The Potential of Novel Bacterial Isolates from Natural Soil for the Control of Brown Rot Disease (Monilinia fructigena) on Apple Fruits
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Pathogen Preparation
2.2. Bacterial Isolates: Preparation and Molecular Identification
2.3. Biochemical Traits of Bacterial Isolates
2.4. Polymerase Chain Reaction (PCR) Detection of the Antibiotic Biosynthetic Genes
2.5. In Vitro Dual Culture Bioassay
2.6. Volatile Products (VOCs) Bioassay
2.7. In Vitro Bacterial Cell-Free Filtrates Effect
2.8. In Vivo Antagonism Experiment
(mm) of the wounds in the control treatment) × 100
2.9. Statistical Analysis
3. Results
3.1. Bacterial Identification
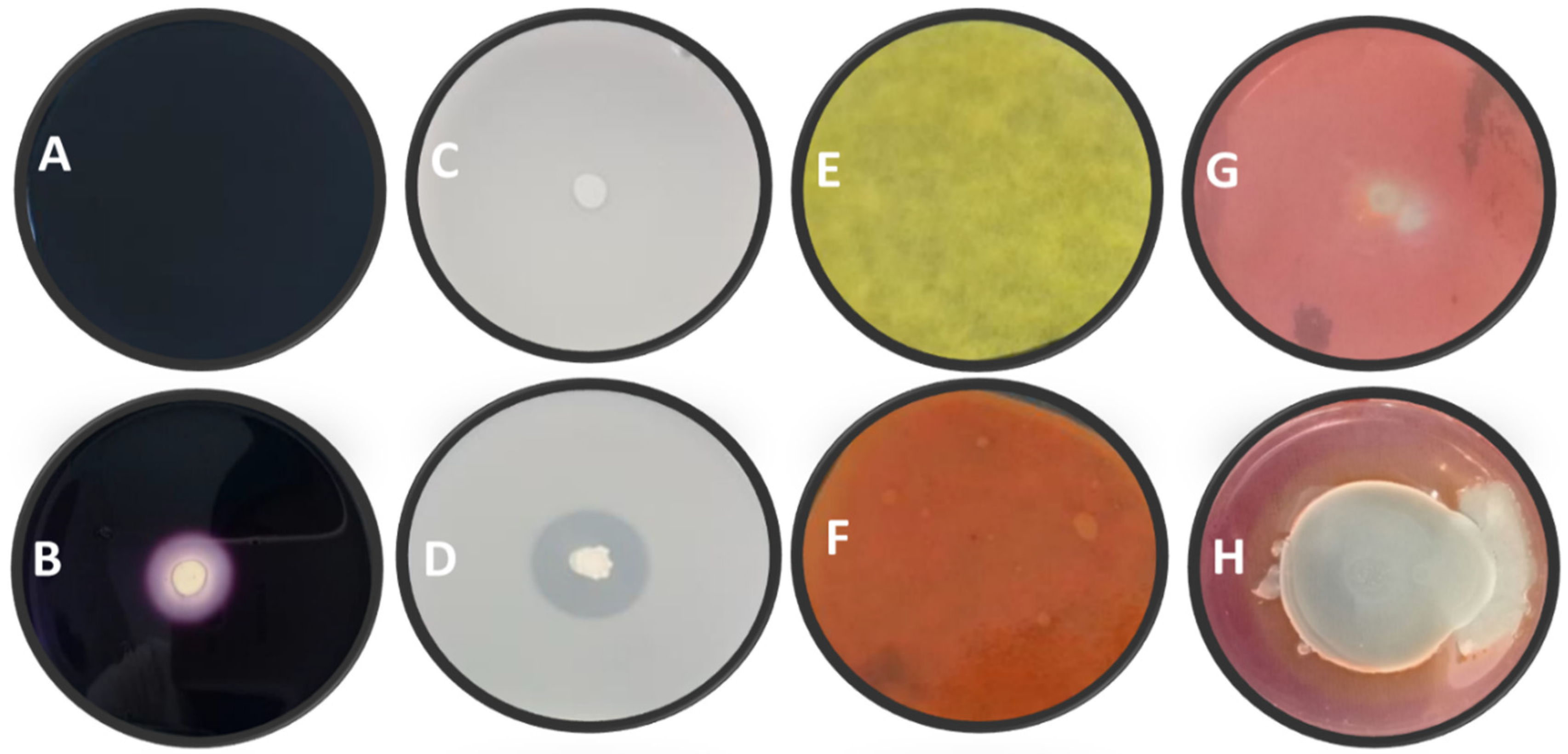
3.2. Biochemical Traits of Bacterial Isolates
3.3. Detection of Lipopeptides by PCR
3.4. In Vitro Dual Confrontation Bioassay
3.5. Effect of Bacteria-Derived VOCs on Mycelial Growth, under In Vitro Conditions
3.6. Effect of Bacterial Cell-Free Filtrates on Mycelial Growth and Spore Germination
3.7. In Vivo Effectiveness of Bacterial Isolates as Affected by Pathogen Pressure
3.7.1. Lower Pathogen Pressure (1 × 103 spores/mL)
3.7.2. Higher Pathogen Pressure (1 × 106 spores/mL)
3.8. Semi-Practical Trials of Selected Bacteria
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lesik, K. Monilinia species causing fruit brown rot, blossom and twig blight in apple orchards in Belarus. Proc. Latv. Acad. Sci. B Nat. Exact Appl. Sci. 2013, 67, 192–194. [Google Scholar]
- Hrustić, J.; Mihajlović, M.; Grahovac, M.; Delibašić, G.; Bulajić, A.; Krstić, B.; Tanovic, B. Genus Monilinia on pome and stone fruit species. Pestic. i Fitomed. 2012, 27, 283–297. [Google Scholar]
- De Oliveira Lino, L. Etude de la Variabilité Génétique de la Sensibilité à la Pourriture Brune au cours du Développement du Fruit Chez la Pêche en Lien avec L’évolution des Caractéristiques Physiques et Biochimiques du Fruit. Ph.D. Thesis, Université d’Avignon et des Pays de Vaucluse, Avignon, France, 2016. [Google Scholar]
- Dam, D.; Guillaume, R. Identification des Facteurs Intervenant dans le Développement du Monilia sur Fleurs et Rameaux d’Abricotiers et Recherche de Résistances Génétiques, sl: Sn. 2017. Available online: https://www.gisfruits.org/content/download/3802/37487/version/1/file/Rapport+D.+DAM.pdf (accessed on 10 August 2017).
