Understanding Molecular Mechanisms of Seed Dormancy for Improved Germination in Traditional Leafy Vegetables: An Overview
Abstract
:1. Introduction
2. Methods
3. Seed Dormancy in Traditional Leafy Vegetables
4. Seed Dormancy Regulation in Plants
4.1. Direct Pathway Regulation
4.2. Hormonal Pathway Regulation
5. Environmental Factors Influencing Seed Dormancy Regulation
6. Seed Coat Components
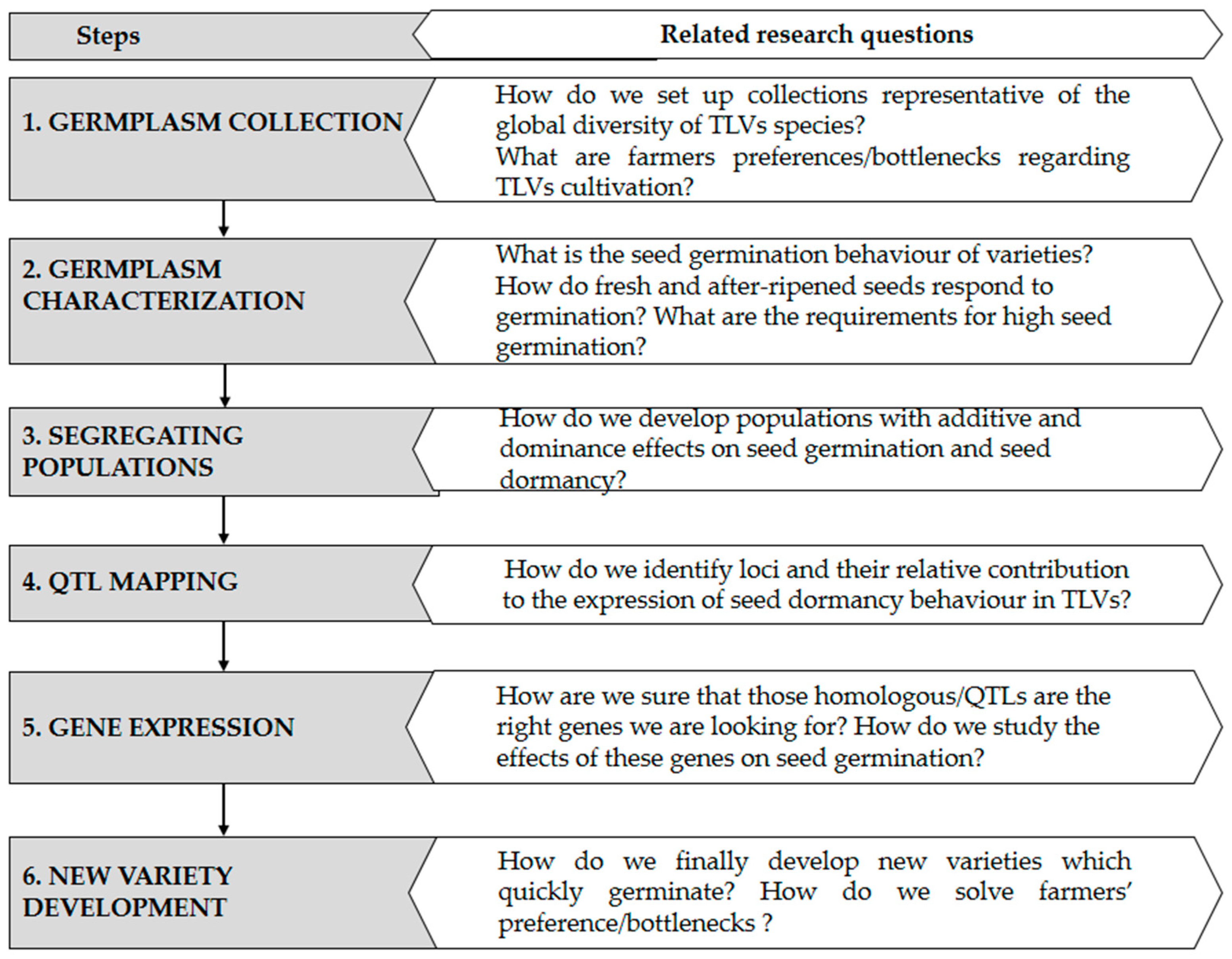
7. Pathway for Dormancy Studies in Traditional Leafy Vegetables
7.1. Germplasm Collection
7.2. Seed Dormancy Characterization
7.3. Development of Mapping Populations for Identification of Candidate Genes Involved in Seed Dormancy
7.4. Tapping into Comparative Genomics to Study Seed Dormancy in TLVs
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Achigan-Dako, E.G.; N’Danikou, S.; Assogba-Komlan, F.; Ambrose-Oji, B.; Ahanchede, A.; Pasquini, M.W. Diversity, geographical, and consumption patterns of traditional vegetables in sociolinguistic communities in Benin: Implications for domestication and utilization. Econ. Bot. 2011, 65, 129–145. [Google Scholar] [CrossRef]
- Kahane, R.; Temple, L.; Brat, P.; De Bon, H. Les légumes feuilles des pays tropicaux: Diversité, richesse économique et valeur santé dans un contexte très fragile. In Proceedings of the Colloque Les légumes: Un Patrimoine à Transmettre et à Valoriser, Angers, France, 7–9 September 2005. [Google Scholar]
- Maundu, P.; Achigan-Dako, E.; Morimoto, Y. Biodiversity of African vegetables. In African Indigenous Vegetables in Urban Agriculture; Shackleton, C.M., Pasquini, M.W., Drescher, A.W., Eds.; Routledge: London, UK, 2009; pp. 97–136. [Google Scholar]
- Towns, A.M.; Shackleton, C. Traditional, Indigenous, or Leafy? A Definition, Typology, and Way Forward for African Vegetables. Econ. Bot. 2018, 72, 461–477. [Google Scholar] [CrossRef]
- Uzilday, B.; Turkan, I.; Sekmen, A.; Ozgur, R.; Karakaya, H. Comparison of ROS formation and antioxidant enzymes in Cleome gynandra (C4) and Cleome spinosa (C3) under drought stress. Plant Sci. 2012, 182, 59–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luoh, J.W.; Begg, C.B.; Symonds, R.C.; Ledesma, D.; Yang, R.-Y. Nutritional yield of African indigenous vegetables in water-deficient and water-sufficient conditions. Food Nutr. Sci. 2014, 5, 812. [Google Scholar] [CrossRef] [Green Version]
- Kansiime, M.K.; Karanja, D.K.; Alokit, C.; Ochieng, J. Derived demand for African indigenous vegetable seed: Implications for farmer-seed entrepreneurship development. Int. Food Agribus. Manag. Rev. 2018, 21, 723–739. [Google Scholar] [CrossRef]
- Abukutsa-Onyango, M. Seed production and support systems for African Leafy Vegetables in three communities In Western Kenya. Afr. J. Food Agric. Nutr. Dev. 2006, 7, 108–116. [Google Scholar]
- Dube, P.; Struik, P.C.; Ngadze, E. Seed health tests of traditional leafy vegetables and pathogenicity in plants. Afr. J. Agric. Res. 2018, 13, 753–770. [Google Scholar]
- Adebooye, O.; Ajayi, S.; Baidu-Forson, J.; Opabode, J. Seed constraint to cultivation and productivity of African indigenous leaf vegetables. Afr. J. Biotechnol. 2005, 4, 1480–1484. [Google Scholar]
- Sogbohossou, E.D.; Achigan-Dako, E.G.; Maundu, P.; Solberg, S.; Deguenon, E.M.; Mumm, R.H.; Hale, I.; Van Deynze, A.; Schranz, M.E. A roadmap for breeding orphan leafy vegetable species: A case study of Gynandropsis gynandra (Cleomaceae). Hortic. Res. 2018, 5, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Baskin, J.M.; Baskin, C.C. A classification system for seed dormancy. Seed Sci. Res. 2004, 14, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Hilhorst, H.W. Standardizing seed dormancy research. In Seed Dormancy; Kermode, A.R., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; Volume 773, pp. 43–52. [Google Scholar]
- Fenner, M.; Thompson, K. The Ecology of Seeds; Cambridge University Press: Cambridge, UK, 2006; p. 260. [Google Scholar]
- Bewley, J.D. Seed germination and dormancy. Plant Cell 1997, 9, 1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bewley, J.D.; Black, M. Seeds. In Seeds Physiology of Development and Germination, 3rd ed.; Springer: Berlin/Heidelberg, Germany; Plenum Press: New York, NY, USA, 1994; pp. 1–33. [Google Scholar]
- Skubacz, A.; Daszkowska-Golec, A. Seed Dormancy: The Complex Process Regulated by Abscisic Acid, Gibberellins, and Other Phytohormones that Makes Seed Germination Work. In Phytohormones-Signaling Mechanisms and Crosstalk in Plant Development and Stress Responses; El-Esawi, M., Ed.; InTech: Vienna, Austria, 2017; pp. 77–100. [Google Scholar]
- Van der Schaar, W.; Alonso-Blanco, C.; Léon-Kloosterziel, K.M.; Jansen, R.C.; Van Ooijen, J.W.; Koornneef, M. QTL analysis of seed dormancy in Arabidopsis using recombinant inbred lines and MQM mapping. Heredity 1997, 79, 190–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, M.; Liu, S.; Zhang, G.; Bai, G. Effects of TaPHS1 and TaMKK3-A Genes on Wheat Pre-Harvest Sprouting Resistance. Agronomy 2018, 8, 210. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Ni, P.; Francki, M.; Hunter, A.; Zhang, Y.; Schibeci, D.; Li, H.; Tarr, A.; Wang, J.; Cakir, M. Genes controlling seed dormancy and pre-harvest sprouting in a rice-wheat-barley comparison. Funct. Integr. Genom. 2004, 4, 84–93. [Google Scholar] [CrossRef]
