Genome-Wide Identification, Structure Characterization, and Expression Profiling of Dof Transcription Factor Gene Family in Wheat (Triticum aestivum L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Classification the Dof Gene Family Members from Wheat
2.2. Phylogenetic Analysis of Dof Genes
2.3. Chromosomal Locations
2.4. Characterization of Predicted TaDof Proteins
2.5. Analysis of Dof Motifs and Gene Structures
2.6. Gene Duplication and Ka/Ks Analyses
2.7. Functional Annotation Sub-Cellular Localization Prediction, and Experimental Confirmation
2.8. Expresssion Pattern Analysis of TaDofs Using Lager-Scale Transcriptome Data
2.9. Identification of miRNA and Cis-Regulatory Element
2.10. Real Time-Quantitative PCR (RT-qPCR)
3. Results
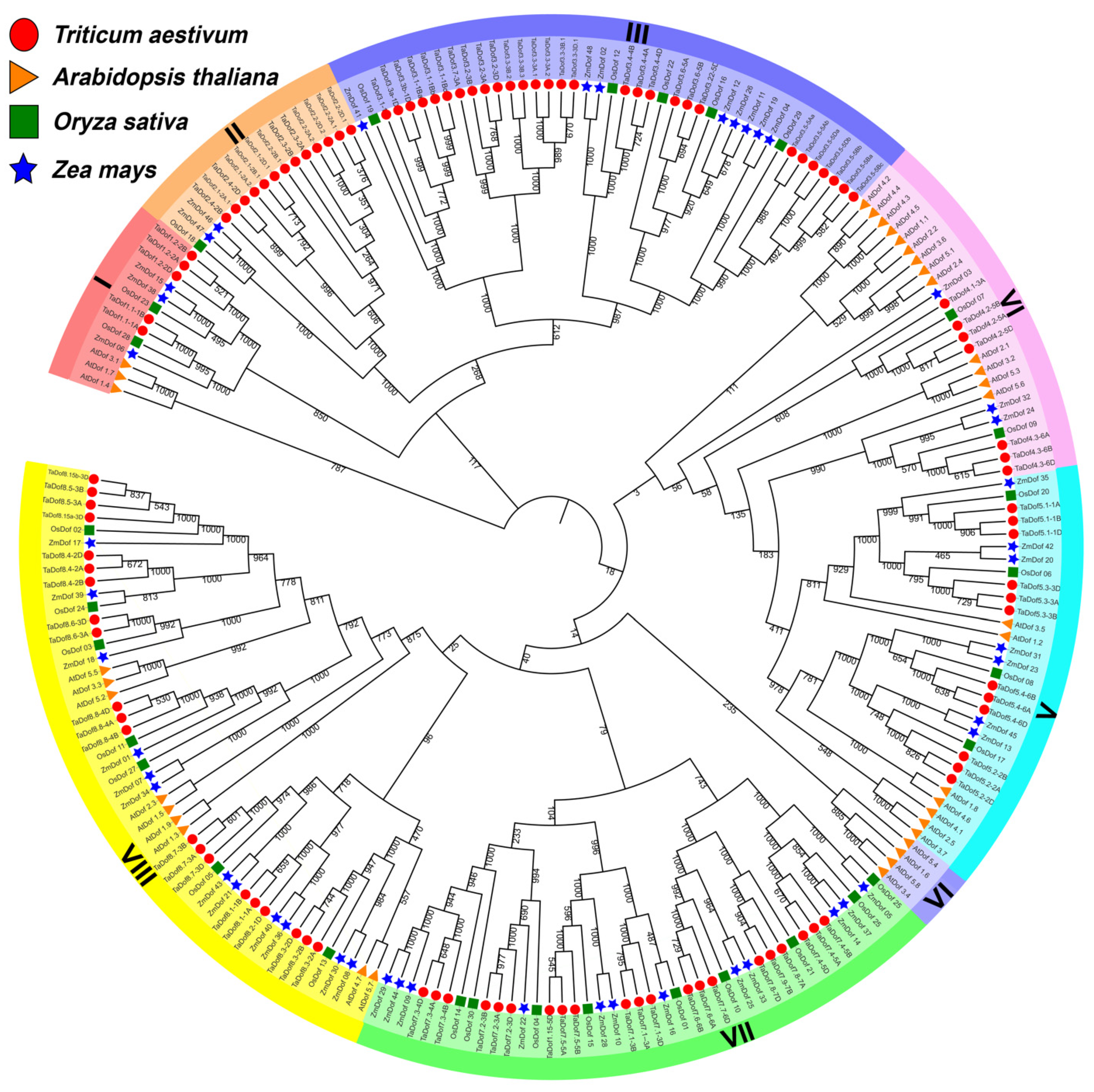
3.1. Identification of 108 TaDofs in Seven Sub-Groups
3.2. The Variable TaDof Proteins Contain a Conserved Zinc-Finger Structure
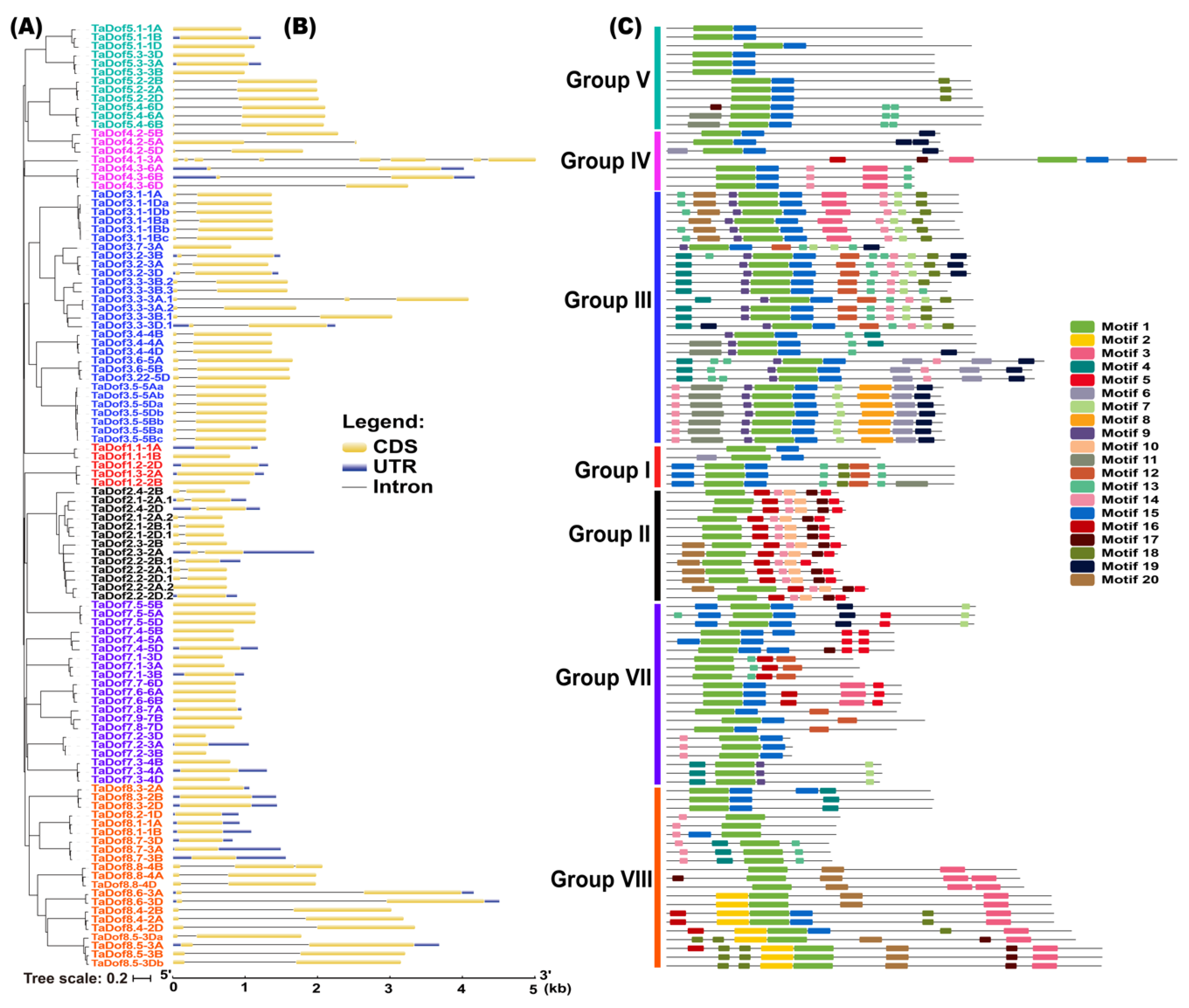
3.3. Gene Structure and Conserved Motifs Are Similar Intragroup but Diverse Intergroup
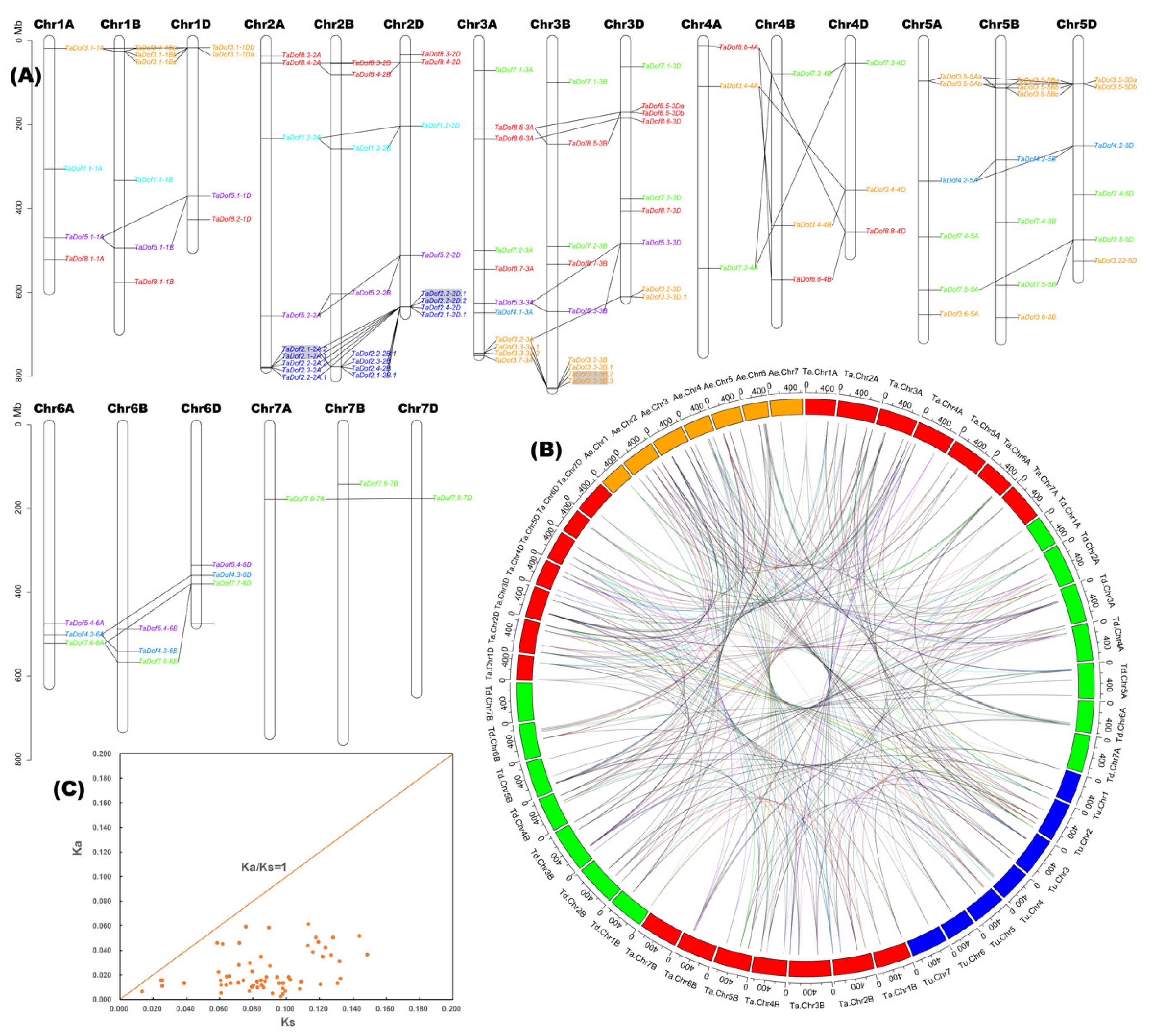
3.4. Polyploidization is the Main Basis of Member Expansion of TaDofs
3.5. Nuclear Localizations of TaDofs Match Their Regulatory Roles during Transcription
3.6. Transcriptome Analysis Revealed Diverse Expression Patterns of TaDofs
3.7. miRNAs and Cis-Regulators Are Essential Players in Molding TaDofs Expression Patterns
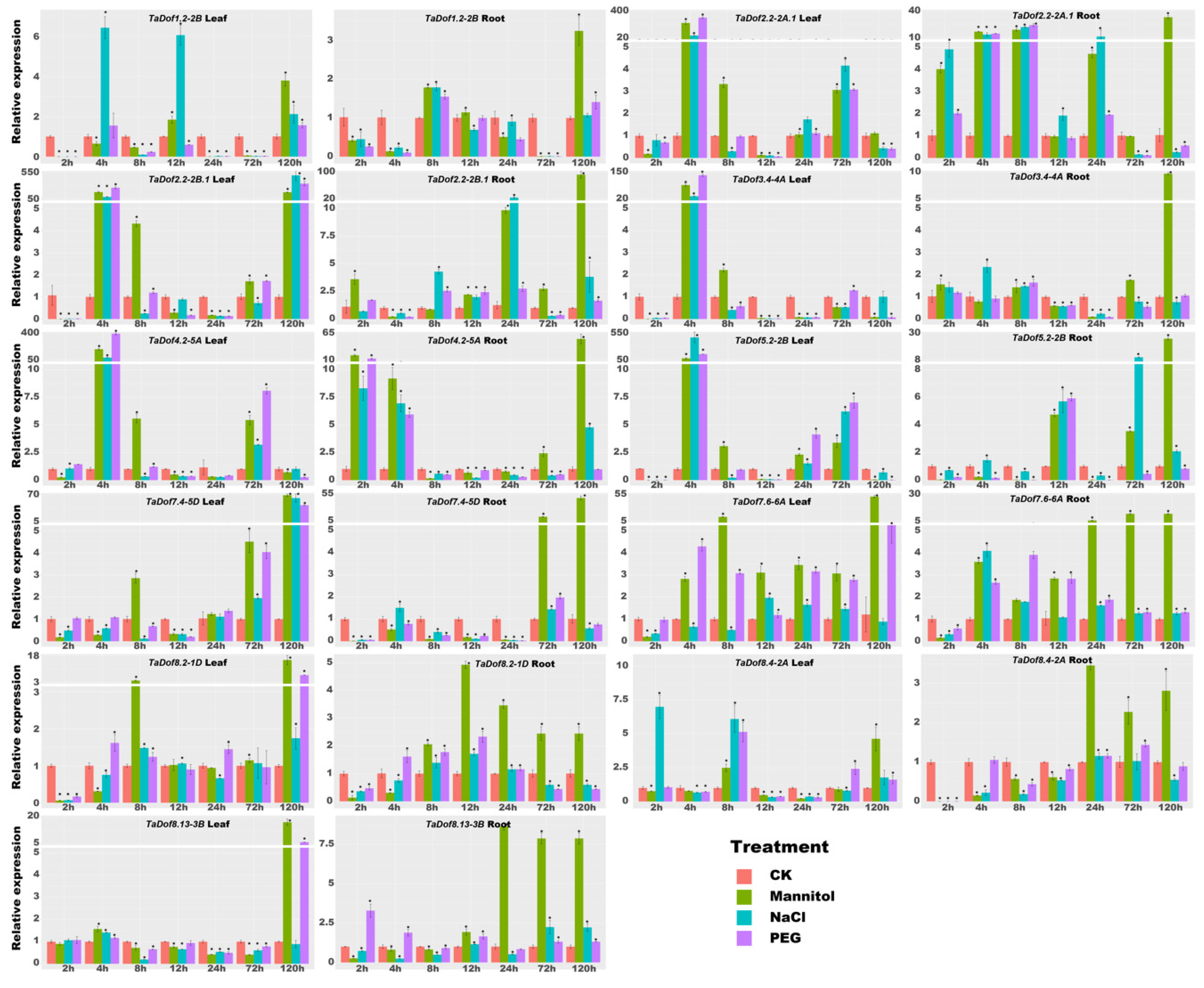
3.8. qRT-PCR Confirmed the Response Capability of TaDofs to Stress Conditions
3.9. Possible Diverse Roles of TaDofs Were Uncovered by BLASTp Searches
4. Discussions
4.1. A Large Number of TaDof Genes Are Present in Common Wheat
4.2. Negatviely Selected TaDof Genes Expand by Polyploidization
4.3. Integration of Expression Profiles, miRNA, and Cis-Elements Uncovers Complex Regulatory Patterns of TaDofs
4.4. A Large Number of TaDof Genes Are Present in Common Wheat
4.5. Functional Clues of Nuclear Localizaed TaDofs Were Uncovered by Customized Annotation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| TaDof | Wheat Dof |
| Dof | DNA binding with one finger |
| aa | Amino acids |
| ML | Maximum Likelihood |
| MW | Molecular weight |
| pI | Isoelectric point |
| GRAVY | Grand average hydropathy |
| Ka | Non-synonymous substitution rate |
| Ks | Synonymous substitution rate |
| Ka/Ks | Ratio of non-synonymous to synonymous mutation rates |
| GO | Gene ontology |
| qRT-PCR | quality real time PCR |
| GFP | Green fluorescent protein |
| TPM | transcripts per kilobase of exon model per million mapped reads |
References
- Boccaccini, A.; Santopolo, S.; Capauto, D.; Lorrai, R.; Minutello, E.; Serino, G.; Costantino, P.; Vittorioso, P. The Dof protein DAG1 and the DELLA protein GAI cooperate in negatively regulating the AtGA3ox1 gene. Mol. Plant 2014, 7, 1486–1489. [Google Scholar] [CrossRef]
- Ma, J.; Li, M.Y.; Wang, F.; Tang, J.; Xiong, A.S. Genome-wide analysis of Dof family transcription factors and their responses to abiotic stresses in Chinese cabbage. BMC Genom. 2015, 16, 33. [Google Scholar] [CrossRef]
- Zhang, X.; Yin, Y.J.; Fan, B.; Li, H.J.; Fei, X.Y.; Cui, X.Y. Advances on the structural characteristics and function of Dof transcription factors in plant. Crops 2016, 2, 14–20. [Google Scholar]
- Liu, L.; White, M.J.; Macrae, T.H. Transcription factors and their genes in higher plants functional domains, evolution and regulation. Eur. J. Biochem. 2010, 262, 247–257. [Google Scholar] [CrossRef]
- Cai, M.; Yuan, T.; Duan, L.; Li, X.; Wang, S. Identification of potential protein regulators bound to the tissue-specific positive and negative cis-acting elements of a green tissue-specific promoter in rice. Plant Biol. 2010, 10, 771–777. [Google Scholar] [CrossRef]
