Variation Analysis of Root System Development in Wheat Seedlings Using Root Phenotyping System
Abstract
1. Introduction
2. Materials and Methods
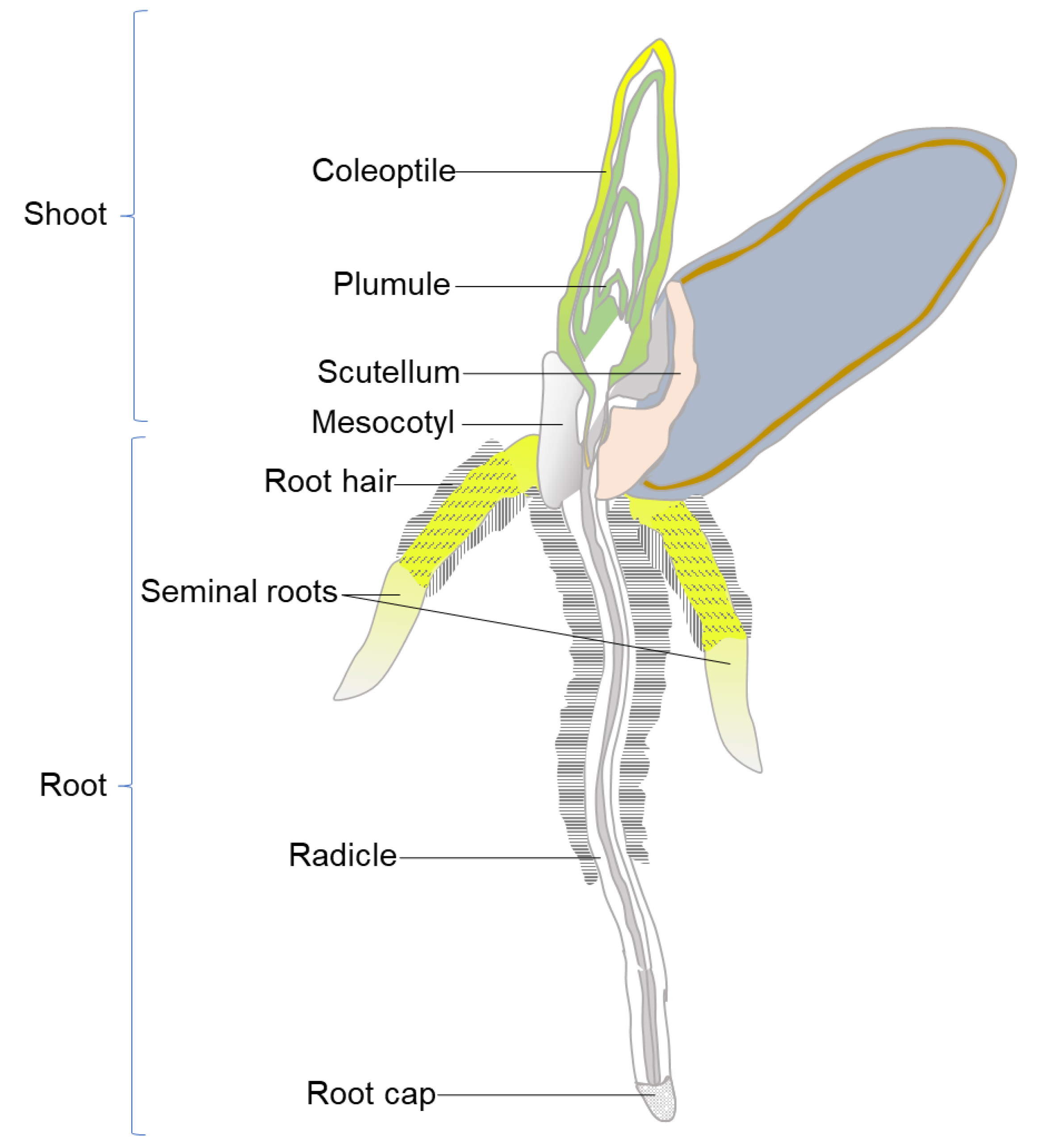
2.1. Experimental Design and Seed Treatment
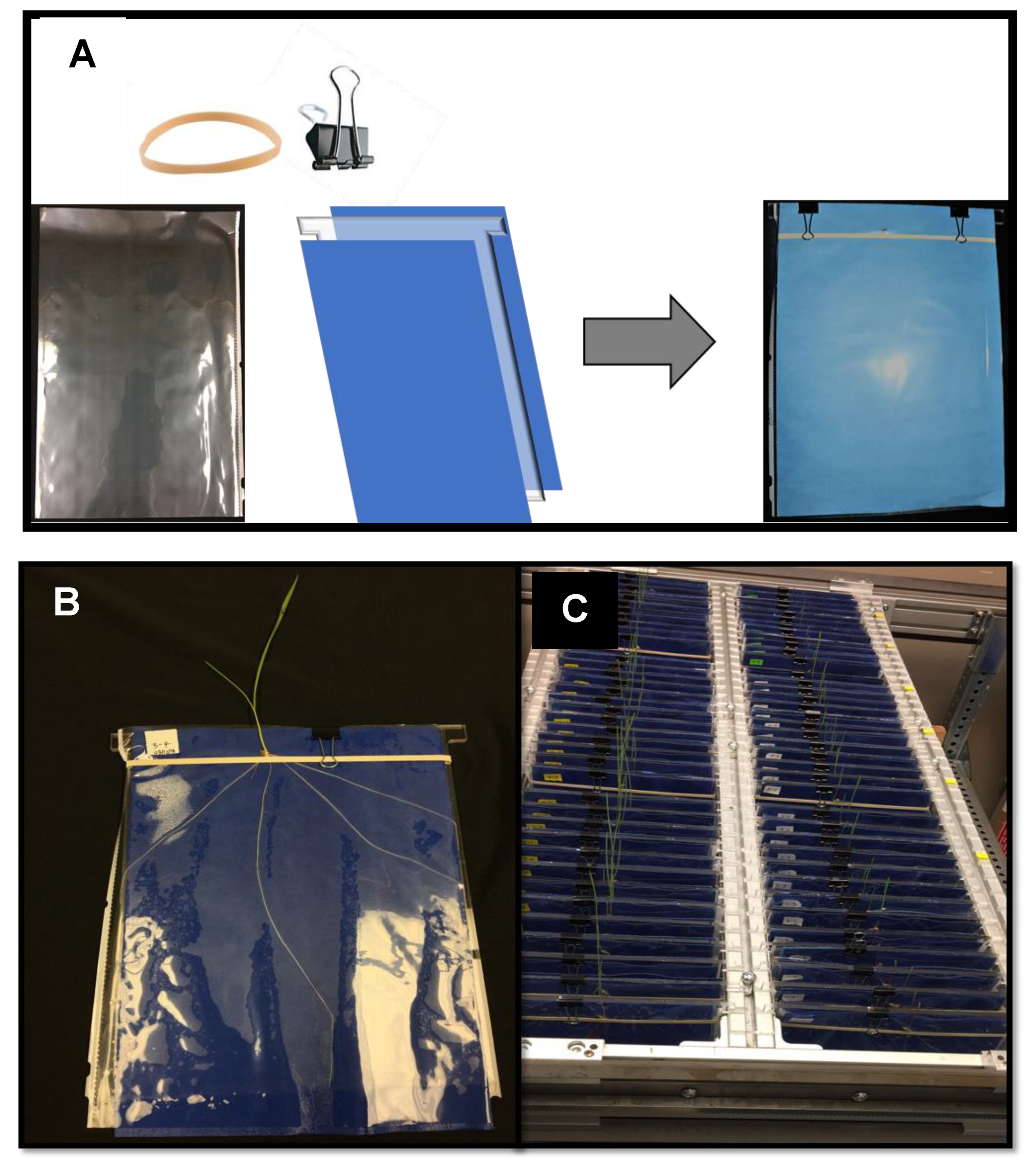
2.2. Design of Experimental Platform
2.3. Imaging and Analysis
3. Results
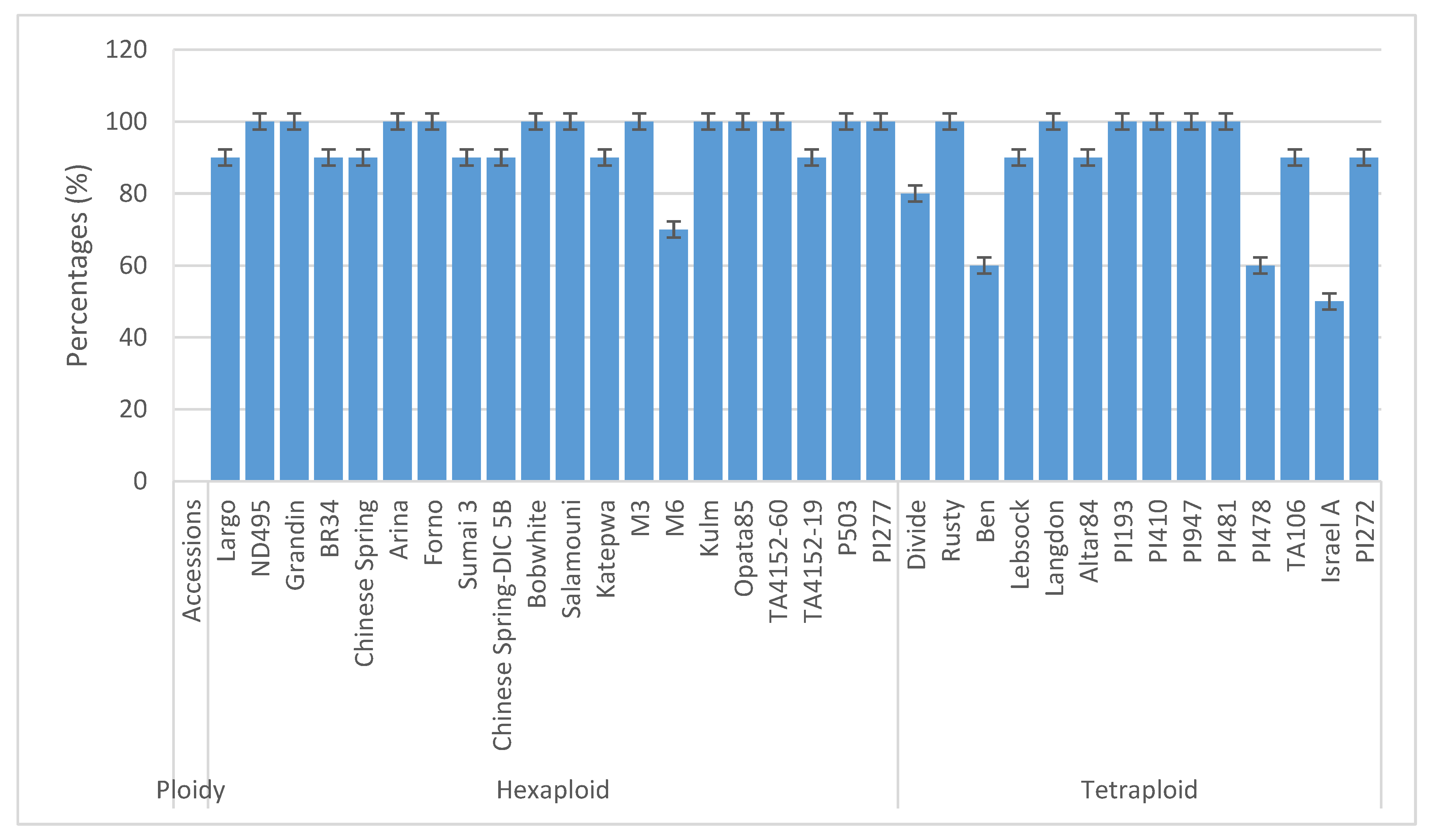
3.1. Frequency Distribution of Germination Potential and Measured Root Traits
3.2. The Hexaploid Wheat Accessions
3.3. The Tetraploid Wheat Accessions
4. Discussion
4.1. The Benefit of the Root System Size
4.2. Kulm and Opata85 May be Useful for RSA Improvement in Hexaploid Wheat
4.3. The Significance of a Rapid Screening Pipeline for Measuring Seedling Root Traits
4.4. Future Work
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| TL | Total length; |