- Debieu, M.; Tang, C.; Stich, B.; Sikosek, T.; Effgen, S.; Josephs, E.; Schmitt, J.; Nordborg, M.; Koornneef, M.; de Meaux, J. Co-variation between seed dormancy, growth rate and flowering time changes with latitude in Arabidopsis thaliana. PLoS ONE 2013, 8, e61075. [Google Scholar] [CrossRef] [Green Version]
- Hawkins, K.; Allen, P.; Meyer, S. Secondary dormancy induction and release in Bromus tectorum seeds: The role of temperature, water potential and hydrothermal time. Seed Sci. Res. 2017, 27, 12–25. [Google Scholar] [CrossRef] [Green Version]
- Koornneef, M.; Alonso-Blanco, C.; Vreugdenhil, D. Naturally occurring genetic variation in Arabidopsis thaliana. Annu. Rev. Plant Biol. 2004, 55, 141–172. [Google Scholar] [CrossRef] [Green Version]
- Zardilis, A.; Hume, A.; Millar, A.J. A multi-model framework for the Arabidopsis life cycle. J. Exp. Bot. 2019, 70, 2463–2477. [Google Scholar] [CrossRef] [Green Version]
- Lu, Q.; Niu, X.; Zhang, M.; Wang, C.; Xu, Q.; Feng, Y.; Yang, Y.; Wang, S.; Yuan, X.; Yu, H. Genome-wide association study of seed dormancy and the genomic consequences of improvement footprints in rice (Oryza sativa L.). Front. Plant Sci. 2018, 8, 2213. [Google Scholar] [CrossRef] [Green Version]
- Ali, A.; Cao, J.; Jiang, H.; Chang, C.; Zhang, H.-P.; Sheikh, S.W.; Shah, L.; Ma, C. Unraveling Molecular and Genetic Studies of Wheat (Triticum aestivum L.) Resistance against Factors Causing Pre-Harvest Sprouting. Agronomy 2019, 9, 117. [Google Scholar] [CrossRef] [Green Version]
- Cota-Sánchez, J.H. Precocious Germination (Vivipary) in Tomato: A Link to Economic Loss? Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2018, 88, 1443–1451. [Google Scholar] [CrossRef]
- Miransari, M.; Smith, D. Plant hormones and seed germination. Environ. Exp. Bot. 2014, 99, 110–121. [Google Scholar] [CrossRef]
- Née, G.; Xiang, Y.; Soppe, W.J. The release of dormancy, a wake-up call for seeds to germinate. Curr. Opin. Plant Biol. 2017, 35, 8–14. [Google Scholar] [CrossRef] [Green Version]
- Baskin, J.M.; Baskin, C.C. New approaches to the study of the evolution of physical and physiological dormancy, the two most common classes of seed dormancy on earth. In The Biology of Seeds: Recent Research Advances; Nicolás, G., Bradford, K., Côme, D., Pritchard, H.W., Eds.; CAB International: Wallingford, UK, 2003; pp. 371–380. [Google Scholar]
- Shilla, O.; Abukutsa-Onyango, M.O.; Dinssa, F.F.; Winkelmann, T. Seed dormancy, viability and germination of Cleome gynandra (L.) BRIQ. Afr. J. Hortic. Sci. 2017, 10, 45–52. [Google Scholar]
- Baskin, C.; Baskin, J. Seeds: Ecology, Biogeography, and Evolution of Dormancy and Germination, 2nd ed.; Elsevier: Amsterdam, The Netherlands; Academic Press: San Diego, CA, USA, 2014. [Google Scholar]
- Motsa, M.; Slabbert, M.; Van Averbeke, W.; Morey, L. Effect of light and temperature on seed germination of selected African leafy vegetables. S. Afr. J. Bot. 2015, 99, 29–35. [Google Scholar] [CrossRef]
- Taab, A.; Andersson, L. Primary dormancy and seedling emergence of black nightshade (Solanum nigrum) and hairy nightshade (Solanum physalifolium). Weed Sci. 2009, 57, 526–532. [Google Scholar] [CrossRef]
- Agble, F. Germination of seeds of Talinum triangulare. Ghana J. Sci. 1970, 10, 29–32. [Google Scholar]
- Kępczynski, J.; Bihun, M.; Kępczynska, E. Ethylene Involvement in the Dormancy of Amaranthus Seeds. In Biology and Biotechnology of the Plant Hormone Ethylene; Kanellis, A.K., Chang, C., Kende, H., Grierson, D., Eds.; Springer: Dordrecht, The Netherlands, 1997; pp. 113–122. [Google Scholar]
- Roberts, H.; Lockett, P.M. Seed dormancy and field emergence in Solanum nigrum L. Weed Res. 1978, 18, 231–241. [Google Scholar] [CrossRef]
- Geneve, R.L. Seed dormancy in commercial vegetable and flower species. Seed Technol. 1998, 236–250. [Google Scholar]
- Ochuodho, J.; Modi, A. Temperature and light requirements for the germination of Cleome gynandra seeds. S. Afr. J. Plant Soil 2005, 22, 49–54. [Google Scholar] [CrossRef]
- Ekpong, B. Effects of seed maturity, seed storage and pre-germination treatments on seed germination of cleome (Cleome gynandra L.). Sci. Hortic. 2009, 119, 236–240. [Google Scholar] [CrossRef]
- Kamotho, G.; Mathenge, P.; Muasya, R.; Dullo, M. Effects of maturity stage, desiccation and storage period on seed quality of cleome (Cleome gynandra L.). Res. Desk 2014, 3, 419–433. [Google Scholar]
- Yepes, J. Study of a weed Cleome gynandra L. Rev. Comalfi 1978, 5, 49–53. [Google Scholar]
- Zharare, G. Differential requirements for breaking seed dormancy in biotypes of Cleome gynandra and two Amaranthus species. Afr. J. Agric. Res. 2012, 7, 5049–5059. [Google Scholar]
- Enayati, V.; Esfandiari, E.; Pourmohammad, A.; Haj Mohammadnia Ghalibaf, K. Evaluation of different methods in seed dormancy breaking and germination of Redroot Pigweed (Amaranthus retroflexus). Iran. J. Seed Res. 2019, 5, 129–137. [Google Scholar] [CrossRef] [Green Version]
- Foley, M.E. Seed dormancy: An update on terminology, physiological genetics, and quantitative trait loci regulating germinability. Weed Sci. 2001, 49, 305–317. [Google Scholar] [CrossRef]
- Alonso-Blanco, C.; Bentsink, L.; Hanhart, C.J.; Blankestijn-de Vries, H.; Koornneef, M. Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics 2003, 164, 711–729. [Google Scholar]