- Umemura, Y.; Ishiduka, T.; Yamamoto, R.; Esaka, M. The Dof domain, a zinc finger DNA-binding domain conserved only in higher plants, truly functions as a Cys2/Cys2 Zn finger domain. Plant J. 2004, 37, 741–749. [Google Scholar] [CrossRef]
- Yanagisawa, S.; Schmidt, R.J. Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. 1999, 17, 209–214. [Google Scholar] [CrossRef]
- Yanagisawa, S.; Izui, K. Molecular cloning of two DNA-binding proteins of maize that are structurally different but interact with the same sequence motif. J. Biol. Chem. 1993, 268, 16028–16036. [Google Scholar]
- Lijavetzky, D.; Carbonero, P.; Vicente-Carbajosa, J. Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol. Biol. 2003, 3, 17. [Google Scholar] [CrossRef]
- Moreno-Risueno, M.A.; Martínez, M.; Vicente-Carbajosa, J.; Carbonero, P. The family of DOF transcription factors: From green unicellular algae to vascular plants. Mol. Genet. Genomic. 2007, 277, 379–390. [Google Scholar] [CrossRef]
- Cai, X.F.; Zhang, Y.Y.; Zhang, C.J.; Zhang, T.Y.; Hu, T.X.; Ye, J.; Zhang, J.H.; Wang, T.T.; Li, H.X.; Ye, Z.B. Genome-wide analysis of plant-specific Dof transcription factor family in tomato. J. Integr. Plant Biol. 2013, 55, 552–566. [Google Scholar] [CrossRef]
- Ge, M.; Lv, Y.; Li, T.; Zhang, T.F.; Zhang, X.L.; Zhao, H. Genome-wide identification and analysis of Dof transcription factor family in maize. Sci. Agric. Sin. 2014, 47, 4563–4572. [Google Scholar]
- Dong, C.; Hu, H.; Xie, J. Genome-wide analysis of the DNA-binding with one zinc finger (Dof) transcription factor family in bananas. Genome 2016, 59, 1085–1100. [Google Scholar] [CrossRef]
- Diaz, I.; Vicente-Carbajosa, J.; Abraham, Z.; Martínez, M.; Isabel-La, M.; Carbonero, P. The GAMYB protein from barley interacts with the Dof transcription factor BPBF and activates endosperm specific genes during seed development. Plant J. 2002, 29, 453–464. [Google Scholar] [CrossRef]
- Krohn, N.M.; Yanagisawa, S.; Grasser, K.D. Specificity of the stimulatory interaction between chromosomal HMBG proteins and the transcription factor Dof2 and its negative regulation by protein kinase CK2 mediated phosphorylation. J. Biol. Chem. 2002, 27736, 32438–32444. [Google Scholar] [CrossRef]
- Noguero, M.; Atif, R.M.; Ochatt, S.; Thompson, R.D. The role of the DNA-binding one zinc finger (DOF) transcription factor family in plants. Plant Sci. 2013, 209, 32–45. [Google Scholar] [CrossRef]
- Yang, J.; Yang, M.F.; Zhang, W.P.; Chen, F.; Shen, S.H. A putative flowering-time-related Dof transcription factor gene, JcDof3, is controlled by the circadian clock in Jatropha curcas. Plant Sci. 2011, 181, 667–674. [Google Scholar] [CrossRef]
- Konishi, M.; Yanagisawa, S. Sequential activation of two Dof transcription factor gene promoters during vascular development in Arabidopsis thaliana. Plant Physiol. Biochem. 2007, 45, 623–629. [Google Scholar] [CrossRef]
- Rueda-Romero, P.; Barrero-Sicilia, C.; Gomez-Cadenas, A.; Carbonero, P.; Onate-Sanchez, L. Arabidopsis thaliana DOF6 negatively affects germination in non-after-ripened seeds and interacts with TCP14. J. Exp. Bot. 2012, 63, 1937–1949. [Google Scholar] [CrossRef]
- Kang, W.H.; Kim, S.; Lee, H.A.; Choi, D.; Yeom, S.I. Genome-wide analysis of Dof transcription factors reveals functional characteristics during development and response to biotic stresses in pepper. Sci. Rep. 2016, 6, 33332. [Google Scholar] [CrossRef]
- Wang, H.; Zhao, S.; Gao, Y.C.; Yang, J. Characterization of Dof transcription factors and their responses to osmotic stress in poplar (Populus trichocarpa). PLoS ONE 2017, 12, e0170210. [Google Scholar] [CrossRef] [PubMed]
