlos, H.; Contreras-Vergara, C.; Muhlia-Almazan, A.; Islas-Osuna, M.; González-Aguilar, G. Expression and enzymatic activity of phenylalanine ammonia-lyase and p-coumarate 3-hydroxylase in mango (Mangifera indica ‘Ataulfo’) during ripening. Genet. Mol. Res. 2014, 13, 3850–3858. [Google Scholar] [CrossRef]
- Boudet, A.-M. Evolution and current status of research in phenolic compounds. Phytochemistry 2007, 68, 2722–2735. [Google Scholar] [CrossRef]
- Han, X.; Lu, W.; Wei, X.; Li, L.; Mao, L.; Zhao, Y. Proteomics analysis to understand the ABA stimulation of wound suberization in kiwifruit. J. Proteom. 2018, 173, 42–51. [Google Scholar] [CrossRef]
- Davison, P.A.; Hunter, C.N.; Horton, P. Overexpression of β-carotene hydroxylase enhances stress tolerance in Arabidopsis. Nature 2002, 418, 203–206. [Google Scholar] [CrossRef]
- Queiroz, C.; Mendes Lopes, M.L.; Fialho, E.; Valente-Mesquita, V.L. Polyphenoloxidase: Characteristics and mechanisms of browning control. Food Rev. Int. 2008, 24, 361–375. [Google Scholar] [CrossRef]
- Khanna-Chopra, R.; Sabarinath, S. Heat-stable chloroplastic Cu/Zn superoxide dismutase in Chenopodium murale. Biochem. Biophys. Res. Commun. 2004, 320, 1187–1192. [Google Scholar] [CrossRef]
- Figueiredo, J.; Silva, M.S.; Figueiredo, A. Subtilisin-like proteases in plant defence: The past, the present and beyond. Mol. Plant Pathol. 2017, 19, 1017–1028. [Google Scholar] [CrossRef]
- Martinoia, E.; Klein, M.; Geisler, M.; Bovet, L.; Forestier, C.; Kolukisaoglu, U.; Müller-Röber, B.; Schulz, B. Multifunctionality of plant ABC transporter—More than just detoxifiers. Planta 2002, 214, 345–355. [Google Scholar] [CrossRef]
- Minas, I.S.; Tanou, G.; Krokida, A.; Karagiannis, E.; Belghazi, M.; Vasilakakis, M.; Papadopoulou, K.K.; Molassiotis, A. Ozone-induced inhibition of kiwifruit ripening is amplified by 1-methylcyclopropene and reversed by exogenous ethylene. BMC Plant Boil. 2018, 18, 358. [Google Scholar] [CrossRef]
- Zeng, L.; Deng, R.; Guo, Z.; Yang, S.; Deng, X.-P. Genome-wide identification and characterization of Glyceraldehyde-3-phosphate dehydrogenase genes family in wheat (Triticum aestivum). BMC Genom. 2016, 17, 240. [Google Scholar] [CrossRef] [Green Version]
- Pilkington, S.; Montefiori, M.; Galer, A.L.; Emery, R.J.N.; Allan, A.C.; Jameson, P.E. Endogenous cytokinin in developing kiwifruit is implicated in maintaining fruit flesh chlorophyll levels. Ann. Bot. 2013, 112, 57–68. [Google Scholar] [CrossRef] [Green Version]
- Ha, S.; Vanková, R.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.-S.P. Cytokinins: Metabolism and function in plant adaptation to environmental stresses. Trends Plant Sci. 2012, 17, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Takasaki, H.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta (BBA)-Bioenerg. 2012, 1819, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Kesari, R.; Trivedi, P.K.; Nath, P. Ethylene-induced ripening in banana evokes expression of defense and stress related genes in fruit tissue. Postharvest Boil. Technol. 2007, 46, 136–143. [Google Scholar] [CrossRef]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta (BBA)-Bioenerg. 2012, 1819, 120–128. [Google Scholar] [CrossRef]