- Kondou, Y.; Higuchi, M.; Takahashi, S.; Sakurai, T.; Ichikawa, T.; Kuroda, H.; Yoshizumi, T.; Tsumoto, Y.; Horii, Y.; Kawashima, M. Systematic approaches to using the FOX hunting system to identify useful rice genes. Plant J. 2009, 57, 883–894. [Google Scholar] [CrossRef]
- Teng, S.; Rognoni, S.; Bentsink, L.; Smeekens, S. The Arabidopsis GSQ5/DOG1 Cvi allele is induced by the ABA-mediated sugar signalling pathway, and enhances sugar sensitivity by stimulating ABI4 expression. Plant J. 2008, 55, 372–381. [Google Scholar] [CrossRef]
- Bentsink, L.; Hanson, J.; Hanhart, C.J.; Blankestijn-de Vries, H.; Coltrane, C.; Keizer, P.; El-Lithy, M.; Alonso-Blanco, C.; de Andrés, M.T.; Reymond, M. Natural variation for seed dormancy in Arabidopsis is regulated by additive genetic and molecular pathways. Proc. Natl. Acad. Sci. USA 2010, 107, 4264–4269. [Google Scholar] [CrossRef] [Green Version]
- Chiang, G.C.; Bartsch, M.; Barua, D.; Nakabayashi, K.; Debieu, M.; Kronholm, I.; Koornneef, M.; Soppe, W.J.; Donohue, K.; de Meaux, J. DOG1 expression is predicted by the seed-maturation environment and contributes to geographical variation in germination in Arabidopsis thaliana. Mol. Ecol. 2011, 20, 3336–3349. [Google Scholar] [CrossRef] [PubMed]
- Kendall, S.L.; Hellwege, A.; Marriot, P.; Whalley, C.; Graham, I.A.; Penfield, S. Induction of dormancy in Arabidopsis summer annuals requires parallel regulation of DOG1 and hormone metabolism by low temperature and CBF transcription factors. Plant Cell 2011, 23, 2568–2580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakabayashi, K.; Bartsch, M.; Xiang, Y.; Miatton, E.; Pellengahr, S.; Yano, R.; Seo, M.; Soppe, W.J. The time required for dormancy release in Arabidopsis is determined by DELAY OF GERMINATION1 protein levels in freshly harvested seeds. Plant Cell 2012, 24, 2826–2838. [Google Scholar] [CrossRef] [Green Version]
- Nonogaki, H. Seed dormancy and germination—Emerging mechanisms and new hypotheses. Front. Plant Sci. 2014, 5, 233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashikawa, I.; Abe, F.; Nakamura, S. DOG1-like genes in cereals: Investigation of their function by means of ectopic expression in Arabidopsis. Plant Sci. 2013, 208, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Bentsink, L.; Jowett, J.; Hanhart, C.J.; Koornneef, M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 17042–17047. [Google Scholar] [CrossRef] [Green Version]
- Sugimoto, K.; Takeuchi, Y.; Ebana, K.; Miyao, A.; Hirochika, H.; Hara, N.; Ishiyama, K.; Kobayashi, M.; Ban, Y.; Hattori, T. Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc. Natl. Acad. Sci. USA 2010, 107, 5792–5797. [Google Scholar] [CrossRef] [Green Version]
- Graeber, K.; Voegele, A.; Büttner-Mainik, A.; Sperber, K.; Mummenhoff, K.; Leubner-Metzger, G. Spatiotemporal seed development analysis provides insight into primary dormancy induction and evolution of the Lepidium DELAY OF GERMINATION1 genes. Plant Physiol. 2013, 161, 1903–1917. [Google Scholar] [CrossRef] [Green Version]
- Ashikawa, I.; Abe, F.; Nakamura, S. Ectopic expression of wheat and barley DOG1-like genes promotes seed dormancy in Arabidopsis. Plant Sci. 2010, 179, 536–542. [Google Scholar] [CrossRef]
- Boccaccini, A.; Santopolo, S.; Capauto, D.; Lorrai, R.; Minutello, E.; Serino, G.; Costantino, P.; Vittorioso, P. The DOF protein DAG1 and the DELLA protein GAI cooperate in negatively regulating the AtGA3ox1 gene. BMC Plant Biol. 2014, 7, 1486–1489. [Google Scholar] [CrossRef] [Green Version]
- Rueda-Romero, P.; Barrero-Sicilia, C.; Gómez-Cadenas, A.; Carbonero, P.; Oñate-Sánchez, L. Arabidopsis thaliana DOF6 negatively affects germination in non-after-ripened seeds and interacts with TCP14. J. Exp. Bot. 2011, 63, 1937–1949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravindran, P.; Verma, V.; Stamm, P.; Kumar, P.P. A novel RGL2–DOF6 complex contributes to primary seed dormancy in Arabidopsis thaliana by regulating a GATA transcription factor. Mol. Plant 2017, 10, 1307–1320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stamm, P.; Ravindran, P.; Mohanty, B.; Tan, E.L.; Yu, H.; Kumar, P.P. Insights into the molecular mechanism of RGL2-mediated inhibition of seed germination in Arabidopsis thaliana. BMC Plant Biol. 2012, 12, 179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gabriele, S.; Rizza, A.; Martone, J.; Circelli, P.; Costantino, P.; Vittorioso, P. The Dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1. Plant J. 2010, 61, 312–323. [Google Scholar] [CrossRef]
- Gualberti, G.; Papi, M.; Bellucci, L.; Ricci, I.; Bouchez, D.; Camilleri, C.; Costantino, P.; Vittorioso, P. Mutations in the Dof zinc finger genes DAG2 and DAG1 influence with opposite effects the germination of Arabidopsis seeds. Plant Cell 2002, 14, 1253–1263. [Google Scholar] [CrossRef] [Green Version]
- Léon-Kloosterziel, K.M.; van de Bunt, G.A.; Zeevaart, J.A.; Koornneef, M. Arabidopsis mutants with a reduced seed dormancy. Plant Physiol. 1996, 110, 233–240. [Google Scholar] [CrossRef] [Green Version]
- Peeters, A.J.; Blankestijn-de Vries, H.; Hanhart, C.J.; Léon-Kloosterziel, K.M.; Zeevaart, J.A.; Koornneef, M. Characterization of mutants with reduced seed dormancy at two novel rdo loci and a further characterization of rdo1 and rdo2 in Arabidopsis. Physiol. Plant 2002, 115, 604–612. [Google Scholar] [CrossRef]