- Corrales, A.-R.; Nebauer, S.G.; Carrillo, L.; Fernández-Nohales, P.; Marqués, J.; Renau-Morata, B.; Granell, A.; Pollmann, S.; Vicente-Carbajosa, J.; Molina, R.-V.; et al. Characterization of tomato cycling Dof factors reveals conserved and new functions in the control of flowering time and abiotic stress responses. J. Exp. Bot. 2014, 65, 995–1012. [Google Scholar] [CrossRef] [PubMed]
- Corrales, A.-R.; Carrillo, L.; Lasierra, P.; Nebauer, S.G.; Dominguez-Figueroa, J.; Renau-Morata, B.; Pollmann, S.; Granell, A.; Molina, R.-V.; Vicente-Carbajosa, J.; et al. Multifaceted role of cycling DOF factor 3 (CDF3) in the regulation of flowering time and abiotic stress responses in Arabidopsis. Plant Cell Environ. 2017, 40, 748–764. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Jia, J.; Lian, Z.; Hu, Y.; Guo, J.; Huo, H.; Zhu, Y.; Gong, H. Silicon enhances the salt tolerance of cucumber through increasing polyamine accumulation and decreasing oxidative damage. Ecotoxicol. Environ. Saf. 2019, 169, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.X.; Gong, H.J.; Yin, J.L. Role of silicon in mediating salt tolerance in plants: A review. Plants 2019, 8, 147. [Google Scholar] [CrossRef]
- Yin, J.L.; Fang, Z.W.; Sun, C.; Zhang, P.; Lu, C.; Wang, S.P.; Ma, D.F.; Zhu, Y.X. Rapid identification of a stripe rust resistant gene in a space-induced wheat mutant using specific locus amplified fragment (SLAF) sequencing. Sci. Rep. 2018, 8, 3086. [Google Scholar] [CrossRef] [PubMed]
- Appels, R.; Eversole, K.; Feuillet, C.; Keller, B.; Rogers, J.; Stein, N.; Ronen, G. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [PubMed]
- Zhu, Y.X.; Yang, L.; Liu, N.; Yang, J.; Zhou, X.K.; Xia, Y.C.; He, Y.; He, Y.Q.; Gong, H.J.; Ma, D.F.; et al. Genome-wide identification, structure characterization, and expression pattern profiling of aquaporin gene family in cucumber. BMC Plant Biol. 2019, 19, 345. [Google Scholar] [CrossRef]
- Jiang, W.; Yang, L.; He, Y.; Zhang, H.; Li, W.; Chen, H.; Ma, D.; Yin, J. Genome-wide identification and transcriptional expression analysis of superoxide dismutase (SOD) family in wheat (Triticum aestivum). PeerJ 2019, 7, e8062. [Google Scholar] [CrossRef]
- Peterson, G.I.; Masel, J. Quantitative prediction of molecular clock and Ka/Ks at short timescales. Mol. Biol. Evol. 2009, 26, 2595–2603. [Google Scholar] [CrossRef]
- Yin, J.; Liu, M.; Ma, D.; Wu, J.; Li, S.; Zhu, Y.; Han, B. Identification of circular RNAs and their targets during tomato fruit ripening. Postharvest Biol. Tec. 2018, 136, 90–98. [Google Scholar] [CrossRef]
- Zhou, R.; Zhu, Y.X.; Zhao, J.; Fang, Z.W.; Wang, S.P.; Yin, J.L.; Chu, Z.H.; Ma, D.F. Transcriptome-wide identification and characterization of potato circular RNAs in response to Pectobacterium carotovorum subspecies brasiliense infection. Int. J. Mol. Sci. 2018, 19, 71. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-González, R.H.; Borrill, P.; Lang, D.; Harrington, S.A.; Brinton, J.; Venturini, L.; Khedikar, Y. The transcriptional landscape of polyploid wheat. Science 2018, 361, eaar6089. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.X.; Jia, J.H.; Yang, L.; Xia, Y.C.; Zhang, H.L.; Jia, J.B.; Zhou, R.; Nie, P.Y.; Yin, J.L.; Ma, D.F.; et al. Identification of cucumber circular RNAs responsive to salt stress. BMC Plant Biol. 2019, 19, 164. [Google Scholar] [CrossRef]
- Song, J.; Ma, D.; Yin, J.; Yang, L.; He, Y.; Zhu, Z.; Tong, H.; Chen, L.; Zhu, G.; Liu, Y.; et al. Genome-wide characterization and expression profiling of squamosa promoter binding protein-like (SBP) transcription factors in wheat (Triticum aestivum L.). Agronomy 2019, 9, 527. [Google Scholar] [CrossRef]