- Van Leeuwen, G.C.M. The Brown Rot Fungi of Fruit Crops (Monilinia spp.), with Special Reference to Monilinia Fructigena (Aderh. & Ruhl.) Honey. Ph.D. Thesis, Wageningen University Wageningen, Wageningen, The Netherlands, 2000. [Google Scholar]
- Bostock, R.M.; Wilcox, S.M.; Wang, G.; Adaskaveg, J.E. Suppression of Monilinia fructicola cutinase production by peach fruit surface phenolic acids. Physiol. Mol. Plant Pathol. 1999, 54, 37–50. [Google Scholar]
- Wade, G.C.; Cruickshank, R.H. Rapid development of resistance of wounds on immature apricot fruit to infection with Monilinia fructicola. J. Phytopathol. 1992, 136, 89–94. [Google Scholar]
- Malandrakis, A.A.; Kavroulakis, N.; Chrysikopoulos, C.V. Use of silver nanoparticles to counter fungicide-resistance in Monilinia fructicola. Sci. Total Environ. 2020, 747, 141287. [Google Scholar]
- Lahlali, R.; Aksissou, W.; Lyousfi, N.; Ezrari, S.; Blenzar, A.; Tahiri, A.; Ennahli, S.; Hrustić, J.; MacLean, D.; Amiri, S. Biocontrol activity and putative mechanism of Bacillus amyloliquefaciens (SF14 and SP10), Alcaligenes faecalis ACBC1, and Pantoea agglomerans ACBP1 against brown rot disease of fruit. Microb. Pathog. 2020, 139, 103914. [Google Scholar]
- Franclet, J. Enquête ANPP-Maladies de conservation des pommes. Arboric. Fruit 1994, 473, 24–26. [Google Scholar]
- Lima, G.; De Curtis, F.; Piedimonte, D.; Spina, A.M.; De Cicco, V. Integration of biocontrol yeast and thiabendazole protects stored apples from fungicide sensitive and resistant isolates of Botrytis cinerea. Postharvest Biol. Technol. 2006, 40, 301–307. [Google Scholar]
- Wellings, P.W. The role of public policy in biological control: Some global trends. Entomophaga 1996, 41, 435–441. [Google Scholar]
- Usall, J.; Casals, C.; Sisquella, M.; Palou, L.; De Cal, A. Alternative technologies to control postharvest diseases of stone fruits. Stewart Postharvest Rev. 2015, 11, 4. [Google Scholar]
- Palou, L.; Ali, A.; Fallik, E.; Romanazzi, G. GRAS, plant-and animal-derived compounds as alternatives to conventional fungicides for the control of postharvest diseases of fresh horticultural produce. Postharvest Biol. Technol. 2016, 122, 41–52. [Google Scholar] [CrossRef]
- Spadaro, D.; Droby, S. Development of biocontrol products for postharvest diseases of fruit: The importance of elucidating the mechanisms of action of yeast antagonists. Trends Food Sci. Technol. 2016, 47, 39–49. [Google Scholar] [CrossRef]
- Calvo, H.; Marco, P.; Blanco, D.; Oria, R.; Venturini, M.E. Potential of a new strain of Bacillus amyloliquefaciens BUZ-14 as a biocontrol agent of postharvest fruit diseases. Food Microbiol. 2017, 63, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Reeleder, R.D.; Sparace, S.A. Influence of nutrients on growth of Epicoccum nigrum. Can. J. Microbiol. 1996, 42, 647–654. [Google Scholar] [CrossRef]
- Madrigal, C.; Melgarejo, P. Mechanisms of action of the antibiotic flavipin on Monilinia laxa and Saccharomyces cerevisiae. Mycol. Res. 1994, 98, 874–878. [Google Scholar] [CrossRef]