| APL | Average seminal length; |
| MW | Maximum width; |
| MD | Maximum depth; |
| WDR | Width-depth ratio; |
| CHA | Convex hull area; |
| APEA | Average seminal emergence area; |
| Cen_X | Horizontal coordinates of centroid; |
| Cen_Y | Vertical coordinates of centroid; |
| PC | Seminal count. |
References
- Khan, M.A.; Gemenet, D.C.; Villordon, A. Root System Architecture and Abiotic Stress Tolerance: Current Knowledge in Root and Tuber Crops. Front Plant Sci. 2016, 7, 1584. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.; De Smet, I. Root system architecture: Insights from Arabidopsis and cereal crops. Philos. Trans R. Soc. B Biol. Sci. 2012, 367, 1441–1452. [Google Scholar] [CrossRef] [PubMed]
- Meister, R.; Rajani, M.S.; Ruzicka, D.; Schachtman, D.P. Challenges of modifying root traits in crops for agriculture. Trends Plant Sci. 2014, 19, 779–788. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.; French, A.P.; Atkinson, J.A.; Wells, D.M.; Bennett, M.; Pridmore, T. Root Architecture and Plant Productivity. Plant Physiol. 1995, 109, 7–13. [Google Scholar] [CrossRef] [PubMed]
- de Dorlodot, S.; Forster, B.; Pagès, L.; Price, A.; Tuberosa, R.; Draye, X. Root system architecture: Opportunities and constraints for genetic improvement of crops. Trends Plant Sci. 2007, 12, 474–481. [Google Scholar] [CrossRef] [PubMed]
- Shewry, P.R. Wheat. J. Exp. Bot. 2009, 60, 1537–1553. [Google Scholar] [CrossRef] [PubMed]
- Mayer, K.X. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 2014, 345, 1251788. [Google Scholar]
- Boyacioglu, H. Global Durum Wheat Use Trending Upward. Available online: https://www.world-grain.com/articles/8777-global-durum-wheat-use-trending-upward (accessed on 31 January 2020).