- Yin, J.; Gu, B.; Huang, G.; Tian, Y.; Quan, J.; Lindqvist-Kreuze, H.; Shan, W. Conserved RXLR effector genes of Phytophthora infestans expressed at the early stage of potato infection are suppressive to host defense. Front. Plant Sci. 2017, 8, 2155. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Qu, L.J.; Zhu, Y.X. Transcription factor families in Arabidopsis: Major progress and outstanding issues for future research. Curr. Opin. Plant Biol. 2006, 9, 544–549. [Google Scholar] [CrossRef]
- Jia, J.; Lu, W.; Zhong, C.; Zhou, R.; Xu, J.; Liu, W.; Gou, X.; Wang, Q.; Yin, J.; Xu, C. The 25–26 nt small RNAs in Phytophthora parasitica are associated with efficient silencing of homologous endogenous genes. Front. Microbiol. 2017, 8, 773. [Google Scholar] [CrossRef]
- Guo, Y.; Qiu, L.J. Genome-wide analysis of the Dof transcription factor gene family reveals soybean-specific duplicable and functional characteristics. PLoS ONE 2013, 8, e76809. [Google Scholar] [CrossRef]
- Venkatesh, J.; Park, S.W. Genome-wide analysis and expression profiling of DNA-binding with one zinc finger (Dof) transcription factor family in potato. Plant Physiol. Biochem. 2015, 94, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Song, A.P.; Gao, T.W.; Li, P.L.; Chen, S.M.; Guan, Z.Y.; Wu, D.; Xin, J.J.; Fan, Q.Q.; Zhao, K.K.; Chen, F.D. Transcriptome-wide identification and expression profiling of the Dof transcription factor gene family in Chrysanthemum morifolium. Front. Plant Sci. 2016, 7, 199. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Liu, B.L.; Liu, P.; Xing, H.R.; Zhang, A.Y.; Zhang, L. Genome-wide analysis and expression profiling of DOF family in Setaria viridis. Mol. Plant Breed. 2018, 13, 4226–4234. [Google Scholar]
- Yanagisawa, S. The Dof family of plant transcription factors. Trends Plant Sci. 2002, 7, 555–560. [Google Scholar] [CrossRef]
- Peng, J.H.; Sun, D.; Nevo, E. Domestication evolution, genetics and genomics in wheat. Mol. Breed. 2011, 28, 281. [Google Scholar] [CrossRef]
- Mena, M.; Vicente-Carbajosa, J.; Schmidt, R.; Carbonero, P. An endosperm-specific DOF protein from barley, highly conserved in wheat, binds to and activates transcription from the prolamin-box of a native B-hordein promoter in barley endosperm. Plant J. 1998, 16, 53–62. [Google Scholar] [CrossRef]
- Washio, K. Identification of Dof proteins with implication in the gibberellin-regulated expression of a peptidase gene following the germination of rice grains. Biochim. Et Biophys. Acta 2001, 1520, 54–62. [Google Scholar] [CrossRef]
- Iwamoto, M.; Higo, K.; Takano, M. Circadian clock- and phytochrome-regulated Dof-like gene, Rdd1, is associated with grain size in rice. Plant Cell Environ. 2009, 32, 592–603. [Google Scholar] [CrossRef]
- Tanaka, M.; Takahata, Y.; Nakayama, H.; Nakatani, M.; Tahara, M. Altered carbohydrate metabolism in the storage roots of sweetpotato plants overexpressing the SRF1 gene, which encodes a Dof zinc finger transcription factor. Planta 2009, 230, 737–746. [Google Scholar] [CrossRef]




| Group | Dof Gene | Gene ID | 1 Length | 2 MW | 3 pI | 4 Ins | 5 Ali | 6 GRAVY |
|---|---|---|---|---|---|---|---|---|
| I | TaDof1.1-1A | TraesCS1A02G171300.1 | 258 | 26.14 | 9.44 | 58.46 | 53.14 | −0.353 |
| TaDof1.1-1B | TraesCS1B02G185900.1 | 264 | 26.54 | 9.36 | 55.33 | 51.93 | −0.329 | |
| TaDof1.2-2A | TraesCS2A02G225900.1 | 356 | 35.6 | 7.69 | 52.59 | 61.69 | −0.229 | |
| TaDof1.2-2B | TraesCS2B02G249200.1 | 355 | 35.68 | 7.71 | 51.38 | 61.01 | −0.262 | |
| TaDof1.2-2D | TraesCS2D02G231600.1 | 356 | 35.71 | 7.7 | 53.47 | 60.81 | −0.265 | |
| II | TaDof2.1-2A.1 | TraesCS2A02G590800.1 | 219 | 23.62 | 7.59 | 75.87 | 47.81 | −0.752 |
| TaDof2.1-2B.1 | TraesCS2B02G592900.1 | 207 | 22.28 | 8.11 | 72.28 | 54.83 | −0.65 | |