- De Cal, A.; Larena, I.; Liñán, M.; Torres, R.; Lamarca, N.; Usall, J.; Domenichini, P.; Bellini, A.; De Eribe, X.; Melgarejo, P. Population dynamics of Epicoccum nigrum, a biocontrol agent against brown rot in stone fruit. J. Appl. Microbiol. 2009, 106, 592–605. [Google Scholar] [CrossRef]
- Wang, Y.; Yuan, Y.; Liu, B.; Zhang, Z.; Yue, T. Biocontrol activity and patulin-removal effects of Bacillus subtilis, Rhodobacter sphaeroides and Agrobacterium tumefaciens against Penicillium expansum. J. Appl. Microbiol. 2016, 121, 1384–1393. [Google Scholar] [CrossRef]
- Hrustić, J.; Delibašić, G.; Stanković, I.; Grahovac, M.; Krstić, B.; Bulajić, A.; Tanović, B. Monilinia spp. causing brown rot of stone fruit in Serbia. Plant Dis. 2015, 99, 709–717. [Google Scholar] [CrossRef]
- Llop, P.; Bonaterra, A.; Peñalver, J.; López, M.M. Development of a highly sensitive nested-PCR procedure using a single closed tube for detection of Erwinia amylovora in asymptomatic plant material. Appl. Environ. Microbiol. 2000, 66, 2071–2078. [Google Scholar] [CrossRef]
- Dinesh, R.; Anandaraj, M.; Kumar, A.; Bini, Y.K.; Subila, K.P.; Aravind, R. Isolation, characterization, and evaluation of multi-trait plant growth promoting rhizobacteria for their growth promoting and disease suppressing effects on ginger. Microbiol. Res. 2015, 173, 34–43. [Google Scholar] [CrossRef]
- Gupta, V.; Peterson, C.B.; Dice, L.T.; Uchiki, T.; Racca, J.; Guo, J.; Xu, Y.; Hettich, R.L.; Zhao, X.; Rothstein, R.; et al. Sml1p is a dimer in solution: Characterization of denaturation and renaturation of recombinant Sml1p. Biochemistry 2004, 43, 8568–8578. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.S.; Ayyadurai, N.; Pandiaraja, P.; Reddy, A.V.; Venkateswarlu, Y.; Prakash, O.; Sakthivel, N. Characterization of antifungal metabolite produced by a new strain Pseudomonas aeruginosa PUPa3 that exhibits broad-spectrum antifungal activity biofertilizing traits. J. Appl. Microbiol. 2005, 98, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Cattelan, A.J.; Hartel, P.G.; Fuhrmann, J.J. Screening for plant growth–promoting rhizobacteria to promote early soybean growth. Soil Sci. Soc. Am. J. 1999, 63, 1670–1680. [Google Scholar] [CrossRef]
- Bakker, A.W.; Schippers, B. Microbial cyanide production in the rhizosphere in relation to potato yield reduction and Pseudomonas spp-mediated plant growth-stimulation. Soil Biol. Biochem. 1987, 19, 451–457. [Google Scholar] [CrossRef]
- Dimkić, I.; Živković, S.; Berić, T.; Ivanović, Ž.; Gavrilović, V.; Stanković, S.; Fira, D. Characterization and evaluation of two Bacillus strains, SS-12.6 and SS-13.1, as potential agents for the control of phytopathogenic bacteria and fungi. Biol. Control 2013, 65, 312–321. [Google Scholar] [CrossRef]
- Ramarathnam, R.; Bo, S.; Chen, Y.; Fernando, W.G.D.; Xuewen, G.; De Kievit, T. Molecular and biochemical detection of fengycin-and bacillomycin D-producing Bacillus spp., antagonistic to fungal pathogens of canola and wheat. Can. J. Microbiol. 2007, 53, 901–911. [Google Scholar] [CrossRef]