- Weiss, E.; Zohary, D. The Neolithic Southwest Asian Founder Crops. Curr. Anthropol. 2011, 52, S237–S254. [Google Scholar] [CrossRef]
- Shu, K.; Liu, X.; Xie, Q.; He, Z.; X-d, L. Two Faces of One Seed: Hormonal Regulation of Dormancy and Germination. Mol. Plant 2016, 9, 34–45. [Google Scholar] [CrossRef]
- Ma, Z.; Bykova, N.V.; Igamberdiev, A.U. Cell signaling mechanisms and metabolic regulation of germination and dormancy in barley seeds. Crop J. 2017, 5, 459–477. [Google Scholar] [CrossRef]
- Atkinson, J.A.; Wingen, L.U.; Griffiths, M.; Pound, M.P.; Gaju, O.; Foulkes, M.J.; Le Gouis, J.; Griffiths, S.; Bennett, M.J.; King, J.; et al. Phenotyping pipeline reveals major seedling root growth QTL in hexaploid wheat. J. Exp. Bot. 2015, 66, 2283–2292. [Google Scholar] [CrossRef] [PubMed]
- Zadoks, J.C.; Chang, T.T.; Konzak, C.F. A decimal code for the growth stages of cereals. Weed Res. 1974, 14, 415–421. [Google Scholar] [CrossRef]
- Fischer, R.A.; Edmeades, G.O. Breeding and Cereal Yield Progress. Crop Sci. 2010, 50, S85. [Google Scholar] [CrossRef]
- Richard, C.; Hickey, L.T.; Fletcher, S.; Jennings, R.; Chenu, K.; Christopher, J.T. High-throughput phenotyping of seminal root traits in wheat. Plant Methods 2015, 11, 13. [Google Scholar] [CrossRef]
- Schroth, G.; Kolbe, D. A method of processing soil core samples for root studies by subsampling. Biol. Fertil. Soils 1994, 18, 60–62. [Google Scholar] [CrossRef]
- Trachsel, S.; Kaeppler, S.M.; Brown, K.M.; Lynch, J.P. Shovelomics: High throughput phenotyping of maize (Zea mays L.) root architecture in the field. Plant Soil 2011, 341, 75–87. [Google Scholar] [CrossRef]
- Xie, Q.; Fernando, K.M.C.; Mayes, S.; Sparkes, D.L. Identifying seedling root architectural traits associated with yield and yield components in wheat. Ann. Bot. 2017, 119, 1115–1129. [Google Scholar] [CrossRef]
- Liu, X.; Li, R.; Chang, X.; Jing, R. Mapping QTLs for seedling root traits in a doubled haploid wheat population under different water regimes. Euphytica 2013, 189, 51–66. [Google Scholar] [CrossRef]
- Cobb, J.N.; DeClerck, G.; Greenberg, A.; Clark, R.; McCouch, S. Next-generation phenotyping: Requirements and strategies for enhancing our understanding of genotype-phenotype relationships and its relevance to crop improvement. Theor. Appl. Genet. 2013, 126, 867–887. [Google Scholar] [CrossRef]
- Hund, A.; Trachsel, S.; Stamp, P. Growth of axile and lateral roots of maize: I development of a phenotying platform. Plant Soil 2009, 325, 335–349. [Google Scholar] [CrossRef]
- Gioia, T.; Galinski, A.; Lenz, H.; Müller, C.; Lentz, J.; Heinz, K.; Briese, C.; Putz, A.; Fiorani, F.; Watt, M.; et al. GrowScreen-PaGe, a non-invasive, high-throughput phenotyping system based on germination paper to quantify crop phenotypic diversity and plasticity of root traits under varying nutrient supply. Funct. Plant Biol. 2017, 44, 76–93. [Google Scholar] [CrossRef]
- Bonser, A.M.; Lynch, J.; Snapp, S. Effect of phosphorus deficiency on growth angle of basal roots in Phaseolus vulgaris. New Phytol. 1996, 132, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Dupuy, L.X.; Wright, G.; Thompson, J.A.; Taylor, A.; Dekeyser, S.; White, C.P.; Thomas, W.T.B.; Nightingale, M.; Hammond, J.P.; Graham, N.S.; et al. Accelerating root system phenotyping of seedlings through a computer-assisted processing pipeline. Plant Methods 2017, 13, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.L.; Dunbabin, V.M.; Diggle, A.J.; Siddique, K.H.M.; Rengel, Z. Development of a novel semi-hydroponic phenotyping system for studying root architecture. Funct. Plant Biol. 2011, 38, 355. [Google Scholar] [CrossRef]