- Zhu, X.; Pan, L.; Xiao, T.; Ren, X.; Liu, Z. Exogenous niacin treatment increases NADPH oxidase in kiwifruit. Braz. J. Boil. 2018, 78, 686–690. [Google Scholar] [CrossRef] [Green Version]
- Horie, Y.; Ito, H.; Kusaba, M.; Tanaka, R.; Tanaka, A. Participation of Chlorophyll b Reductase in the Initial Step of the Degradation of Light-harvesting Chlorophyll a/b-Protein Complexes in Arabidopsis. J. Boil. Chem. 2009, 284, 17449–17456. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Huang, R.; Quan, R. Mutation in Mg-Protoporphyrin IX Monomethyl Ester Cyclase Decreases Photosynthesis Capacity in Rice. PLoS ONE 2017, 12, e0171118. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Nishizawa, T.; Takemoto, M.; Kumazaki, K.; Yamashita, K.; Hirata, K.; Minoda, A.; Nagatoishi, S.; Tsumoto, K.; Ishitani, R.; et al. Structure of the triose-phosphate/phosphate translocator reveals the basis of substrate specificity. Nat. Plants 2017, 3, 825–832. [Google Scholar] [CrossRef]
- Pilkington, S.; Montefiori, M.; Jameson, P.E.; Allan, A.C. The control of chlorophyll levels in maturing kiwifruit. Planta 2012, 236, 1615–1628. [Google Scholar] [CrossRef]
- Keown, J.; Griffin, M.D.W.; Mertens, H.D.T.; Pearce, F. Small Oligomers of Ribulose-bisphosphate Carboxylase/Oxygenase (Rubisco) Activase Are Required for Biological Activity. J. Boil. Chem. 2013, 288, 20607–20615. [Google Scholar] [CrossRef] [Green Version]
- Boggio, S.B.; Palatnik, J.F.; Heldt, H.W.; Valle, E. Changes in amino acid composition and nitrogen metabolizing enzymes in ripening fruits of Lycopersicon esculentum Mill. Plant Sci. 2000, 159, 125–133. [Google Scholar] [CrossRef]
- Binder, S. Branched-Chain Amino Acid Metabolism in Arabidopsis thaliana. Arab. Book 2010, 8, e0137. [Google Scholar] [CrossRef] [Green Version]
- Ringer, K.L.; Davis, E.M.; Croteau, R. Monoterpene metabolism. Cloning, expression, and characterization of (−)-isopiperitenol/(−)-carveol dehydrogenase of peppermint and spearmint. Plant Physiol. 2005, 137, 863–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Lin-Wang, K.; Deng, C.; Warran, B.; Wang, L.; Yu, B.; Yang, H.; Wang, J.; Espley, R.; Zhang, J.; et al. Comparative Transcriptome Analysis of White and Purple Potato to Identify Genes Involved in Anthocyanin Biosynthesis. PLoS ONE 2015, 10, 0129148. [Google Scholar] [CrossRef] [PubMed]
- Onik, J.C.; Hu, X.; Lin, Q.; Wang, Z. Comparative Transcriptomic Profiling to Understand Pre- and Post-Ripening Hormonal Regulations and Anthocyanin Biosynthesis in Early Ripening Apple Fruit. Molecules 2018, 23, 1908. [Google Scholar] [CrossRef] [Green Version]
- Li, S.-B.; Xie, Z.-Z.; Hu, C.-G.; Zhang, J.-Z. A Review of Auxin Response Factors (ARFs) in Plants. Front. Plant Sci. 2016, 7, 137. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Sample Name | Run Format | Max Read Length | Total Reads | Mapped Reads | Mapping Rate |
|---|---|---|---|---|---|
| Kiwi Control | 101 × 2 | 101 | 51,331,356 | 41,747,134 | 81.33 |
| Kiwi Ethylene | 101 × 2 | 101 | 50,615,434 | 41,801,951 | 82.59 |
| Average | 101 × 2 | 101 | 50,973,395 | 41,774,542 | 81.96 |
| Sample Name | Yield (Bases) | Reads | % of ≥Q30 Bases (PF) | Mean Quality Score (PF) |
|---|---|---|---|---|
| Kiwi Control | 3,102,225,807 | 30,715,107 | 94.915 | 35.155 |