- Hsieh, F.C.; Li, M.C.; Lin, T.C.; Kao, S.S. Rapid detection and characterization of surfactin-producing Bacillus subtilis and closely related species based on PCR. Curr. Microbiol. 2004, 49, 186–191. [Google Scholar] [CrossRef]
- Lahlali, R.; Bajii, M.; Jijakli, M. Isolation and evaluation of bacteria and fungi as biological control agents against Rhizoctonia solani. Commun. Agric. Appl. Biol. Sci. 2007, 72, 973–982. [Google Scholar]
- Guevara-Avendaño, E.; Carrillo, J.D.; Ndinga-Muniania, C.; Moreno, K.; Méndez-Bravo, A.; Guerrero-Analco, J.A.; Eskalen, A.; Reverchon, F. Antifungal activity of avocado rhizobacteria against Fusarium euwallaceae and Graphium spp., associated with Euwallacea spp. nr. fornicatus, and Phytophthora cinnamomi. Antonie Van Leeuwenhoek 2018, 111, 563–572. [Google Scholar] [CrossRef]
- Li, Z.; Guo, B.; Wan, K.; Cong, M.; Huang, H.; Ge, Y. Effects of bacteria-free filtrate from Bacillus megaterium strain L2 on the mycelium growth and spore germination of Alternaria alternata. Biotechnol. Biotechnol. Equip. 2015, 29, 1062–1068. [Google Scholar] [CrossRef]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.-P.; Clément, C.; Ouhdouch, Y.; Van Wezel, G.P. Taxonomy, Physiology, and Natural Products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [PubMed]
- Compant, S.; Duffy, B.; Nowak, J.; Clément, C.; Barka, E.A. Use of plant growth-promoting bacteria for biocontrol of plant diseases: Principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 2005, 71, 4951–4959. [Google Scholar] [CrossRef] [PubMed]
- Aiello, D.; Restuccia, C.; Stefani, E.; Vitale, A.; Cirvilleri, G. Postharvest biocontrol ability of Pseudomonas synxantha against Monilinia fructicola and Monilinia fructigena on stone fruit. Postharvest Biol. Technol. 2019, 149, 83–89. [Google Scholar] [CrossRef]
- Yánez-Mendizábal, V.; Zeriouh, H.; Viñas, I.; Torres, R.; Usall, J.; de Vicente, A.; Pérez-García, A.; Teixidó, N. Biological control of peach brown rot (Monilinia spp.) by Bacillus subtilis CPA-8 is based on production of fengycin-like lipopeptides. Eur. J. Plant Pathol. 2012, 132, 609–619. [Google Scholar] [CrossRef]
- Gotor-Vila, A.; Teixidó, N.; Di Francesco, A.; Usall, J.; Ugolini, L.; Torres, R.; Mari, M. Antifungal effect of volatile organic compounds produced by Bacillus amyloliquefaciens CPA-8 against fruit pathogen decays of cherry. Food Microbiol. 2017, 64, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Schneider, K.E.; Li, X.Z. Development of biocontrol agents from food microbial isolates for controlling post-harvest peach brown rot caused by Monilinia fructicola. Int. J. Food Microbiol. 2008, 126, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhou, T.; He, D.; Li, X.Z.; Wu, H.; Liu, W.; Gao, X. Functions of lipopeptides bacillomycin D and fengycin in antagonism of Bacillus amyloliquefaciens C06 towards Monilinia fructicola. J. Mol. Microbiol. Biotechnol. 2011, 20, 43–52. [Google Scholar] [CrossRef]