| TaDof2.1-2D.1 | TraesCS2D02G563400.1 | 207 | 22.26 | 8.5 | 74.11 | 50.14 | −0.717 | |
| TaDof2.1-2A.2 | TraesCS2A02G590700.1 | 201 | 21.7 | 8.17 | 77.18 | 52.59 | −0.655 | |
| TaDof2.2-2A.1 | TraesCS2A02G591200.1 | 214 | 22.81 | 6.12 | 82.89 | 48.5 | −0.61 | |
| TaDof2.2-2B.1 | TraesCS2B02G592600.1 | 186 | 20.07 | 6.89 | 83.66 | 43.17 | −0.84 | |
| TaDof2.2-2D.1 | TraesCS2D02G563000.1 | 217 | 23.22 | 6.07 | 83.52 | 46.04 | −0.751 | |
| TaDof2.2-2A.2 | TraesCS2A02G591000.1 | 249 | 26.58 | 8.08 | 80.88 | 50.32 | −0.681 | |
| TaDof2.2-2D.2 | TraesCS2D02G563100.1 | 225 | 23.98 | 6.57 | 73.35 | 53.51 | −0.644 | |
| TaDof2.3-2A | TraesCS2A02G591100.1 | 212 | 22.46 | 8.8 | 74.08 | 51.04 | −0.621 | |
| TaDof2.3-2B | TraesCS2B02G592700.1 | 222 | 23.63 | 5.93 | 74.09 | 51.62 | −0.637 | |
| TaDof2.4-2B | TraesCS2B02G592800.1 | 212 | 23.17 | 6.88 | 68.66 | 49.43 | −0.799 | |
| TaDof2.4-2D | TraesCS2D02G563200.1 | 212 | 23.36 | 8.17 | 69.1 | 51.86 | −0.578 | |
| III | TaDof3.1-1A | TraesCS1A02G035200.1 | 361 | 37.2 | 8.72 | 62.47 | 52.91 | −0.393 |
| TaDof3.1-1Ba | TraesCS1B02G045000.1 | 356 | 36.34 | 8.91 | 56.93 | 55.03 | −0.337 | |
| TaDof3.1-1Bb | TraesCS1B02G045000.2 | 362 | 37.08 | 8.91 | 56.45 | 54.39 | −0.372 | |
| TaDof3.1-1Bc | TraesCS1B02G045000.3 | 367 | 37.71 | 8.61 | 56.13 | 54.71 | −0.378 | |
| TaDof3.1-1Da | TraesCS1D02G036700.1 | 361 | 36.96 | 8.77 | 58.66 | 54.02 | −0.381 | |
| TaDof3.1-1Db | TraesCS1D02G036700.2 | 366 | 37.59 | 8.4 | 58.31 | 54.34 | −0.387 | |
| TaDof 3.2-3A | TraesCS3A02G532000.1 | 372 | 38.54 | 8.63 | 61.58 | 34.57 | −0.732 | |
| TaDof 3.2-3B | TraesCS3B02G608800.1 | 376 | 38.77 | 8.48 | 61.85 | 34.2 | −0.746 | |
| TaDof 3.2-3D | TraesCS3D02G537400.1 | 376 | 38.63 | 8.48 | 61.16 | 34.2 | −0.716 | |
| TaDof 3.3-3A.1 | TraesCS3A02G532100.1 | 379 | 41 | 9.06 | 56.61 | 53.35 | −0.596 | |
| TaDof 3.3-3A.2 | TraesCS3A02G532200.1 | 355 | 38.35 | 8.36 | 53.49 | 49.24 | −0.616 | |
| TaDof 3.3-3A.3 | TraesCS3A02G539000.1 | 269 | 28.74 | 6.88 | 54.81 | 42.94 | −0.611 | |
| TaDof 3.3-3B.1 | TraesCS3B02G608900.1 | 355 | 37.7 | 8.91 | 55.31 | 51.44 | −0.521 | |
| TaDof 3.3-3B.2 | TraesCS3B02G609000.1 | 352 | 37.65 | 8.69 | 58.15 | 42.44 | −0.651 | |
| TaDof 3.3-3B.3 | TraesCS3B02G609100.1 | 347 | 36.89 | 6.95 | 55.21 | 48.41 | −0.554 | |
| TaDof 3.3-3D.1 | TraesCS3D02G537500.1 | 382 | 41.6 | 8.86 | 56.46 | 43.48 | −0.804 | |
| TaDof3.4-4A | TraesCS4A02G097800.1 | 383 | 39.72 | 9.42 | 66.88 | 47.6 | −0.565 | |
| TaDof3.4-4B | TraesCS4B02G206800.1 | 379 | 39.35 | 9.5 | 67.07 | 48.47 | −0.557 | |
| TaDof3.4-4D | TraesCS4D02G207600.1 | 381 | 39.47 | 9.62 | 65.82 | 47.09 | −0.557 | |
| TaDof3.5-5Aa | TraesCS5A02G078100.1 | 342 | 35.27 | 8.91 | 54.6 | 57.49 | −0.338 | |
| TaDof3.5-5Ab | TraesCS5A02G078100.2 | 339 | 34.88 | 8.91 | 54.65 | 57.99 | −0.326 | |
| TaDof3.5-5Ba | TraesCS5B02G087600.1 | 340 | 34.93 | 8.91 | 55.34 | 58.97 | −0.332 | |
| TaDof3.5-5Bb | TraesCS5B02G087600.2 | 341 | 35.06 | 8.91 | 55.77 | 58.8 | −0.341 | |
| TaDof3.5-5Bc | TraesCS5B02G087600.3 | 344 | 35.44 | 8.91 | 55.71 | 58.28 | −0.353 | |
| TaDof3.5-5Da | TraesCS5D02G093800.1 | 341 | 35.42 | 8.91 | 52.08 | 57.03 | −0.377 | |
| TaDof3.5-5Db | TraesCS5D02G093800.2 | 345 | 35.68 | 8.91 | 51.62 | 56.7 | −0.379 | |
| TaDof3.6-5A | TraesCS5A02G479400.1 | 467 | 47.91 | 9.43 | 44.63 | 56.75 | −0.673 | |
| TaDof3.6-5B | TraesCS5B02G492600.1 | 452 | 46.27 | 9.3 | 59.89 | 45.66 | −0.588 | |
| TaDof3.6-5D | TraesCS5D02G493000.1 | 455 | 46.47 | 9.3 | 55.86 | 44.53 | −0.594 | |
| IV | TaDof4.1-3A | TraesCS3A02G403500.1 | 631 | 69.62 | 8.19 | 55.97 | 67.29 | −0.5 |
| TaDof4.2-5A | TraesCS5A02G155900.1 | 338 | 34.78 | 8.09 | 42.25 | 53.67 | −0.435 | |
| TaDof4.2-5B | TraesCS5B02G154100.1 | 338 | 35.21 | 8.78 | 43.57 | 50.8 | −0.464 | |