- Liu, Y.; Geyer, R.; Van Zanten, M.; Carles, A.; Li, Y.; Hörold, A.; van Nocker, S.; Soppe, W.J. Identification of the Arabidopsis REDUCED DORMANCY 2 gene uncovers a role for the polymerase associated factor 1 complex in seed dormancy. PLoS ONE 2011, 6, 22241. [Google Scholar] [CrossRef] [Green Version]
- Grasser, M.; Kane, C.M.; Merkle, T.; Melzer, M.; Emmersen, J.; Grasser, K.D. Transcript elongation factor TFIIS is involved in Arabidopsis seed dormancy. J. Mol. Biol. 2009, 386, 598–611. [Google Scholar] [CrossRef]
- Liu, Y.; Koornneef, M.; Soppe, W.J. The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell 2007, 19, 433–444. [Google Scholar] [CrossRef] [Green Version]
- Yazdanpanah, F.; Hanson, J.; Hilhorst, H.W.; Bentsink, L. Differentially expressed genes during the imbibition of dormant and after-ripened seeds—A reverse genetics approach. BMC Plant Biol. 2017, 17, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R.R.; Gampala, S.S.; Rock, C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell Online 2002, 14, S15–S45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flematti, G.R.; Ghisalberti, E.L.; Dixon, K.W.; Trengove, R.D. A compound from smoke that promotes seed germination. Science 2004, 305, 977. [Google Scholar] [CrossRef] [PubMed]
- Kanno, Y.; Jikumaru, Y.; Hanada, A.; Nambara, E.; Abrams, S.R.; Kamiya, Y.; Seo, M. Comprehensive hormone profiling in developing Arabidopsis seeds: Examination of the site of ABA biosynthesis, ABA transport and hormone interactions. Plant Cell Physiol. 2010, 51, 1988–2001. [Google Scholar] [CrossRef] [Green Version]
- Nambara, E.; Marion-Poll, A. ABA action and interactions in seeds. Trends Plant Sci. 2003, 8, 213–217. [Google Scholar] [CrossRef]
- Frey, A.; Effroy, D.; Lefebvre, V.; Seo, M.; Perreau, F.; Berger, A.; Sechet, J.; To, A.; North, H.M.; Marion-Poll, A. Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J. 2012, 70, 501–512. [Google Scholar] [CrossRef]
- Shu, K.; Zhang, H.; Wang, S.; Chen, M.; Wu, Y.; Tang, S.; Liu, C.; Feng, Y.; Cao, X.; Xie, Q. ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLoS Genet. 2013, 9, e1003577. [Google Scholar] [CrossRef] [Green Version]
- Shu, K.; Liu, X.-D.; Xie, Q.; He, Z.-H. Two faces of one seed: Hormonal regulation of dormancy and germination. Mol. Plant 2016, 9, 34–45. [Google Scholar] [CrossRef] [Green Version]
- Kim, W.; Lee, Y.; Park, J.; Lee, N.; Choi, G. HONSU, a protein phosphatase 2C, regulates seed dormancy by inhibiting ABA signaling in Arabidopsis. Plant Cell Physiol. 2013, 54, 555–572. [Google Scholar] [CrossRef] [Green Version]
- Umezawa, T.; Sugiyama, N.; Mizoguchi, M.; Hayashi, S.; Myouga, F.; Yamaguchi-Shinozaki, K.; Ishihama, Y.; Hirayama, T.; Shinozaki, K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 17588–17593. [Google Scholar] [CrossRef] [Green Version]
- Kucera, B.; Cohn, M.A.; Leubner-Metzger, G. Plant hormone interactions during seed dormancy release and germination. Seed Sci. Res. 2005, 15, 281–307. [Google Scholar] [CrossRef]
- Finkelstein, R.; Reeves, W.; Ariizumi, T.; Steber, C. Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 2008, 59, 387–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gómez-Cadenas, A.; Zentella, R.; Walker-Simmons, M.K.; Ho, T.-H.D. Gibberellin/abscisic acid antagonism in barley aleurone cells: Site of action of the protein kinase PKABA1 in relation to gibberellin signaling molecules. Plant Cell 2001, 13, 667–679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Footitt, S.; Slocombe, S.P.; Larner, V.; Kurup, S.; Wu, Y.; Larson, T.; Graham, I.; Baker, A.; Holdsworth, M. Control of germination and lipid mobilization by COMATOSE, the Arabidopsis homologue of human ALDP. EMBO J. 2002, 21, 2912–2922. [Google Scholar] [CrossRef] [Green Version]
- Cao, D.; Hussain, A.; Cheng, H.; Peng, J. Loss of function of four DELLA genes leads to light-and gibberellin-independent seed germination in Arabidopsis. Planta 2005, 223, 105–113. [Google Scholar] [CrossRef]
- Tyler, L.; Thomas, S.G.; Hu, J.; Dill, A.; Alonso, J.M.; Ecker, J.R.; Sun, T.-P. DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 2004, 135, 1008–1019. [Google Scholar] [CrossRef] [Green Version]
- Graeber, K.; Linkies, A.; Steinbrecher, T.; Mummenhoff, K.; Tarkowská, D.; Turečková, V.; Ignatz, M.; Sperber, K.; Voegele, A.; de Jong, H. DELAY OF GERMINATION1 mediates a conserved coat-dormancy mechanism for the temperature-and gibberellin-dependent control of seed germination. Proc. Natl. Acad. Sci. USA 2014, 111, 3571–3580. [Google Scholar] [CrossRef] [Green Version]
- Cantoro, R.; Crocco, C.D.; Benech-Arnold, R.L.; Rodríguez, M.V. In vitro binding of Sorghum bicolor transcription factors ABI4 and ABI5 to a conserved region of a GA 2-OXIDASE promoter: Possible role of this interaction in the expression of seed dormancy. J. Exp. Bot. 2013, 64, 5721–5735. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Kim, Y.-S.; Kim, S.-G.; Jung, J.-H.; Woo, J.-C.; Park, C.-M. Integration of auxin and salt signals by the NAC transcription factor NTM2 during seed germination in Arabidopsis. Plant Physiol. 2011, 156, 537–549. [Google Scholar] [CrossRef] [Green Version]
- Ramaih, S.; Guedira, M.; Paulsen, G.M. Relationship of indoleacetic acid and tryptophan to dormancy and preharvest sprouting of wheat. Funct. Plant Biol. 2003, 30, 939–945. [Google Scholar] [CrossRef]