- Willetts, H.J.; Byrde, R.J.W.; Fielding, A.H.; Wong, A.-L. The taxonomy of the brown rot fungi (Monilinia spp.) related to their extracellular cell wall-degrading enzymes. Microbiology 1977, 103, 77–83. [Google Scholar] [CrossRef]
- Falconi, C.J.; Mendgen, K. Epiphytic fungi on apple leaves and their value for control of the postharvest pathogens Botrytis cinerea, Monilinia fructigena and Penicillium expansum. J. Plant Dis. Prot. 1994, 101, 38–47. [Google Scholar]
- Larena, I.; Linan, M.; Melgarejo, P. Antibiotic production of the biocontrol agents epicoccum nigrum and candida sake. Plant Prot. Sci. 2003, 38, 205–208. [Google Scholar] [CrossRef]
- Esmaeel, Q.; Miotto, L.; Rondeau, M.; Leclère, V.; Clément, C.; Jacquard, C.; Sanchez, L.; Ait Barka, E. Paraburkholderia phytofirmans PsJN-Plants Interaction: From Perception to the Induced Mechanisms. Front. Microbiol. 2018, 9, 2093. [Google Scholar] [CrossRef] [PubMed]
- Sethi, S.K.; Mukherjee, A.K. Screening of biocontrol potential of indigenous Bacillus spp. isolated from rice rhizosphere against, R. solani, S. oryzae, S. rolfsii and response towards growth of rice. J. Pure Appl. Microbiol. 2018, 12, 41–53. [Google Scholar]
- Liu, Y.; Yao, S.; Deng, L.; Ming, J.; Zeng, K. Different mechanisms of action of isolated epiphytic yeasts against Penicillium digitatum and Penicillium italicum on citrus fruit. Postharvest Biol. Technol. 2019, 152, 100–110. [Google Scholar] [CrossRef]
- Sindhu, S.S.; Dadarwal, K.R. Chitinolytic and cellulolytic Pseudomonas sp. antagonistic to fungal pathogens enhances nodulation by Mesorhizobium sp. Cicer in chickpea. Microbiol. Res. 2001, 156, 353–358. [Google Scholar] [CrossRef]
- Richter, B.S.; Ivors, K.; Shi, W.; Benson, D.M. Cellulase activity as a mechanism for suppression of Phytophthora root rot in mulches. Phytopathology 2011, 101, 223–230. [Google Scholar] [CrossRef]
- Downer, A.J.; Menge, J.A.; Pond, E. Effects of cellulytic enzymes on Phytophthora cinnamomi. Phytopathology 2001, 91, 839–846. [Google Scholar] [CrossRef]
- Durairaj, K.; Velmurugan, P.; Park, J.H.; Chang, W.S.; Park, Y.J.; Senthilkumar, P.; Choi, K.-M.; Lee, J.H.; Oh, B.-T. Characterization and assessment of two biocontrol bacteria against Pseudomonas syringae wilt in Solanum lycopersicum and its genetic responses. Microbiol. Res. 2018, 206, 43–49. [Google Scholar] [CrossRef]
- Kim, Y.S.; Balaraju, K.; Jeon, Y. Effects of rhizobacteria Paenibacillus polymyxa APEC136 and Bacillus subtilis APEC170 on biocontrol of postharvest pathogens of apple fruits. J. Zhejiang Univ. Sci. B 2016, 17, 931–940. [Google Scholar] [CrossRef]
- Mardanova, A.M.; Fanisovna Hadieva, G.; Tafkilevich Lutfullin, M.; Valer’evna Khilyas, I.; Farvazovna Minnullina, L.; Gadelevna Gilyazeva, A.; Bogomolnaya, L.M.; Sharipova, M.R. Bacillus subtilis Strains with antifungal activity against the phytopathogenic Fungi. Agric. Sci. 2017, 8, 1–20. [Google Scholar]
- Choubane, S.; Cheba, B.A.; Benourrad, A. Screening and phenotypic diversity of amylase producing rhizospheric bacteria from some north african plants. Procedia Technol. 2016, 22, 1197–1204. [Google Scholar] [CrossRef][Green Version]