| Kiwi Ethylene | 3,119,780,718 | 30,888,918 | 95.395 | 35.240 |
| Selection Criteria | Upregulation | Downregulation | Total |
|---|---|---|---|
| 2-fold | 13,361 | 15,221 | 28,582 |
| 2-fold and p value < 0.05 | 1682 | 855 | 2537 |
| 2-fold, p value < 0.05 and FDR < 0.1 | 9 | 3 | 12 |
| Locus/Accession | Gene Description | Log Fold Change | p value |
|---|---|---|---|
| CM009673.1 | Methanol O-anthraniloyltransferase | 7.13 | 0.041 |
| CM009677.1 | Transcription factor MYB1R1-like | 7.08 | 0.049 |
| CM009663.1 | Oligopeptide transporters (OPTs) | 6.82 | 0.041 |
| CM009661.1 | Phenylalanine ammonia-lyase (PAL) | 6.79 | 0.001 |
| CM009676.1 | Peptide methionine sulfoxide reductase (MsrA) | 6.74 | 0.041 |
| CM009657.1 | L-Ascorbate oxidase | 6.70 | 0.006 |
| CM009654.1 | Pectate lyase (PL) | 6.41 | 0.049 |
| CM009668.1 | β-Amylase | 6.30 | 0.001 |
| CM009674.1 | Universal stress protein A-like protein | 6.24 | 0.007 |
| CM009660.1 | 3-Ketoacyl-CoA synthase | 6.23 | 0.001 |
| CM009656.1 | Polygalacturonase (PG) | 6.16 | 0.005 |
| CM009682.1 | Gibberellin 2-beta-dioxygenase | 6.16 | 0.007 |
| CM009656.1 | β-Carotene hydroxylase | 6.11 | 0.005 |
| CM009666.1 | Xyloglucan endotransglucosylase/hydrolase (XTH) protein-like | 5.38 | 0.044 |
| CM009680.1 | Peroxidase | 5.16 | 0.042 |
| CM009680.1 | Galacturonosyltransferase (GAUT) | 5.02 | 0.004 |
| CM009660.1 | L-Gulonolactone oxidase | 4.95 | 0.005 |
| CM009673.1 | 1-Aminocyclopropane-1-carboxylate oxidase (ACCO) | 4.81 | 0.002 |
| CM009671.1 | Polyphenol oxidase (PPO) | 4.81 | 0.001 |
| CM009657.1 | Ethylene-responsive transcription factor RAP2-10-like | 4.57 | 0.003 |
| CM009656.1 | Aspartate aminotransferase (AST) | 4.53 | 0.005 |
| CM009664.1 | β-Galactosidase (β-gal) | 4.50 | 0.011 |
| CM009676.1 | Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | 4.29 | 0.002 |
| CM009666.1 | Ethylene-responsive transcription factor RAP2-3-like | 4.06 | 0.007 |
| CM009679.1 | Cinnamoyl-CoA reductase (CCR) | 3.75 | 0.044 |
| CM009680.1 | Adenylate isopentenyltransferase | 3.71 | 0.007 |
| CM009659.1 | NAC transcription factor | 3.65 | 0.015 |
| CM009672.1 | Prolyl 4-hydroxylase (P4H) | 3.64 | 0.012 |
| CM009659.1 | β-1,4-Mannosyl-glycoprotein-like | 3.58 | 0.018 |
| CM009668.1 | WRKY transcription factor | 3.47 | 0.018 |
| CM009680.1 | Ribose-5-phosphate isomerase (Rpi) | 3.35 | 0.023 |
| CM009680.1 | UDP-glucose 6-dehydrogenase | 3.34 | 0.024 |
| CM009662.1 | Glutamate dehydrogenase B (GLDH) | 3.20 | 0.023 |
| CM009670.1 | Ethylene-responsive transcription factor 4-like | 3.00 | 0.010 |
| CM009675.1 | Pyruvate kinase | 2.99 | 0.020 |
| CM009669.1 | Arogenate dehydrogenase | 2.91 | 0.046 |
| CM009659.1 | NADPH oxidase | 2.77 | 0.020 |
| CM009680.1 | S-Adenosylmethionine decarboxylase beta chain-like | 2.75 | 0.041 |
| CM009667.1 | Trehalase | 2.63 | 0.026 |
| CM009678.1 | Pectin acetylesterase (PAE) | 2.60 | 0.026 |
| CM009659.1 | 6-Phosphogluconolactonase (6PGL) | 2.48 | 0.046 |
| CM009666.1 | Sucrose-phosphate synthase | 2.38 | 0.043 |
| Locus/Accession | Gene Description | Log Fold Change | p value |
|---|---|---|---|
| CM009675.1 | Chlorophyll a-b binding protein-like | −8.26 | 0.038 |
| CM009671.1 | Chlorophyll a-b binding protein 3Cprecursor | −8.00 | 0.002 |