- Liu, A.; Gao, F.; Kanno, Y.; Jordan, M.C.; Kamiya, Y.; Seo, M.; Ayele, B.T. Regulation of wheat seed dormancy by after-ripening is mediated by specific transcriptional switches that induce changes in seed hormone metabolism and signaling. PLoS ONE 2013, 8, e56570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Zhang, H.; Zhao, Y.; Feng, Z.; Li, Q.; Yang, H.-Q.; Luan, S.; Li, J.; He, Z.-H. Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 15485–15490. [Google Scholar] [CrossRef] [Green Version]
- Belin, C.; Megies, C.; Hauserová, E.; Lopez-Molina, L. Abscisic acid represses growth of the Arabidopsis embryonic axis after germination by enhancing auxin signaling. Plant Cell 2009, 21, 2253–2268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arc, E.; Sechet, J.; Corbineau, F.; Rajjou, L.; Marion-Poll, A. ABA crosstalk with ethylene and nitric oxide in seed dormancy and germination. Front. Plant Sci. 2013, 4, 1–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corbineau, F.; Xia, Q.; Bailly, C.; El-Maarouf-Bouteau, H. Ethylene, a key factor in the regulation of seed dormancy. Front. Plant Sci. 2014, 5, 539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subbiah, V.; Reddy, K.J. Interactions between ethylene, abscisic acid and cytokinin during germination and seedling establishment in Arabidopsis. J. BioSci. (Bangalore) 2010, 35, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Cao, H.; Sun, Y.; Li, X.; Chen, F.; Carles, A.; Li, Y.; Ding, M.; Zhang, C.; Deng, X. Arabidopsis paired amphipathic helix proteins SNL1 and SNL2 redundantly regulate primary seed dormancy via abscisic acid–ethylene antagonism mediated by histone deacetylation. Plant Cell 2013, 25, 149–166. [Google Scholar] [CrossRef] [Green Version]
- Steber, C.M.; McCourt, P. A role for brassinosteroids in germination in Arabidopsis. Plant Physiol. 2001, 125, 763–769. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Yu, D. BRASSINOSTEROID INSENSITIVE2 interacts with ABSCISIC ACID INSENSITIVE5 to mediate the antagonism of brassinosteroids to abscisic acid during seed germination in Arabidopsis. Plant Cell 2014, 26, 4394–4408. [Google Scholar] [CrossRef] [Green Version]
- Chitnis, V.R.; Gao, F.; Yao, Z.; Jordan, M.C.; Park, S.; Ayele, B.T. After-ripening induced transcriptional changes of hormonal genes in wheat seeds: The cases of brassinosteroids, ethylene, cytokinin and salicylic acid. PLoS ONE 2014, 9, 87543. [Google Scholar] [CrossRef] [Green Version]
- Nambara, E.; Okamoto, M.; Tatematsu, K.; Yano, R.; Seo, M.; Kamiya, Y. Abscisic acid and the control of seed dormancy and germination. Seed Sci. Res. 2010, 20, 55–67. [Google Scholar] [CrossRef]
- Jacobsen, J.V.; Barrero, J.M.; Hughes, T.; Julkowska, M.; Taylor, J.M.; Xu, Q.; Gubler, F. Roles for blue light, jasmonate and nitric oxide in the regulation of dormancy and germination in wheat grain (Triticum aestivum L.). Planta 2013, 238, 121–138. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Truong, T.T.; Barrero, J.M.; Jacobsen, J.V.; Hocart, C.H.; Gubler, F. A role for jasmonates in the release of dormancy by cold stratification in wheat. J. Exp. Bot. 2016, 67, 3497–3508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Z.; Zhang, Z.-L.; Hanzlik, S.; Cook, E.; Shen, Q.J. Salicylic acid inhibits gibberellin-induced alpha-amylase expression and seed germination via a pathway involving an abscisic-acid-inducible WRKY gene. Plant Mol. Biol. 2007, 64, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, S.G.; Park, C.M. Salicylic acid promotes seed germination under high salinity by modulating antioxidant activity in Arabidopsis. New Phytol. 2010, 188, 626–637. [Google Scholar] [CrossRef] [PubMed]
- Dewar, J.; Taylor, J.; Berjak, P. Changes in selected plant growth regulators during germination in sorghum. Seed Sci. Res. 1998, 8, 1–8. [Google Scholar] [CrossRef]
- Cook, C.; Whichard, L.P.; Turner, B.; Wall, M.E.; Egley, G.H. Germination of witchweed (Striga lutea Lour.): Isolation and properties of a potent stimulant. Science 1966, 154, 1189–1190. [Google Scholar] [CrossRef]
- López-Ráez, J.A.; Charnikhova, T.; Fernández, I.; Bouwmeester, H.; Pozo, M.J. Arbuscular mycorrhizal symbiosis decreases strigolactone production in tomato. J. Plant Physiol. 2011, 168, 294–297. [Google Scholar] [CrossRef]
- Stanga, J.P.; Smith, S.M.; Briggs, W.R.; Nelson, D.C. SUPPRESSOR OF MORE AXILLARY GROWTH2 1 (SMAX2) controls seed germination and seedling development in Arabidopsis. Plant Physiol. 2013, 163, 318–330. [Google Scholar] [CrossRef] [Green Version]
- Lefebvre, V.; North, H.; Frey, A.; Sotta, B.; Seo, M.; Okamoto, M.; Nambara, E.; Marion-Poll, A. Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 2006, 45, 309–319. [Google Scholar] [CrossRef]
- Martínez-Andújar, C.; Ordiz, M.I.; Huang, Z.; Nonogaki, M.; Beachy, R.N.; Nonogaki, H. Induction of 9-cis-epoxycarotenoid dioxygenase in Arabidopsis thaliana seeds enhances seed dormancy. Proc. Natl. Acad. Sci. USA 2011, 108, 17225–17229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saez, A.; Apostolova, N.; Gonzalez-Guzman, M.; Gonzalez-Garcia, M.P.; Nicolas, C.; Lorenzo, O.; Rodriguez, P.L. Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J. 2004, 37, 354–369. [Google Scholar] [CrossRef] [PubMed]