- El-Sayed, W.S.; Akhkha, A.; El-Naggar, M.Y.; Elbadry, M. In vitro antagonistic activity, plant growth promoting traits and phylogenetic affiliation of rhizobacteria associated with wild plants grown in arid soil. Front. Microbiol. 2014, 5, 651. [Google Scholar] [CrossRef] [PubMed]
- Dunne, C.; Crowley, J.J.; Moënne-Loccoz, Y.; Dowling, D.N.; O’Gara, F. Biological control of Pythium ultimum by Stenotrophomonas maltophilia W81 is mediated by an extracellular proteolytic activity. Microbiology 1997, 143, 3921–3931. [Google Scholar] [CrossRef]
- Budi, S.W.; van Tuinen, D.; Arnould, C.; Dumas-Gaudot, E.; Gianinazzi-Pearson, V.; Gianinazzi, S. Hydrolytic enzyme activity of Paenibacillus sp. strain B2 and effects of the antagonistic bacterium on cell integrity of two soil-borne pathogenic fungi. Appl. Soil Ecol. 2000, 15, 191–199. [Google Scholar] [CrossRef]
- Mounia, Y.-A.; Noreddine, K.C.; Laid, D.; Insaf, B.; Mounira, K.A.; Hlne, C.; Philippe, T. Antifungal activity and bioactive compounds produced by Bacillus mojavensis and Bacillus subtilis. Afr. J. Microbiol. Res. 2014, 8, 476–484. [Google Scholar] [CrossRef]
- Cipollone, R.; Ascenzi, P.; Tomao, P.; Imperi, F.; Visca, P. Enzymatic detoxification of cyanide: Clues from Pseudomonas aeruginosa rhodanese. J. Mol. Microbiol. Biotechnol. 2008, 15, 199–211. [Google Scholar] [CrossRef] [PubMed]
- Samuel, S.; Muthukkaruppan, S.M. Characterization of plant growth promoting rhizobacteria and fungi associated with rice, mangrove and effluent contaminated soil. Curr Bot. 2011, 2, 22–25. [Google Scholar]
- Gautam, S. Characterization of Plant Growth Promoting Rhizobacteria and Evaluation of Their Biocontrol Potential Against Tomato Bacterial Canker. Master’s Thesis, University of Horticulture and Forestry, Nauni Solan, India, 2014; 151p. [Google Scholar]
- Laville, J.; Blumer, C.; Von Schroetter, C.; Gaia, V.; Défago, G.; Keel, C.; Haas, D. Characterization of the hcnABC gene cluster encoding hydrogen cyanide synthase and anaerobic regulation by ANR in the strictly aerobic biocontrol agent Pseudomonas fluorescens CHA0. J. Bacteriol. 1998, 180, 3187–3196. [Google Scholar] [CrossRef]
- Voisard, C.; Keel, C.; Haas, D.; Dèfago, G. Cyanide production by Pseudomonas fluorescens helps suppress black root rot of tobacco under gnotobiotic conditions. EMBO J. 1989, 8, 351–358. [Google Scholar] [CrossRef]
- Mari, M.; Martini, C.; Guidarelli, M.; Neri, F. Postharvest biocontrol of Monilinia laxa, Monilinia fructicola and Monilinia fructigena on stone fruit by two Aureobasidium pullulans strains. Biol. Control 2012, 60, 132–140. [Google Scholar] [CrossRef]
- Droby, S.; Wisniewski, M.; El Ghaouth, A.; Wilson, C. Influence of food additives on the control of postharvest rots of apple and peach and efficacy of the yeast-based biocontrol product Aspire. Postharvest Biol. Technol. 2003, 27, 127–135. [Google Scholar] [CrossRef]