| CM009665.1 | Chlorophyll a-b binding protein 37precursor | −6.23 | 0.001 |
| CM009681.1 | Auxin response factor-like | −6.19 | 0.035 |
| CM009681.1 | Linoleate 13S-lipoxygenase | −5.74 | 0.002 |
| CM009674.1 | Glucose-1-phosphate adenylyltransferase large subunit 2/amyloplastic-like | −5.70 | 0.033 |
| CM009659.1 | Aquaporin PIP2-7-like | −5.68 | 0.002 |
| CM009657.1 | Carboxylesterase | −5.55 | 0.037 |
| CM009666.1 | LRR receptor-like serine/threonine-protein kinase | −5.55 | 0.039 |
| CM009673.1 | Zinc finger protein-like | −5.50 | 0.034 |
| CM009681.1 | ABC transporter G family member 3-like | −5.32 | 0.041 |
| CM009676.1 | UDP-glycosyltransferase | −5.31 | 0.001 |
| CM009678.1 | Phosphoenolpyruvate carboxylase (PEP carboxylase) | −5.26 | 0.039 |
| CM009677.1 | Ribulose bisphosphate carboxylase/oxygenase activase (RuBisCO) | −4.86 | 0.006 |
| CM009662.1 | Ethylene-responsive transcription factor TINY-like | −4.79 | 0.046 |
| CM009674.1 | Receptor-like protein kinase | −4.64 | 0.049 |
| CM009667.1 | Cellulose synthase A catalytic subunit 8 (UDP-forming)-like | −4.61 | 0.003 |
| CM009678.1 | Methyltransferase | −4.38 | 0.003 |
| CM009675.1 | Triose phosphate/phosphate translocator TPT-like | −4.35 | 0.007 |
| CM009682.1 | Expansin-A1-like | −4.08 | 0.046 |
| CM009657.1 | Subtilisin-like protease | −3.97 | 0.007 |
| CM009673.1 | 3-Hydroxyisobutyrate dehydrogenase-like | −3.78 | 0.009 |
| CM009674.1 | Mannan endo-1,4-beta-mannosidase | −3.65 | 0.017 |
| CM009656.1 | Caffeoylshikimate esterase (CSE) | −3.62 | 0.014 |
| CM009678.1 | α-Glucosidase | −3.59 | 0.010 |
| CM009661.1 | Linoleate 9S-lipoxygenase | −3.36 | 0.017 |
| CM009673.1 | Superoxide dismutase (SOD) | −3.35 | 0.016 |
| CM009669.1 | Flavonoid 3’-monooxygenase (F3’H) | −3.29 | 0.023 |
| CM009665.1 | Chlorophyll(ide) b reductase | −2.99 | 0.030 |
| CM009669.1 | (-)-Isopiperitenol/(-)-carveol dehydrogenase | −2.81 | 0.048 |
| CM009656.1 | Magnesium-protoporphyrin IX monomethyl ester (oxidative) cyclase | −2.76 | 0.017 |
| CM009677.1 | Aquaporin PIP1-4-like | −2.58 | 0.050 |
| CM009674.1 | 4-Coumarate-CoA ligase-like | −2.27 | 0.027 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tilahun, S.; Choi, H.R.; Kwon, H.; Park, S.M.; Park, D.S.; Jeong, C.S. Transcriptome Analysis of ‘Haegeum’ Gold Kiwifruit Following Ethylene Treatment to Improve Postharvest Ripening Quality. Agronomy 2020, 10, 487. https://doi.org/10.3390/agronomy10040487
Tilahun S, Choi HR, Kwon H, Park SM, Park DS, Jeong CS. Transcriptome Analysis of ‘Haegeum’ Gold Kiwifruit Following Ethylene Treatment to Improve Postharvest Ripening Quality. Agronomy. 2020; 10(4):487. https://doi.org/10.3390/agronomy10040487
Chicago/Turabian StyleTilahun, Shimeles, Han Ryul Choi, Hyok Kwon, Sung Min Park, Do Su Park, and Cheon Soon Jeong. 2020. "Transcriptome Analysis of ‘Haegeum’ Gold Kiwifruit Following Ethylene Treatment to Improve Postharvest Ripening Quality" Agronomy 10, no. 4: 487. https://doi.org/10.3390/agronomy10040487
APA StyleTilahun, S., Choi, H. R., Kwon, H., Park, S. M., Park, D. S., & Jeong, C. S. (2020). Transcriptome Analysis of ‘Haegeum’ Gold Kiwifruit Following Ethylene Treatment to Improve Postharvest Ripening Quality. Agronomy, 10(4), 487. https://doi.org/10.3390/agronomy10040487