- Mehrotra, R.; Bhalothia, P.; Bansal, P.; Basantani, M.K.; Bharti, V.; Mehrotra, S. Abscisic acid and abiotic stress tolerance–Different tiers of regulation. J. Plant Physiol. 2014, 171, 486–496. [Google Scholar] [CrossRef] [PubMed]
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 2016, 7, 571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karssen, C.; Hilhorst, H.; Koornneef, M. The benefit of biosynthesis and response mutants to the study of the role of abscisic acid in plants. In Plant Growth Substances 1988; Springer: Berlin, Germany, 1990; pp. 23–31. [Google Scholar]
- Bentsink, L.; Koornneef, M. Seed dormancy and germination. Arab. Book 2008, 6, 119. [Google Scholar] [CrossRef] [Green Version]
- Jones, H.D.; Kurup, S.; Peters, N.C.; Holdsworth, M.J. Identification and analysis of proteins that interact with the Avena fatua homologue of the maize transcription factor VIVIPAROUS1. Plant J. 2000, 21, 133–142. [Google Scholar] [CrossRef] [Green Version]
- Nambara, E.; Naito, S.; McCourt, P. A mutant of Arabidopsis which is defective in seed development and storage protein accumulation is a new abi3 allele. Plant J. 1992, 2, 435–441. [Google Scholar] [CrossRef]
- Ng, D.W.; Chandrasekharan, M.B.; Hall, T.C. The 5′ UTR negatively regulates quantitative and spatial expression from the ABI3 promoter. Plant Mol. Biol. 2004, 54, 25–38. [Google Scholar] [CrossRef]
- Beaudoin, N.; Serizet, C.; Gosti, F.; Giraudat, J. Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 2000, 12, 1103–1115. [Google Scholar] [CrossRef] [Green Version]
- Chiwocha, S.D.; Cutler, A.J.; Abrams, S.R.; Ambrose, S.J.; Yang, J.; Ross, A.R.; Kermode, A.R. The etr1-2 mutation in Arabidopsis thaliana affects the abscisic acid, auxin, cytokinin and gibberellin metabolic pathways during maintenance of seed dormancy, moist-chilling and germination. Plant J. 2005, 42, 35–48. [Google Scholar] [CrossRef] [Green Version]
- Cadman, C.S.; Toorop, P.E.; Hilhorst, H.W.; Finch-Savage, W.E. Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J. 2006, 46, 805–822. [Google Scholar] [CrossRef] [PubMed]
- Millar, A.A.; Jacobsen, J.V.; Ross, J.J.; Helliwell, C.A.; Poole, A.T.; Scofield, G.; Reid, J.B.; Gubler, F. Seed dormancy and ABA metabolism in Arabidopsis and barley: The role of ABA 8′-hydroxylase. Plant J. 2006, 45, 942–954. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Gong, Z.; Rock, C.D.; Subramanian, S.; Guo, Y.; Xu, W.; Galbraith, D.; Zhu, J.-K. Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev. Cell 2001, 1, 771–781. [Google Scholar] [CrossRef] [Green Version]
- Parcy, F.; Valon, C.; Kohara, A.; Miséra, S.; Giraudat, J. The ABSCISIC ACID-INSENSITIVE3, FUSCA3, and LEAFY COTYLEDON1 loci act in concert to control multiple aspects of Arabidopsis seed development. Plant Cell 1997, 9, 1265–1277. [Google Scholar]
- Parcy, F.; Valon, C.; Raynal, M.; Gaubier-Comella, P.; Delseny, M.; Giraudat, J. Regulation of gene expression programs during Arabidopsis seed development: Roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 1994, 6, 1567–1582. [Google Scholar]
- Ding, Z.J.; Yan, J.Y.; Li, G.X.; Wu, Z.C.; Zhang, S.Q.; Zheng, S.J. WRKY41 controls Arabidopsis seed dormancy via direct regulation of ABI3 transcript levels not downstream of ABA. Plant J. 2014, 79, 810–823. [Google Scholar] [CrossRef]
- Barrero, J.M.; Millar, A.A.; Griffiths, J.; Czechowski, T.; Scheible, W.R.; Udvardi, M.; Reid, J.B.; Ross, J.J.; Jacobsen, J.V.; Gubler, F. Gene expression profiling identifies two regulatory genes controlling dormancy and ABA sensitivity in Arabidopsis seeds. Plant J. 2010, 61, 611–622. [Google Scholar] [CrossRef]
- Vaistij, F.E.; Gan, Y.; Penfield, S.; Gilday, A.D.; Dave, A.; He, Z.; Josse, E.-M.; Choi, G.; Halliday, K.J.; Graham, I.A. Differential control of seed primary dormancy in Arabidopsis ecotypes by the transcription factor SPATULA. Proc. Natl. Acad. Sci. USA 2013, 110, 10866–10871. [Google Scholar] [CrossRef] [Green Version]
- Yamagishi, K.; Tatematsu, K.; Yano, R.; Preston, J.; Kitamura, S.; Takahashi, H.; McCourt, P.; Kamiya, Y.; Nambara, E. CHOTTO1, a double AP2 domain protein of Arabidopsis thaliana, regulates germination and seedling growth under excess supply of glucose and nitrate. Plant Cell Physiol. 2008, 50, 330–340. [Google Scholar] [CrossRef] [Green Version]
- Yano, R.; Kanno, Y.; Jikumaru, Y.; Nakabayashi, K.; Kamiya, Y.; Nambara, E. CHOTTO1, a putative double APETALA2 repeat transcription factor, is involved in abscisic acid-mediated repression of gibberellin biosynthesis during seed germination in Arabidopsis. Plant Physiol. 2009, 151, 641–654. [Google Scholar] [CrossRef] [Green Version]
- Skubacz, A.; Daszkowska-Golec, A.; Szarejko, I. The role and regulation of ABI5 (ABA-Insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front. Plant Sci. 2016, 7, 1884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griffiths, J.; Kohji, M.; Rieu, I.; Zentella, R.; Zhang, Z.-L.; Powers, S.J.; Gong, F.; Phillips, A.L.; Hedden, P.; Sun, T.-P.; et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 2007, 18, 3399–3414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueguchi-Tanaka, M.; Nakajima, M.; Katoh, E.; Ohmiya, H.; Asano, K.; Saji, S. Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 2007, 19, 2140–2155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dill, A.; Thomas, S.G.; Hu, J.; Steber, C.M.; Sun, T.-P. The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 2004, 16, 1392–1405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, X.; Richards, D.E.; Fleck, B.; Xie, D.; Burton, N.; Harberd, N.P. The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 2004, 16, 1406–1418. [Google Scholar] [CrossRef] [Green Version]