- Conway, W.S.; Leverentz, B.; Janisiewicz, W.J.; Saftner, R.A.; Camp, M.J. Improving biocontrol using antagonist mixtures with heat and/or sodium bicarbonate to control postharvest decay of apple fruit. Postharvest Biol. Technol. 2005, 36, 235–244. [Google Scholar] [CrossRef]
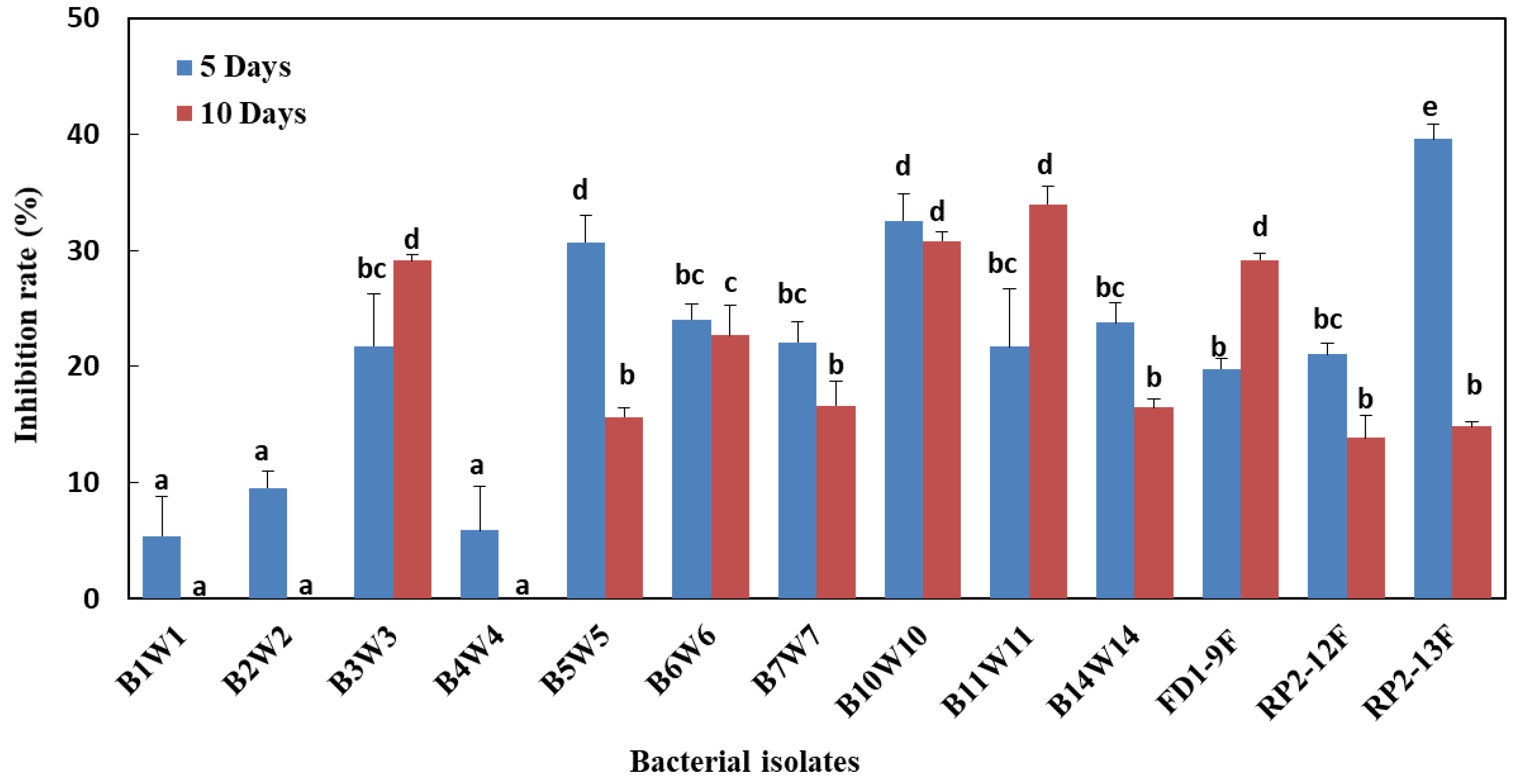
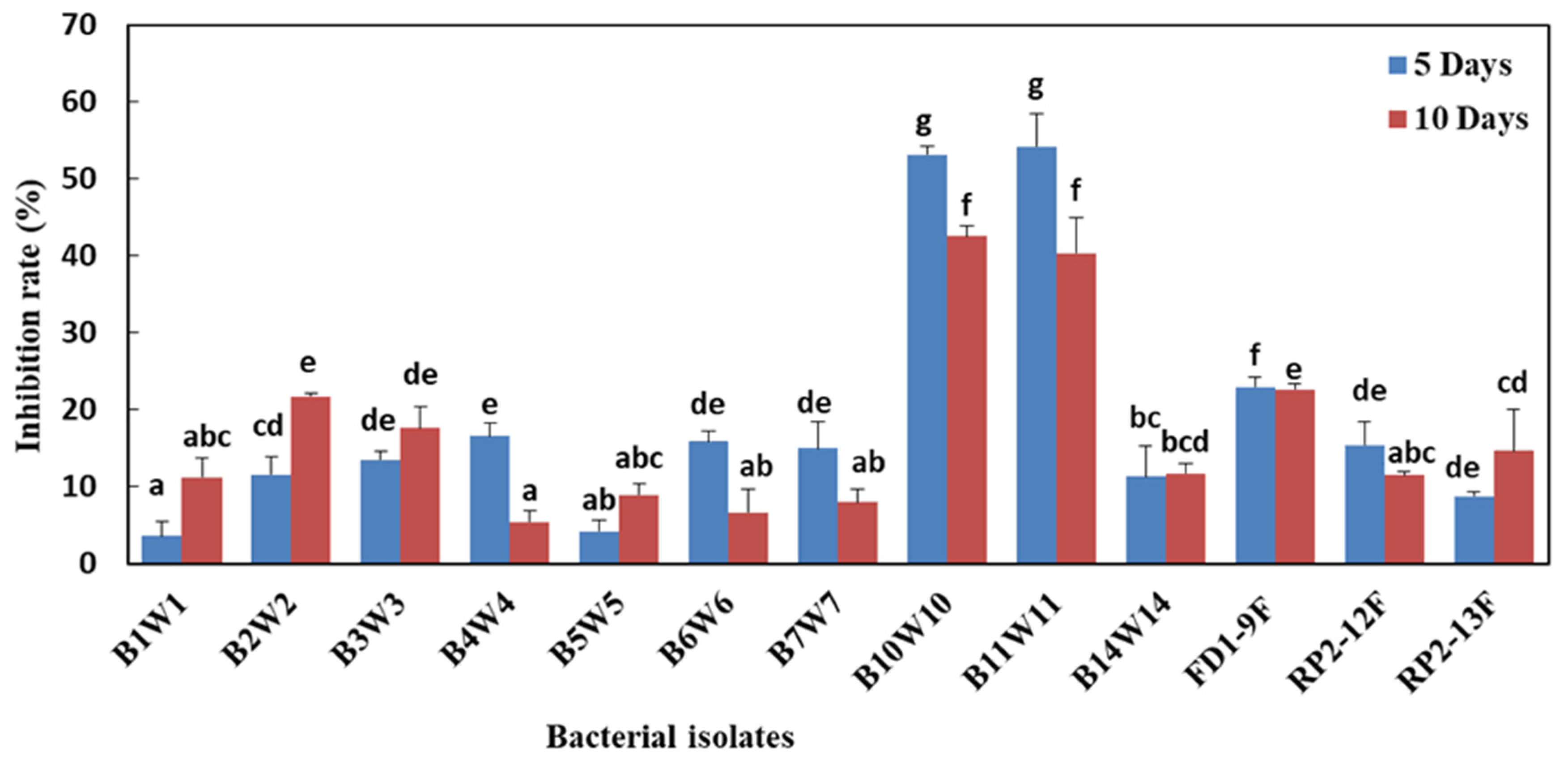
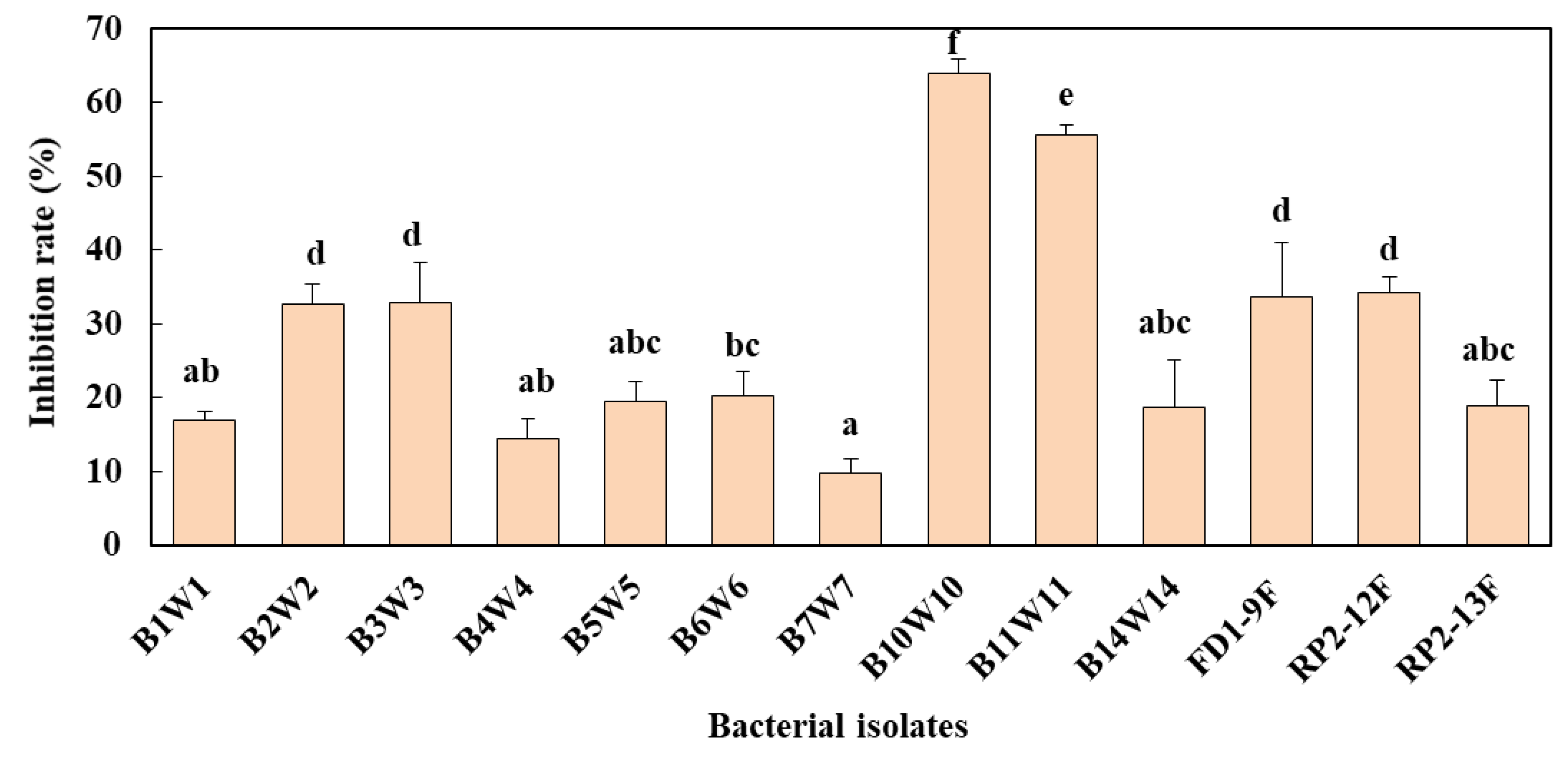

| Bacterial Isolates | Molecular Identification | In Vitro Inhibition Rate (%) * | ||
|---|---|---|---|---|
| Blast Research Results | Accession | 5 Days | 10 Days | |
| B1W1 | Bacillus amyloliquefaciens | MH727534 | 33.16 d | 80.27 e |
| B4W4 | Bacillus amyloliquefaciens | MK106157 | 30.01 c | 78.69 c |
| B7W7 | Bacillus amyloliquefaciens | MK106668 | 36.92 f | 82.86 f |
| B10W10 | Bacillus amyloliquefaciens | MT939665 | 36.37 f | 82.64 f |
| B14W14 | Bacillus siamensis | MK122722 | 33.13 d | 80.11 cd |
| B2W2 | Bacillus subtilis | MT940642 | 31.92 cd | 78.95 cd |
| B3W3 | Bacillus subtilis | MK106034 | 35.81 ef | 82.11 f |
| B5W5 | Bacillus subtilis | MK507777 | 35.93 f | 82.10 f |
| RP2-12F | Bacillus subtilis | MK518068 | 19.46 b | 72.70 b |
| RP2-13F | Bacillus subtilis | MK518364 | 15.50 a | 43.10 a |
| B6W6 | Pseudomonas sp. | MT940644 | 35.94 f | 82.41 f |
| B11W11 | Pseudomonas sp. | MK106058 | 33.75 de | 80.62 e |
| FD1-9F | Pseudomonas sp. | MK516189 | 30.38 c | 78.57 c |
| Lipopeptides | Design Primer | Primer Sequence | Product Length (bp) | Annealing T° | Reference |
|---|---|---|---|---|---|
| Bacillomycin | BACC1F/BACC1R | GAAGGACACGGCAGAGAGTC/ CGCTGATGACTGTTCATGCT | 875 bp | 60 °C | [29] |
| Fengycin | FEND1F/ FEND1R | TTTGGCAGCAGGAGAAGTT/ GCTGTCCGTTCTGCTTTTTC | 964 bp | 62 °C | [29] |
| Iturin | ITUP1F/ ITUP2R | AGCTTAGGGAACAATTGTCATCGGGGCTTC/ TCAGATAGGCCGCCATATCGGAATGATTCG | 2 kb | 45 °C | [28] |
| Surfactin | P17/ P18 | ATGAAGATTTACGGAATTTA/ TTATAAAAGCTCTTCGTACG | 675 bp | 53 °C | [30] |
| Bacterial Strains | Amylase | Protease | Cellulase | HCN |
|---|---|---|---|---|
| Bacillus amyloliquefaciens B1W1 | 1.02 b | 1.26 bc | 1.02 b | − |
| Bacillus amyloliquefaciens B4W4 | 1.37 f | 2.44 f | 1.05 b | + |
| Bacillus amyloliquefaciens B7W7 | 1.31 ef | 1.17 b | 0 a | − |
| Bacillus amyloliquefaciens B10W10 | 1.18 d | 1.70 d | 0 a | − |
| Bacillus siamensis B14W14 | 1.15 cd | 1.16 b | 1.21 def | + |
| Bacillus subtilis B2W2 | 1.1 bc | 1.98 e | 1.23 ef | − |
| Bacillus subtilis B3W3 | 1.06 b | 1.12 b | 1.18 cde | − |
| Bacillus subtilis B5W5 | 1.1 bc | 1.14 b | 1.38 g | − |
| Bacillus subtilis RP2-12F | 1.06 b | 1.95 e | 1.29 fg | − |
| Bacillus subtilis RP2-13F | 1.04 b | 1.26 bc | 1.11 bcd | − |
| Pseudomonas sp. B6W6 | 1.29 e | 1.68 d | 0 a | − |
| Pseudomonas sp. B11W11 | 1.17 cd | 0 a | 1.07 bc | − |
| Pseudomonas sp. FD1-9F | 0 a | 0 a | 1.30 fg | − |
| Strain | Bacillomycin | Fengycin | Iturin | Surfactin |
|---|---|---|---|---|
| Bacillus amyloliquefaciens B1W1 | + | − | + | − |
| Bacillus amyloliquefaciens B4W4 | + | − | + | − |
| Bacillus amyloliquefaciens B7W7 | − | − | + | − |