| TaDof4.2-5D | TraesCS5D02G161000.1 | 342 | 35.47 | 8.59 | 47.71 | 56.17 | −0.421 | |
| TaDof4.3-6A | TraesCS6A02G274000.1 | 306 | 32.64 | 8.05 | 48.23 | 61.41 | −0.355 | |
| TaDof4.3-6B | TraesCS6B02G301500.1 | 306 | 32.49 | 8.32 | 46.64 | 61.76 | −0.329 | |
| TaDof4.3-6D | TraesCS6D02G254200.1 | 306 | 32.46 | 8.32 | 48.68 | 60.78 | −0.342 | |
| V | TaDof5.1-1A | TraesCS1A02G275000.1 | 316 | 33.51 | 5.3 | 54.35 | 62.82 | −0.399 |
| TaDof5.1-1B | TraesCS1B02G284300.1 | 316 | 33.59 | 5.39 | 51.1 | 61.87 | −0.406 | |
| TaDof5.1-1D | TraesCS1D02G274700.1 | 377 | 40.2 | 6.45 | 58.09 | 70.5 | −0.284 | |
| TaDof5.2-2A | TraesCS2A02G402200.1 | 378 | 38.35 | 8.65 | 47.94 | 44.37 | −0.499 | |
| TaDof5.2-2B | TraesCS2B02G420400.1 | 376 | 38.11 | 8.65 | 48.39 | 44.6 | −0.508 | |
| TaDof5.2-2D | TraesCS2D02G399500.1 | 378 | 38.3 | 8.65 | 47.62 | 43.6 | −0.52 | |
| TaDof5.3-3A | TraesCS3A02G377000.1 | 331 | 35.23 | 4.68 | 53.93 | 60.18 | −0.343 | |
| TaDof5.3-3B | TraesCS3B02G409600.1 | 331 | 35.22 | 4.68 | 53.49 | 60.48 | −0.349 | |
| TaDof5.3-3D | TraesCS3D02G370100.1 | 331 | 35.23 | 4.66 | 53.5 | 58.7 | −0.393 | |
| TaDof5.4-6A | TraesCS6A02G255500.1 | 392 | 40.23 | 8.45 | 51.62 | 44.46 | −0.64 | |
| TaDof5.4-6B | TraesCS6B02G270100.1 | 389 | 40.12 | 8.45 | 50.75 | 45.78 | −0.624 | |
| TaDof5.4-6D | TraesCS6D02G236700.1 | 391 | 40.05 | 8.45 | 50.35 | 44.32 | −0.635 | |
| VII | TaDof7.1-3A | TraesCS3A02G106500.1 | 238 | 25.31 | 9.41 | 65.44 | 72.69 | −0.221 |
| TaDof7.1-3B | TraesCS3B02G125100.1 | 230 | 24.43 | 9.21 | 66.26 | 70.09 | −0.28 | |
| TaDof7.1-3D | TraesCS3D02G108600.1 | 230 | 24.47 | 9.38 | 61.2 | 70.09 | −0.259 | |
| TaDof7.2-3A | TraesCS3A02G271700.1 | 155 | 16 | 10.46 | 70.93 | 48.62 | −0.541 | |
| TaDof7.2-3B | TraesCS3B02G305400.1 | 154 | 15.89 | 10.46 | 67.58 | 48.57 | −0.523 | |
| TaDof7.2-3D | TraesCS3D02G271300.1 | 152 | 15.77 | 10.46 | 74.47 | 45.92 | −0.597 | |
| TaDof7.3-4A | TraesCS4A02G234000.1 | 266 | 28.04 | 9.46 | 54.37 | 61.47 | −0.306 | |
| TaDof7.3-4B | TraesCS4B02G081500.1 | 265 | 27.85 | 9.3 | 50.92 | 64.64 | −0.272 | |
| TaDof7.3-4D | TraesCS4D02G080100.1 | 263 | 27.72 | 9.27 | 55.2 | 62.93 | −0.283 | |
| TaDof7.4-5A | TraesCS5A02G251800.1 | 281 | 28.69 | 4.89 | 62.93 | 44.77 | −0.557 | |
| TaDof7.4-5B | TraesCS5B02G249800.1 | 281 | 28.84 | 4.93 | 59.96 | 46.83 | −0.507 | |
| TaDof7.4-5D | TraesCS5D02G259700.1 | 281 | 28.86 | 4.86 | 64.28 | 45.84 | −0.576 | |
| TaDof7.5-5A | TraesCS5A02G401800.1 | 381 | 39.47 | 9.42 | 64.7 | 54.67 | −0.448 | |
| TaDof7.5-5B | TraesCS5B02G406500.1 | 382 | 39.51 | 9.42 | 66.42 | 54.53 | −0.442 | |
| TaDof7.5-5D | TraesCS5D02G412000.1 | 380 | 39.26 | 9.33 | 66.94 | 55.58 | −0.422 | |
| TaDof7.6-6A | TraesCS6A02G287700.1 | 291 | 30.38 | 5.57 | 71.43 | 50.52 | −0.42 | |
| TaDof7.6-6B | TraesCS6B02G317100.1 | 289 | 30.31 | 5.92 | 68.33 | 52.53 | −0.401 | |
| TaDof7.7-6D | TraesCS6D02G268400.1 | 290 | 30.21 | 5.91 | 69.89 | 50.34 | −0.417 | |
| TaDof7.8-7A | TraesCS7A02G213400.1 | 284 | 29.15 | 6.81 | 45.64 | 53.87 | −0.251 | |
| TaDof7.8-7D | TraesCS7D02G215300.1 | 284 | 29.02 | 6.71 | 49.17 | 56.3 | −0.222 | |
| TaDof7.9-7B | TraesCS7B02G120600.1 | 319 | 32.92 | 8.62 | 49.72 | 54.39 | −0.378 | |
| VIII | TaDof8.1-1A | TraesCS1A02G334100.1 | 209 | 22.24 | 9.6 | 50.11 | 57.51 | −0.494 |
| TaDof8.1-1B | TraesCS1B02G347400.1 | 209 | 22.27 | 9.6 | 53.56 | 58.9 | −0.476 | |
| TaDof8.2-1D | TraesCS1D02G336600.1 | 214 | 23.01 | 9.32 | 48.04 | 59.35 | −0.516 | |
| TaDof8.3-2A | TraesCS2A02G079200.1 | 326 | 32.88 | 9.11 | 62.13 | 50.46 | −0.372 | |
| TaDof8.3-2B | TraesCS2B02G094000.1 | 330 | 33.27 | 9.16 | 59.68 | 48.67 | −0.376 | |
| TaDof8.3-2D | TraesCS2D02G076600.1 | 328 | 33.04 | 9.24 | 63.44 | 48.96 | −0.383 | |