- McGinnis, K.M.; Thomas, S.G.; Soule, J.D.; Strader, L.C.; Zale, J.M.; Sun, T.-P.; Steber, C.M. The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 2003, 15, 1120–1130. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, A.; Itoh, H.; Gomi, K.; Ueguchi-Tanaka, M.; Ishiyama, K.; Kobayashi, M.; Jeong, D.-H.; An, G.; Kitano, H.; Ashikari, M. Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 2003, 299, 1896–1898. [Google Scholar] [CrossRef] [Green Version]
- Magome, H.; Yamaguchi, S.; Hanada, A.; Kamiya, Y.; Oda, K. The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. Plant J. 2008, 56, 613–626. [Google Scholar] [CrossRef]
- Yaish, M.W.; El-kereamy, A.; Zhu, T.; Beatty, P.H.; Good, A.G.; Bi, Y.-M. The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet. 2010, 6, 1001098. [Google Scholar] [CrossRef] [Green Version]
- Narsai, R.; Law, S.R.; Carrie, C.; Xu, L.; Whelan, J.; Law, S.R.; Carrie, C.; Xu, L.; Whelan, J. In-depth temporal transcriptome profiling reveals a crucial developmental switch with roles for RNA processing and organelle metabolism that are essential for germination in Arabidopsis. Plant Physiol. 2011, 157, 1342–1362. [Google Scholar] [CrossRef] [Green Version]
- Wilson, R.L.; Kim, H.; Bakshi, A.; Binder, B.M. The ethylene receptors ETHYLENE RESPONSE1 and ETHYLENE RESPONSE2 have contrasting roles in seed germination of Arabidopsis during salt stress. Plant Physiol. 2014, 165, 1353–1366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xi, W.; Liu, C.; Hou, X.; Yu, H. MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 2010, 22, 1733–1748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xi, W.; Yu, H. MOTHER OF FT AND TFL1 regulates seed germination and fertility relevant to the brassinosteroid signaling pathway. Plant Signal. Behav. 2010, 5, 1315–1317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dave, A.; Vaistij, F.E.; Gilday, A.D.; Penfield, S.D.; Graham, I.A. Regulation of Arabidopsis thaliana seed dormancy and germination by 12-oxo-phytodienoic acid. J. Exp. Bot. 2016, 67, 2277–2284. [Google Scholar] [CrossRef] [Green Version]
- Finch-Savage, W.E.; Cadman, C.S.; Toorop, P.E.; Lynn, J.R.; Hilhorst, W.H. Seed dormancy release in Arabidopsis Cvi by dry after-ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. Plant J. 2007, 51, 60–78. [Google Scholar] [CrossRef]
- Simpson, G.M. Seed Dormancy in Grasses; Cambridge University Press: Cambridge, UK, 2007; pp. 197–206. [Google Scholar]
- Footitt, S.; Douterelo-Soler, I.; Clay, H.; Finch-Savage, W.E. Dormancy cycling in Arabidopsis seeds is controlled by seasonally distinct hormone-signaling pathways. Proc. Natl. Acad. Sci. USA 2011, 108, 20236–20241. [Google Scholar] [CrossRef] [Green Version]
- Buriro, M.; Oad, F.C.; Keerio, M.I.; Tunio, S.; Gandahi, A.W.; Hassan, S.W.U.; Oad, S.M. Wheat seed germination under the influence of temperature regimes. Sarhad J. Agric. 2011, 27, 539–543. [Google Scholar]
- Nyachiro, J.; Clarke, F.; DePauw, R.; Knox, R.; Armstrong, K. Temperature effects on seed germination and expression of seed dormancy in wheat. Euphytica 2002, 126, 123–127. [Google Scholar] [CrossRef]
- Reddy, L.; Metzger, R.; Ching, T. Effect of Temperature on Seed Dormancy of Wheat. Crop Sci. 1985, 25, 455–458. [Google Scholar] [CrossRef]
- Lim, S.; Park, J.; Lee, N.; Jeong, J.; Toh, S.; Watanabe, A.; Kim, J.; Kang, H.; Kim, D.H.; Kawakami, N. ABA-INSENSITIVE3, ABA-INSENSITIVE5, and DELLAs interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell 2013, 25, 4863–4878. [Google Scholar] [CrossRef] [Green Version]
- Contreras, S.; Bennett, M.A.; Metzger, J.D.; Tay, D. Maternal light environment during seed development affects lettuce seed weight, germinability, and storability. HortScience 2008, 43, 845–852. [Google Scholar] [CrossRef] [Green Version]
- Barrero, J.M.; Downie, A.B.; Xu, Q.; Gubler, F. A role for barley CRYPTOCHROME1 in light regulation of grain dormancy and germination. Plant Cell 2014, 26, 1094–1104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gubler, F.; Hughes, T.; Waterhouse, P.; Jacobsen, J. Regulation of dormancy in barley by blue light and after-ripening: Effects on abscisic acid and gibberellin metabolism. Plant Physiol. 2008, 147, 886–896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ochuodho, J.O.; Modi, A.T. Light-induced transient dormancy in Cleome gynandra L. seeds. Afr. J. Agric. Res. 2007, 2, 587–591. [Google Scholar]
- Derkx, M.; Karssen, C. Effects of light and temperature on seed dormancy and gibberellin-stimulated germination in Arabidopsis thaliana: Studies with gibberellin-deficient and-insensitive mutants. Physiol. Plant 1993, 89, 360–368. [Google Scholar] [CrossRef]
- Debeaujon, I.; Lepiniec, L.; Pourcel, L.; Routaboul, J.-M. Seed coat development and dormancy. Annu. Plant Rev. 2007, 27, 25–49. [Google Scholar]