- Zhou, T.; Wang, L.; Li, S.; Gao, Y.; Du, Y.; Zhao, L.; Liu, W.; Yang, W. Interactions Between Light Intensity and Phosphorus Nutrition Affect the P Uptake Capacity of Maize and Soybean Seedling in a Low Light Intensity Area. Front Plant Sci. 2019, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Joshi, D.C.; Singh, V.; Hunt, C.; Mace, E.; van Oosterom, E.; Sulman, R.; Jordan, D.; Hammer, G. Development of a phenotyping platform for high throughput screening of nodal root angle in sorghum. Plant Methods 2017, 13, 56. [Google Scholar] [CrossRef]
- Pound, M.P.; French, A.P.; Atkinson, J.A.; Wells, D.M.; Bennett, M.J.; Pridmore, T. RootNav: Navigating images of complex root architectures. Plant Physiol. 2013, 162, 1802–1814. [Google Scholar] [CrossRef]
- Li, A.; Liu, D.; Yang, W.; Kishii, M.; Mao, L. Synthetic Hexaploid Wheat: Yesterday, Today, and Tomorrow. Engineering 2018, 4, 552–558. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Hoagland, D.R.; Arnon, D.I. The Water-Culture Method for Growing Plants without Soil. Circ. Calif. Agric. Exp. Stn. 1950, 347, 32. [Google Scholar]
- Cao, P.; Ren, Y.; Zhang, K.; Teng, W.; Zhao, X.; Dong, Z.; Liu, X.; Qin, H.; Li, Z.; Wang, D.; et al. Further genetic analysis of a major quantitative trait locus controlling root length and related traits in common wheat. Mol. Breed. 2014, 33, 975–985. [Google Scholar] [CrossRef]
- Mergoum, M.; Frohberg, R.C.; Rasmussen, J.W.; Friesen, T.L.; Hareland, G.; Simsek, S. Breeding for CLEARFIELD Herbicide Tolerance: Registration of ‘ND901CL’ Spring Wheat. J. Plant Regist. 2009, 3, 170–174. [Google Scholar] [CrossRef]
- Ghaffary, S.M.T.; Faris, J.D.; Friesen, T.L.; Visser, R.G.F.; van der Lee, T.A.J.; Robert, O.; Kema, G.H.J. New broad-spectrum resistance to septoria tritici blotch derived from synthetic hexaploid wheat. Theor. Appl. Genet. 2012, 124, 125–142. [Google Scholar] [CrossRef] [PubMed]
- Faris, J.D.; Zhang, Z.; Lu, H.; Lu, S.; Reddy, L.; Cloutier, S.; Fellers, J.P.; Meinhardt, S.W.; Rasmussen, J.B.; Xu, S.S.; et al. A unique wheat disease resistance-like gene governs effector-triggered susceptibility to necrotrophic pathogens. Proc. Natl. Acad. Sci. USA 2010, 107, 13544–13549. [Google Scholar] [CrossRef]
- Borner, A.; Schumann, E.; Furste, A.; Coster, H.; Leithold, B.; Roder, M.; Weber, W. Mapping of quantitative trait loci determining agronomic important characters in hexaploid wheat (Triticum aestivum L.). Theor. Appl. Genet. 2002, 105, 921–936. [Google Scholar] [CrossRef]
- Kumar, N.; Kulwal, P.L.; Balyan, H.S.; Gupta, P.K. QTL mapping for yield and yield contributing traits in two mapping populations of bread wheat. Mol. Breed. 2007, 19, 163–177. [Google Scholar] [CrossRef]
- Kulwal, P.; Roy, J.; Balyan, H.; Gupta, P. QTL mapping for growth and leaf characters in bread wheat. Plant Sci. 2003, 164, 267–277. [Google Scholar] [CrossRef]
- Yu, M.; Mao, S.; Chen, G.; Liu, Y.; Li, W.; Wei, Y.; Liu, C.; Zheng, Y. QTLs for Waterlogging Tolerance at Germination and Seedling Stages in Population of Recombinant Inbred Lines Derived from a Cross Between Synthetic and Cultivated Wheat Genotypes. J. Integr. Agric. 2014, 13, 31–39. [Google Scholar] [CrossRef]
- Yu, M.; Chen, G.-Y.; Pu, Z.-E.; Zhang, L.-Q.; Liu, D.-C.; Lan, X.-J.; Wei, Y.-M.; Zheng, Y.-L. Quantitative trait locus mapping for growth duration and its timing components in wheat. Mol. Breed. 2015, 35, 44. [Google Scholar] [CrossRef]
- Ingram, P.A.; Zhu, J.; Shariff, A.; Davis, I.W.; Benfey, P.N.; Elich, T. High-throughput imaging and analysis of root system architecture in Brachypodium distachyon under differential nutrient availability. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012, 367, 1559–1569. [Google Scholar] [CrossRef]
- Shorinola, O.; Kaye, R.; Golan, G.; Peleg, Z.; Kepinski, S.; Uauy, C. Genetic screening for mutants with altered seminal root numbers in hexaploid wheat using a high-throughput root phenotyping platform. G3 2019, 9, 2799–2809. [Google Scholar] [CrossRef]



| Accession | PI/CItr | Common Name | Taxon | Subspecies | Ploidy | Origin |
|---|---|---|---|---|---|---|
| Largo | CItr 17895 | Synthetic hexaploid wheat | Triticum turgidum × Aegiliops tauschii | Synthetic | 6× | U.S., North Dakota |