| TaDof8.4-2A | TraesCS2A02G100800.1 | 427 | 51.04 | 5.62 | 59.09 | 50.19 | −0.659 | |
| TaDof8.4-2B | TraesCS2B02G118000.1 | 479 | 50.92 | 5.88 | 53.28 | 51 | −0.629 | |
| TaDof8.4-2D | TraesCS2D02G100300.1 | 501 | 53.35 | 6.27 | 53.28 | 51.68 | −0.615 | |
| TaDof8.5-3A | TraesCS3A02G180600.1 | 539 | 58.12 | 5.06 | 54.91 | 51.45 | −0.815 | |
| TaDof8.5-3B | TraesCS3B02G210300.1 | 539 | 58.13 | 5.14 | 58.7 | 51.47 | −0.806 | |
| TaDof8.5-3Da | TraesCS3D02G185500.1 | 506 | 55.03 | 5.73 | 52.48 | 54.8 | −0.72 | |
| TaDof8.5-3Db | TraesCS3D02G185500.2 | 538 | 57.94 | 5.1 | 55.61 | 51.75 | −0.798 | |
| TaDof8.6-3A | TraesCS3A02G189600.1 | 476 | 51.91 | 7.14 | 56.86 | 67.21 | −0.544 | |
| TaDof8.6-3D | TraesCS3D02G193100.1 | 476 | 51.91 | 6.54 | 54.31 | 64.75 | −0.555 | |
| TaDof8.7-3A | TraesCS3A02G306800.1 | 202 | 22.05 | 9.78 | 66.46 | 49.95 | −0.732 | |
| TaDof8.7-3B | TraesCS3B02G329700.1 | 204 | 22.19 | 9.64 | 68.25 | 50.39 | −0.692 | |
| TaDof8.7-3D | TraesCS3D02G295100.1 | 200 | 21.8 | 9.91 | 72.17 | 47.05 | −0.774 | |
| TaDof8.8-4A | TraesCS4A02G017700.1 | 437 | 47.27 | 8.15 | 52.77 | 51.94 | −0.75 | |
| TaDof8.8-4B | TraesCS4B02G286400.1 | 433 | 46.78 | 8.45 | 48.55 | 55.75 | −0.663 | |
| TaDof8.8-4D | TraesCS4D02G285100.1 | 442 | 47.62 | 8.15 | 48.52 | 55.48 | −0.689 |
| ID | Species | Function | Hits of Wheat TaDof Genes |
|---|---|---|---|
| NtBBF1 | Tobacco | Auxin response | TaDof3.4-4A, TaDof3.15-4B, TaDof3.4-4D, TaDof4.3-6A, TaDof5.1-1D, TaDof7.6-6A, TaDof7.6-6B, TaDof7.7-6D, TaDof8.3-2A, TaDof8.3-2B, TaDof8.3-2D |
| OsDof3 | Rice | Gibberellin response | TaDof4.3-6B, TaDof4.3-6D |
| AtDof3.4/OBP1 | Arabidopsis | Cell cycle, defense response | TaDof7.6-6A, TaDof7.6-6B, TaDof7.7-6D |
| AtDof4.1 | Arabidopsis | Transcriptional repressor | TaDof5.1-1A, TaDof5.4-6A, TaDof5.4-6B, TaDof5.4-6D, TaDof5.1-1B, TaDof5.1-1D, TaDof5.2-2A, TaDof5.2-2B, TaDof5.2-2D |
| GmDof17-1 | Maize | Growth inhibition | TaDof5.3-3A, TaDof5.3-3B, TaDof5.3-3D |
| AtDof2.4 | Arabidopsis | Vascular development | TaDof3.6-5A, TaDof4.3-6A, TaDof4.3-6B, TaDof4.3-6D, TaDof7.1-3D |
| AtDof5.1 | Arabidopsis | Leaf axial patterning | TaDof1.2-2A, TaDof1.2-2B, TaDof1.2-2D, TaDof3.4-4A, TaDof3.15-4B, TaDof3.4-4D, TaDof3.6-5A, TaDof3.5-5B, TaDof3.3-3B.1, TaDof4.3-6A, TaDof4.3-6B, TaDof4.3-6D, TaDof8.3-2A, TaDof8.3-2B, TaDof8.3-2D |
| AtDof5.4/OBP4 | Arabidopsis | Root hair growth | TaDof5.1-1B, TaDof5.1-1D, TaDof5.3-3A, TaDof5.3-3B, TaDof5.3-3D, TaDof7.5-5A, TaDof7.5-5B, TaDof7.5-5D, TaDof7.1-3B |
| StDof1 | Potato | Guard cell specificity | TaDof1.2-2A, TaDof1.2-2B, TaDof1.2-2D, TaDof3.6-5A, TaDof3.6-5B, TaDof3.22, TaDof4.1, TaDof4.3-6B, TaDof4.3-6D, TaDof7.5-5A, TaDof7.5-5B, TaDof7.5-5D, TaDof7.8-7A, TaDof7.9-7B, TaDof7.8-7D, TaDof8.3-2A, TaDof8.3-2B, TaDof8.3-2D |
| BPBF | Barley | Endosperm specificity | TaDof4.2-5A, TaDof4.2-5B, TaDof4.2-5D |
| WPBF | Wheat | Endosperm specificity | TaDof4.2-5A, TaDof4.2-5B, TaDof4.2-5D |
| COG1 | Arabidopsis | Seed germination pathway mediated by light | TaDof8.5-3A, TaDof8.5-3B, TaDof8.5-3D, TaDof8.4-2A, TaDof8.4-2B, TaDof8.4-2D |
| DGA2 | Arabidopsis | Seed germinating | TaDof3.5-5A, TaDof3.5-5B, TaDof3.5-5D, TaDof5.3-3A, TaDof5.3-3B, TaDof5.3-3D, TaDof7.5-5A, TaDof7.5-5B, TaDof7.5-5D |
| SAD | Barley | Seed germinating | TaDof4.3-6A, TaDof4.3-6B, TaDof4.3-6D, TaDof5.1-1A, TaDof5.4-6A, TaDof5.4-6B, TaDof5.4-6D, TaDof5.1-1B, TaDof5.1-1D, TaDof5.3-3B, TaDof5.3-3D |
| MaDof23 | Banana | Fruit ripening | TaDof4.3-6A, TaDof4.3-6B, TaDof4.3-6D, TaDof7.5-5A, TaDof7.5-5D, TaDof8.3-2A, TaDof8.3-2B, TaDof8.3-2D |