| Bacillus amyloliquefaciens B10W10 | − | − | + | − |
| Bacillus siamensis B14W14 | + | − | + | − |
| Bacillus subtilis B2W2 | + | − | + | − |
| Bacillus subtilis B3W3 | + | − | + | − |
| Bacillus subtilis B5W5 | − | − | + | − |
| Bacillus subtilis RP2-12F | + | + | + | − |
| Bacillus subtilis RP2-13F | + | − | + | − |
| Pseudomonas sp. B6W6 | + | − | − | + |
| Pseudomonas sp. B11W11 | + | + | − | − |
| Pseudomonas sp. FD1-9F | + | + | + | − |
| Bacterial Isolates | Pathogen Concentration | |||
|---|---|---|---|---|
| 1 × 103 spores/mL | 1 × 106 spores/mL | |||
| 5 Days | 10 Days | 5 Days | 10 Days | |
| Bacillus amyloliquefaciens B1W1 | 61.44 cx | 37.52 abc | 71.81 g | 76.84 c |
| Bacillus amyloliquefaciens B4W4 | 59.91 bc | 26.22 ab | 70.43 g | 73.76 c |
| Bacillus amyloliquefaciens B7W7 | 64.09 c | 41.75 abcd | 50.22 de | 66.84 bc |
| Bacillus amyloliquefaciens B10W10 | 0.00 a | 21.73 a | 0.00 a | 43.59 a |
| Bacillus siamensis B14W14 | 68.76 c | 43.85 abcd | 64.33 fg | 66.84 bc |
| Bacillus subtilis B2W2 | 60.34 c | 35.25 abc | 44.01 cd | 71.52 bc |
| Bacillus subtilis B3W3 | 63.09 c | 49.85 bcd | 69.08 g | 76.16 c |
| Bacillus subtilis B5W5 | 77.91 c | 51.23 cd | 46.93 cde | 69.78 bc |
| Bacillus subtilis RP2-12F | 63.73 c | 51.83 cd | 69.81 g | 70.53 bc |
| Bacillus subtilis RP2-13F | 54.19 bc | 65.34 e | 66.67 fg | 70.15 bc |
| Pseudomonas sp. B6W6 | 64.79 c | 59.37 d | 69.64 g | 71.79 bc |
| Pseudomonas sp. B11W11 | 0.00 a | 28.99 abc | 0.00 a | 51.09 ab |
| Pseudomonas sp. FD1-9F | 41.01 abc | 24.54 a | 36.84 c | 69.23 bc |
| Fungicide (TM) y | 9.24 ab | 34.95 abc | 24.86 b | 40.23 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lahlali, R.; Mchachti, O.; Radouane, N.; Ezrari, S.; Belabess, Z.; Khayi, S.; Mentag, R.; Tahiri, A.; Barka, E.A. The Potential of Novel Bacterial Isolates from Natural Soil for the Control of Brown Rot Disease (Monilinia fructigena) on Apple Fruits. Agronomy 2020, 10, 1814. https://doi.org/10.3390/agronomy10111814
Lahlali R, Mchachti O, Radouane N, Ezrari S, Belabess Z, Khayi S, Mentag R, Tahiri A, Barka EA. The Potential of Novel Bacterial Isolates from Natural Soil for the Control of Brown Rot Disease (Monilinia fructigena) on Apple Fruits. Agronomy. 2020; 10(11):1814. https://doi.org/10.3390/agronomy10111814
Chicago/Turabian StyleLahlali, Rachid, Ouafae Mchachti, Nabil Radouane, Said Ezrari, Zineb Belabess, Slimane Khayi, Rachid Mentag, Abdessalem Tahiri, and Essaid Ait Barka. 2020. "The Potential of Novel Bacterial Isolates from Natural Soil for the Control of Brown Rot Disease (Monilinia fructigena) on Apple Fruits" Agronomy 10, no. 11: 1814. https://doi.org/10.3390/agronomy10111814
APA StyleLahlali, R., Mchachti, O., Radouane, N., Ezrari, S., Belabess, Z., Khayi, S., Mentag, R., Tahiri, A., & Barka, E. A. (2020). The Potential of Novel Bacterial Isolates from Natural Soil for the Control of Brown Rot Disease (Monilinia fructigena) on Apple Fruits. Agronomy, 10(11), 1814. https://doi.org/10.3390/agronomy10111814