| ND495 | N/A | Common wheat | Triticum aestivum | aestivum | 6× | U.S., North Dakota |
| Grandin | PI 531005 | Common wheat | Triticum aestivum | aestivum | 6× | U.S., North Dakota |
| BR34 | N/A | Common wheat | Triticum aestivum | aestivum | 6× | Brazil |
| Chinese Spring | CItr 14108 | Common wheat | Triticum aestivum | aestivum | 6× | China |
| Arina | N/A | Common wheat | Triticum aestivum | aestivum | 6× | Switzerland |
| Forno | N/A | Common wheat | Triticum aestivum | aestivum | 6× | Switzerland |
| Sumai 3 | PI 481542 | Common wheat | Triticum aestivum | aestivum | 6× | China |
| Chinese Spring-DIC 5B | N/A | Common wheat | Triticum aestivum | aestivum | 6× | U.S., Missouri |
| Bobwhite | PI 520554 | Common wheat | Triticum aestivum | aestivum | 6× | Mexico, CIMMYT |
| Salamouni | PI 182673 | Common wheat | Triticum aestivum | aestivum | 6× | Lebanon |
| Katepwa | N/A | Common wheat | Triticum aestivum | aestivum | 6× | Canada |
| M3 | N/A | Synthetic hexaploid wheat | Triticum turgidum × Aegiliops tauschii | Synthetic | 6× | Mexico, CIMMYT |
| PI277 | PI 277012 | Spelt wheat | Triticum aestivum | spelta | 6× | Spain |
| M6 | N/A | Synthetic hexaploid wheat | Triticum turgidum × Aegiliops tauschii | Synthetic | 6× | Mexico, CIMMYT |
| Kulm | PI 590576 | Common wheat | Triticum aestivum | aestivum | 6× | U.S., North Dakota |
| Opata85 | PI 591776 | Common wheat | Triticum aestivum | aestivum | 6× | Mexico, CIMMYT |
| TA4152-60 | N/A | Synthetic hexaploid wheat | Triticum turgidum × Aegiliops tauschii | Synthetic | 6× | Mexico, CIMMYT |
| TA4152-19 | N/A | Synthetic hexaploid wheat | Triticum turgidum × Aegiliops tauschii | Synthetic | 6× | Mexico, CIMMYT |
| P503 | N/A | Spelt wheat | Triticum aestivum | spelta | 6× | Iran |
| Divide | N/A | Durum wheat | Triticum turgidum | durum | 4× | U.S., North Dakota |
| Rusty | PI 639869 | Durum wheat | Triticum turgidum | durum | 4× | U.S., North Dakota |
| Ben | N/A | Durum wheat | Triticum turgidum | durum | 4× | U.S., North Dakota |
| Lebsock | N/A | Durum wheat | Triticum turgidum | durum | 4× | U.S., North Dakota |
| Langdon | N/A | Durum wheat | Triticum turgidum | durum | 4× | U.S., North Dakota |
| Altar84 | N/A | Durum wheat | Triticum turgidum | durum | 4× | Mexico, CIMMYT |
| PI193 | PI 193833 | Cultivated emmer | Triticum turgidum | dicoccum | 4× | Ethiopia |
| PI410 | PI 41025 | Cultivated emmer | Triticum turgidum | dicoccum | 4× | Russia |
| PI947 | PI 94749 | Persian wheat | Triticum turgidum | carthlicum | 4× | Georgia |
| PI481 | PI 481521 | Wild emmer | Triticum turgidum | dicoccoides | 4× | Israel |
| PI478 | PI 478742 | Wild emmer | Triticum turgidum | dicoccoides | 4× | Israel |
| TA106 | N/A | Wild emmer | Triticum turgidum | dicoccoides | 4× | Israel |
| Israel A | N/A | Wild emmer | Triticum turgidum | dicoccoides | 4× | Israel |
| PI272 | PI 272527 | Cultivated emmer | Triticum turgidum | dicoccum | 4× | Hungary |
| Total Length | Seminal Length | Seminal Count | Maximum Width | Maximum Depth | Width-Depth Ratio | Convex Hull Area | Seminal Emergence Angle | Centroid_X | Centroid_Y | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accession | Mean | STD | Mean | STD | Mean | STD | Mean | STD | Mean | STD | Mean | STD | Mean | STD | Mean | STD | Mean | STD | Mean | STD |
| Largo | 1407.054 | 444.8384 | 400.8567 | 127.1285 | 3.56 | 0.726 | 332.22 | 179.31 | 751.89 | 182 | 0.4569 | 0.26354 | 105875 | 66594.71 | 22.6833 | 7.0495 | −0.008 | 67.68921 | 193.0758 | 63.93469 |
| ND495 | 1991.549 | 383.8933 | 532.204 | 207.123 | 4 | 0.816 | 378.8 | 260.032 | 1017.9 | 187.838 | 0.3512 | 0.19509 | 188694.2 | 162351.9 | 29.39 | 9.58095 | 3.6871 | 43.52455 | 279.4124 | 100.8176 |
| Salamouni | 2628.581 | 1295.576 | 611.519 | 269.4623 | 4.1 | 1.449 | 416.2 | 256.94 | 944.5 | 319.004 | 0.4251 | 0.24228 | 226789.1 | 154967.4 | 23.224 | 27.50345 | 76.3194 | 87.27794 | 277.9139 | 125.8842 |