| OsDof15 | Rice | Root meristem cell proliferation | TaDof3.4-4A, TaDof3.15-4B, TaDof3.4-4D, TaDof3.5-5A, TaDof3.5-5B, TaDof3.5-5D, TaDof5.1-1A, TaDof5.1-1B, TaDof5.1-1D, TaDof5.3-3A, TaDof5.3-3B, TaDof5.3-3D, TaDof7.6-6A, TaDof7.6-6B, TaDof7.7-6D, TaDof7.8-7D, TaDof8.3-2A, TaDof8.3-2B, TaDof8.3-2D |
| OsDof24 | Rice | Determining flowering time | TaDof1.2-2A, TaDof1.2-2D, TaDof4.3-6A, TaDof4.3-6B, TaDof4.3-6D, TaDof5.1-1A, TaDof5.1-1B, TaDof5.1-1D, TaDof5.3-3A, TaDof5.3-3B, TaDof5.3-3D, TaDof7.1-3A, TaDof7.5-5A, TaDof7.5-5B, TaDof7.5-5D, TaDof7.6-6A, TaDof7.6-6B, TaDof7.7-6D, TaDof7.9-7B, TaDof7.8-7D, TaDof7.1-3B, TaDof7.1-3D, TaDof7.3-4A, TaDof7.3-4B, TaDof7.3-4D, TaDof8.3-2A, TaDof8.3-2B, TaDof8.3-2D |
| Rdd1 | Rice | Photoperiodic flowering | TaDof8.5-3B |
| SCAP1 | Arabidopsis | Development of functional stomata | TaDof7.1-3A, TaDof7.1-3D |
| AtDof1.1/OBP2 | Arabidopsis | Glucosinolate biosynthesis | TaDof4.3-6A, TaDof4.3-6B, TaDof4.3-6D, TaDof5.3-3A, TaDof5.3-3B, TaDof5.3-3D |
| ZmDof1 | Maize | Pollen development, light response, carbon metabolism, nitrogen assimilation | TaDof1.2-2A, TaDof1.2-2B, TaDof4.3-6A, TaDof4.3-6B, TaDof4.3-6D, TaDof5.3-3A, TaDof5.3-3B, TaDof5.3-3D, TaDof7.1-3A, TaDof7.5-5A, TaDof7.5-5B, TaDof7.5-5D, TaDof7.6-6A, TaDof7.6-6B, TaDof7.7-6D, TaDof7.9-7B, TaDof7.1-3B, TaDof7.1-3D |
| OsDOF18 | Rice | Carbon and nitrogen metabolism | TaDof1.2-2A, TaDof1.2-2D, TaDof4.3-6A, TaDof4.3-6B, TaDof4.3-6D, TaDof5.1-1A, TaDof5.1-1B, TaDof5.1-1D, TaDof5.3-3A, TaDof5.3-3B, TaDof5.3-3D, TaDof7.1, TaDof7.5-5A, TaDof7.5-5B, TaDof7.5-5D, TaDof7.6-6A, TaDof7.6-6B, TaDof7.7-6D, TaDof7.9-7B, TaDof7.8-7D, TaDof7.1-3B, TaDof7.1-3D, TaDof7.3-4A, TaDof7.3-4B, TaDof7.3-4D, TaDof8.3-2A, TaDof8.3-2B, TaDof8.3-2D |
| SRF1 | Sweetpotato | Carbon metabolism | TaDof8.1-1A |
| ZmDOF36 | Maize | Starch synthesis | TaDof1.2-2D, TaDof2.1-2A.2, TaDof2.2-2D.1, TaDof2.4-2D, TaDof2.1-2D.1, TaDof2.1-2A.2, TaDof2.4-2A, TaDof2.2-2B.1, TaDof2.4-2B, TaDof2.1-2B.1, TaDof3.6-5A, TaDof3.6-5B, TaDof7.8-7A, TaDof7.9-7B, TaDof7.8-7D, TaDof7.1-3D, TaDof8.3-2A, TaDof8.3-2B, TaDof8.3-2D |
| OsDof25 | Rice | C4 photosynthesis | TaDof4.3-6A, TaDof4.3-6B, TaDof4.3-6D, TaDof5.1-1A, TaDof5.4-6A, TaDof5.4-6B, TaDof5.4-6D, TaDof5.1-1B, TaDof5.2-2A, TaDof5.2-2B, TaDof5.2-2D, TaDof5.3-3A, TaDof5.3-3B, TaDof5.3-3D, TaDof7.1-3A, TaDof7.5-5A, TaDof7.5-5B, TaDof7.5-5D, TaDof7.8-7A, TaDof7.9-7B, TaDof7.8-7D, TaDof7.1-3B, TaDof7.1-3D, TaDof7.3-4A, TaDof7.3-4B, TaDof7.3-4D, TaDof8.3-2A, TaDof8.3-2B, TaDof8.3-2D |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, Z.; Jiang, W.; He, Y.; Ma, D.; Liu, Y.; Wang, S.; Zhang, Y.; Yin, J. Genome-Wide Identification, Structure Characterization, and Expression Profiling of Dof Transcription Factor Gene Family in Wheat (Triticum aestivum L.). Agronomy 2020, 10, 294. https://doi.org/10.3390/agronomy10020294
Fang Z, Jiang W, He Y, Ma D, Liu Y, Wang S, Zhang Y, Yin J. Genome-Wide Identification, Structure Characterization, and Expression Profiling of Dof Transcription Factor Gene Family in Wheat (Triticum aestivum L.). Agronomy. 2020; 10(2):294. https://doi.org/10.3390/agronomy10020294
Chicago/Turabian StyleFang, Zhengwu, Wenqiang Jiang, Yiqin He, Dongfang Ma, Yike Liu, Shuping Wang, Yingxin Zhang, and Junliang Yin. 2020. "Genome-Wide Identification, Structure Characterization, and Expression Profiling of Dof Transcription Factor Gene Family in Wheat (Triticum aestivum L.)" Agronomy 10, no. 2: 294. https://doi.org/10.3390/agronomy10020294
APA StyleFang, Z., Jiang, W., He, Y., Ma, D., Liu, Y., Wang, S., Zhang, Y., & Yin, J. (2020). Genome-Wide Identification, Structure Characterization, and Expression Profiling of Dof Transcription Factor Gene Family in Wheat (Triticum aestivum L.). Agronomy, 10(2), 294. https://doi.org/10.3390/agronomy10020294