| Katepwa | 3251.233 | 668.9902 | 644.2344 | 221.368 | 5.22 | 0.833 | 572.44 | 285.073 | 1035.33 | 210.224 | 0.5466 | 0.23586 | 333568.1 | 213121.5 | 31.5122 | 15.02541 | 9.21678 | 111.8648 | 278.9432 | 89.18816 |
| M3 | 2051.728 | 955.6283 | 531.802 | 145.6446 | 3.7 | 1.059 | 224.3 | 148.072 | 989.8 | 271.476 | 0.2196 | 0.12853 | 126481.6 | 117850.5 | 19.818 | 13.55339 | −4.9703 | 51.40844 | 284.2637 | 93.55736 |
| M6 | 1701.343 | 876.2018 | 504.2471 | 196.2748 | 3.29 | 1.254 | 278.57 | 178.362 | 860.29 | 241.249 | 0.3064 | 0.19161 | 120445.6 | 85442.86 | 23.2343 | 23.46807 | 26.79043 | 50.84016 | 239.2451 | 95.57134 |
| Kulm | 4071.759 | 780.7794 | 800.521 | 166.8296 | 5 | 0.471 | 1075 | 240.575 | 1274.8 | 297.392 | 0.8693 | 0.21052 | 671934.4 | 282573.1 | 30.945 | 3.86432 | 34.0647 | 110.5345 | 354.779 | 87.14734 |
| Opata | 4372.535 | 947.7119 | 786.908 | 224.1664 | 5.3 | 0.675 | 803.7 | 329.924 | 1355.4 | 202.447 | 0.595 | 0.2199 | 611183.9 | 348976.6 | 23.772 | 6.56108 | 0.5184 | 83.79973 | 383.1734 | 75.31793 |
| TA60 | 3453.33 | 739.9634 | 872.307 | 305.9635 | 4.1 | 0.876 | 632.8 | 283.294 | 1351.1 | 293.192 | 0.4748 | 0.20016 | 451817.1 | 236567.4 | 19.944 | 7.12346 | 43.7792 | 88.52216 | 433.4055 | 133.3076 |
| TA19 | 2331.48 | 441.3384 | 680.5678 | 183.436 | 3.56 | 0.726 | 221.22 | 186.238 | 1052.78 | 147.466 | 0.2186 | 0.21618 | 116801.6 | 74324.72 | 20.8933 | 6.69294 | 1.45578 | 49.8508 | 356.9752 | 70.49687 |
| Grandin | 3649.392 | 689.1213 | 727.775 | 157.4818 | 5.1 | 0.738 | 902.9 | 368.004 | 1177.09 | 294.013 | 0.803 | 0.41375 | 536945.6 | 253719 | 39.773 | 12.66347 | 9.7508 | 60.9755 | 334.9114 | 75.75706 |
| P503 | 2635.32 | 451.2482 | 815.313 | 167.5469 | 3.3 | 0.675 | 567.6 | 341.986 | 1288.2 | 244.35 | 0.4742 | 0.33644 | 336351.3 | 164600.2 | 35.085 | 18.39818 | 28.2041 | 62.06401 | 390.4914 | 94.16569 |
| BR34 | 2486.309 | 621.315 | 625.36 | 193.1676 | 4 | 0.866 | 505.11 | 162.079 | 1114.33 | 325.571 | 0.4858 | 0.17914 | 277661.6 | 117046.7 | 25.4289 | 7.82762 | 44.02322 | 37.9747 | 322.6331 | 110.6642 |
| CSpring | 2526.974 | 787.4891 | 599.1433 | 226.2535 | 4.33 | 1.225 | 366.78 | 197.488 | 1106.89 | 265.426 | 0.3302 | 0.16149 | 212942.8 | 131228.1 | 35.7667 | 6.43028 | 1.78556 | 73.44133 | 297.4512 | 127.6352 |
| Arina | 3211.019 | 799.5607 | 818.916 | 197.3953 | 4 | 0.816 | 623.2 | 283.085 | 1307.3 | 213.307 | 0.491 | 0.23865 | 430168.5 | 185150.7 | 24.287 | 10.61435 | 70.9476 | 114.6787 | 403.6996 | 86.95764 |
| Forno | 2856.862 | 514.3885 | 891.16 | 116.4221 | 3.2 | 0.422 | 302.1 | 170.187 | 1499.6 | 281.496 | 0.2054 | 0.10853 | 246348 | 136554.2 | 24.115 | 15.79232 | 62.0981 | 91.38127 | 489.6822 | 94.50695 |
| Sumai3 | 3292.031 | 700.3864 | 700.5822 | 186.8713 | 4.78 | 0.667 | 332.11 | 130.143 | 1330.11 | 191.472 | 0.2538 | 0.10798 | 252744.3 | 128540.6 | 19.75 | 10.36522 | 8.46689 | 64.34472 | 394.5089 | 85.76959 |
| CSpringDIC | 3409.904 | 589.5194 | 704.0922 | 127.4161 | 4.89 | 0.601 | 532.78 | 243.404 | 1327.78 | 172.431 | 0.3948 | 0.16589 | 401419.2 | 188296.1 | 23.9867 | 10.9366 | 26.17744 | 51.21258 | 373.2856 | 67.95206 |
| Bobwhite | 2354.442 | 708.3413 | 723.972 | 213.3244 | 3.3 | 0.675 | 452 | 333.689 | 1155.6 | 244.926 | 0.3768 | 0.26688 | 248701.9 | 195832.2 | 17.424 | 7.47542 | 33.4653 | 65.00261 | 346.2481 | 85.93454 |
| Total Length | Seminal Length | Seminal Count | Maximum Width | Maximum Depth | Width-Depth Ratio | Convex Hull Area | Seminal Emergence Angle | Centroid_X | Centroid_Y | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accession | Mean | STD | Mean | STD | Mean | STD | Mean | STD | Mean | STD | Mean | STD | Mean | STD | Mean | STD | Mean | STD | Mean | STD |
| Divide | 2451.268 | 1990.837 | 556.8063 | 377.8616 | 3.75 | 1.909 | 179.5 | 183.808 | 968 | 467.962 | 0.163 | 0.11099 | 132953.1 | 141900.5 | 24.745 | 11.29471 | 62.338 | 54.1888 | 285.8023 | 183.9844 |
| Rusty | 1390.807 | 1255.677 | 314.033 | 226.8649 | 3.9 | 1.101 | 289.9 | 323.018 | 668.2 | 296.794 | 0.355 | 0.30589 | 117461.6 | 178758.6 | 19.006 | 10.84459 | 1.5814 | 46.94676 | 148.5815 | 108.4252 |
| Ben | 3226.34 | 1606.199 | 613.275 | 250.9193 | 4.67 | 1.862 | 325.5 | 176.039 | 1041.17 | 364.806 | 0.2894 | 0.17678 | 192996.1 | 100061.9 | 17.0567 | 6.29637 | −7.84817 | 31.76269 | 322.1412 | 136.3388 |
| Lebsock | 2709.894 | 1156.688 | 782.76 | 384.2838 | 3.56 | 1.13 | 352.78 | 215.12 | 1064.44 | 330.105 | 0.3325 | 0.18338 | 234695.5 | 203638.2 | 12.5878 | 12.2572 | 82.69922 | 67.46444 | 361.4402 | 171.0809 |
| Langdon | 3344.535 | 910.3358 | 647.958 | 140.2428 | 5.1 | 0.568 | 541.6 | 228.824 | 1119.8 | 198.192 | 0.4879 | 0.19377 | 319992.9 | 119248.3 | 31.939 | 15.33947 | −9.1897 | 60.98329 | 328.3687 | 72.69708 |
| Altar | 2509.834 | 1114.787 | 529.8867 | 216.3208 | 4.78 | 1.202 | 559.33 | 327.655 | 951.56 | 284.677 | 0.5769 | 0.27336 | 261532.6 | 218210.9 | 25.2367 | 12.27078 | 3.87411 | 44.27495 | 230.5439 | 106.7623 |
| PI193 | 3673.169 | 1406.628 | 745.731 | 256.8887 | 4.9 | 0.738 | 589.9 | 298.531 | 1266 | 337.197 | 0.4442 | 0.17098 | 484674.4 | 318533.1 | 25.628 | 12.4758 | −71.1325 | 67.57832 | 385.4912 | 132.6479 |
| PI410 | 3583.227 | 1531.398 | 814.01 | 338.0744 | 4.5 | 1.08 | 494.5 | 248.758 | 1211.7 | 369.315 | 0.4068 | 0.14769 | 351557.5 | 262529.1 | 15.345 | 6.84556 | 95.9932 | 142.1959 | 373.7213 | 141.8784 |
| PI947 | 3931.771 | 1070.18 | 772.071 | 210.8183 | 5.1 | 0.568 | 647.8 | 167.364 | 1125.7 | 280.126 | 0.6101 | 0.23353 | 389343.7 | 188785.1 | 34.931 | 8.18785 | 54.9579 | 125.2904 | 334.0316 | 96.79933 |
| PI481 | 1971.452 | 905.4402 | 707.311 | 291.7558 | 2.8 | 0.422 | 283.2 | 176.585 | 1045.6 | 315.264 | 0.2587 | 0.1009 | 162980 | 161573.2 | 17.845 | 13.14568 | −1.4536 | 44.00917 | 340.9898 | 134.9614 |
| PI478 | 1060.158 | 414.0258 | 449.97 | 99.81476 | 2.33 | 0.816 | 230 | 138.466 | 801.83 | 138.077 | 0.2761 | 0.16724 | 75115.56 | 49265.76 | 15.71 | 9.12903 | −1.32917 | 29.95209 | 214.8288 | 41.66894 |
| TA106 | 1789.02 | 682.8475 | 596.3411 | 227.6161 | 3 | 0 | 515 | 274.868 | 944.22 | 258.074 | 0.5188 | 0.18626 | 226387.3 | 185876.3 | 17.5533 | 7.23385 | −3.023 | 18.78727 | 275.3341 | 94.02308 |
| Israel | 2400.58 | 1152.096 | 814.988 | 352.7714 | 2.8 | 0.447 | 552.8 | 345.331 | 1215.6 | 395.11 | 0.4045 | 0.19579 | 331445.6 | 247329.2 | 18.316 | 9.29823 | 7.3922 | 39.28395 | 391.6284 | 161.1772 |
| PI277 | 2494.331 | 756.1297 | 586.835 | 250.0628 | 4.4 | 0.699 | 769.1 | 268.301 | 1056.8 | 313.966 | 0.7469 | 0.24797 | 388875.3 | 231645.1 | 27.434 | 9.31469 | 52.6313 | 47.89209 | 289.165 | 126.5571 |
| PI272 | 3406.442 | 1182.352 | 726.5822 | 217.0046 | 4.67 | 0.707 | 623 | 258.412 | 1232.33 | 278.285 | 0.5107 | 0.18407 | 418640.8 | 225100.3 | 26.1411 | 12.37334 | 62.85044 | 61.72506 | 342.3118 | 119.2106 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adeleke, E.; Millas, R.; McNeal, W.; Faris, J.; Taheri, A. Variation Analysis of Root System Development in Wheat Seedlings Using Root Phenotyping System. Agronomy 2020, 10, 206. https://doi.org/10.3390/agronomy10020206
Adeleke E, Millas R, McNeal W, Faris J, Taheri A. Variation Analysis of Root System Development in Wheat Seedlings Using Root Phenotyping System. Agronomy. 2020; 10(2):206. https://doi.org/10.3390/agronomy10020206
Chicago/Turabian StyleAdeleke, Ekundayo, Reneth Millas, Waymon McNeal, Justin Faris, and Ali Taheri. 2020. "Variation Analysis of Root System Development in Wheat Seedlings Using Root Phenotyping System" Agronomy 10, no. 2: 206. https://doi.org/10.3390/agronomy10020206
APA StyleAdeleke, E., Millas, R., McNeal, W., Faris, J., & Taheri, A. (2020). Variation Analysis of Root System Development in Wheat Seedlings Using Root Phenotyping System. Agronomy, 10(2), 206. https://doi.org/10.3390/agronomy10020206