- Groos, C.; Gay, G.; Perretant, M.-R.; Gervais, L.; Bernard, M.; Dedryver, F.; Charmet, G. Study of the relationship between pre-harvest sprouting and grain color by quantitative trait loci analysis in a white× red grain bread-wheat cross. Theor. Appl. Genet. 2002, 104, 39–47. [Google Scholar] [CrossRef]
- Sweeney, M.T.; Thomson, M.J.; Pfeil, B.E.; McCouch, S. Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 2006, 18, 283–294. [Google Scholar] [CrossRef] [Green Version]
- Adebo, H.O.; Ahoton, L.E.; Quenum, F.; Ezin, V. Agro-morphological characterization of Corchorus olitorius cultivars of Benin. Annu. Res. Rev. Biol. 2015, 7, 229–240. [Google Scholar] [CrossRef]
- Stetter, M.G.; Vidal-Villarejo, M.; Schmid, K.J. Convergent seed color adaptation during repeated domestication of an ancient new world grain. BioRxiv 2019. BioRxiv:547943. [Google Scholar]
- Akubugwo, I.; Obasi, A.; Ginika, S. Nutritional potential of the leaves and seeds of black nightshade-Solanum nigrum L. Var virginicum from Afikpo-Nigeria. Pak. J. Nutr. 2007, 6, 323–326. [Google Scholar] [CrossRef]
- Paśko, P.; Sajewicz, M.; Gorinstein, S.; Zachwieja, Z. Analysis of selected phenolic acids and flavonoids in Amaranthus cruentus and Chenopodium quinoa seeds and sprouts by HPLC. Acta Chromatogr. 2008, 20, 661–672. [Google Scholar] [CrossRef] [Green Version]
- Yang, R.-Y.; Lin, S.; Kuo, G. Content and distribution of flavonoids among 91 edible plant species. Asia Pac. J. Clin. Nutr. 2008, 17, 275–279. [Google Scholar] [PubMed]
- Foley, M.E.; Fennimore, S.A. Genetic basis for seed dormancy. Seed Sci. Res. 1998, 8, 173–182. [Google Scholar] [CrossRef]
- Nguyen, T.-P.; Keizer, P.; van Eeuwijk, F.; Smeekens, S.; Bentsink, L. Natural variation for seed longevity and seed dormancy are negatively correlated in Arabidopsis thaliana. Plant Physiol. 2012, 160, 2083–2092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Schmitt, J.; Dorn, L.; Griffith, C.; Effgen, S.; Takao, S.; Koornneef, M.; Donohue, K.J.M.E. The earliest stages of adaptation in an experimental plant population: Strong selection on QTLS for seed dormancy. Mol. Ecol. 2010, 19, 1335–1351. [Google Scholar] [CrossRef]
- Clerkx, E.J.; El-Lithy, M.E.; Vierling, E.; Ruys, G.J.; Blankestijn-De Vries, H.; Groot, S.P.; Vreugdenhil, D.; Koornneef, M. Analysis of natural allelic variation of Arabidopsis seed germination and seed longevity traits between the accessions Landsberg erecta and Shakdara, using a new recombinant inbred line population. Plant Physiol. 2004, 135, 432–443. [Google Scholar] [CrossRef] [Green Version]
- Keurentjes, J.J.; Willems, G.; van Eeuwijk, F.; Nordborg, M.; Koornneef, M. A comparison of population types used for QTL mapping in Arabidopsis thaliana. Plant Genet. Resour. 2011, 9, 185–188. [Google Scholar] [CrossRef] [Green Version]
- Alonso-Blanco, C.; Peeters, A.J.; Koornneef, M.; Lister, C.; Dean, C.; van den Bosch, N.; Pot, J.; Kuiper, M.T. Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J. 1998, 14, 259–271. [Google Scholar] [CrossRef]
- Qin, X.; Zeevaart, J.A. Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiol. 2002, 128, 544–551. [Google Scholar] [CrossRef]
- Thompson, A.J.; Jackson, A.C.; Symonds, R.C.; Mulholland, B.J.; Dadswell, A.R.; Blake, P.S.; Burbidge, A.; Taylor, I.B. Ectopic expression of a tomato 9-cis-epoxycarotenoid dioxygenase gene causes over-production of abscisic acid. Plant J. 2000, 23, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y. Molecular Plant Breeding; CAB International: Oxfordshire, UK, 2010; pp. 195–247. [Google Scholar]
- Kearsey, M.; Farquhar, A. QTL analysis in plants; where are we now? Heredity 1998, 80, 137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bove, J.; Jullien, M.; Grappin, P. Functional genomics in the study of seed germination. Genome Biol. 2001, 3, 1002.1. [Google Scholar] [CrossRef] [PubMed]
- Holdsworth, M.J.; Finch-Savage, W.E.; Grappin, P.; Job, D. Post-genomics dissection of seed dormancy and germination. Trends Plant Sci. 2008, 13, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Chibani, K.; Ali-Rachedi, S.; Job, C.; Job, D.; Jullien, M.; Grappin, P. Proteomic analysis of seed dormancy in Arabidopsis. Plant Physiol. 2006, 142, 1493–1510. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Wu, X.; Tsang, E.; Cutler, A.J. Transcriptional profiling of imbibed Brassica napus seed. Genomics 2005, 86, 718–730. [Google Scholar] [CrossRef]
- Fait, A.; Angelovici, R.; Less, H.; Ohad, I.; Urbanczyk-Wochniak, E.; Fernie, A.R.; Galili, G. Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant Physiol. 2006, 142, 839–854. [Google Scholar] [CrossRef] [Green Version]
- Gao, F.; Ayele, B.T. Functional genomics of seed dormancy in wheat: Advances and prospects. Front. Plant Sci. 2014, 5, 458. [Google Scholar] [CrossRef] [Green Version]
- Harada, J.J.; Pelletier, J. Genome-wide analyses of gene activity during seed development. Seed Sci. Res. 2012, 22, S15–S22. [Google Scholar] [CrossRef]
- Schranz, M.E.; Song, B.-H.; Windsor, A.J.; Mitchell-Olds, T. Comparative genomics in the Brassicaceae: A family-wide perspective. Curr. Opin. Plant Biol. 2007, 10, 168–175. [Google Scholar] [CrossRef]
- Ayenan, M.A.T.; Sodedji, K.A.F.; Nwankwo, C.I.; Olodo, K.F.; Alladassi, M.E.B. Harnessing genetic resources and progress in plant genomics for fonio (Digitaria spp.) improvement. Genet. Resour. Crop Evol. 2018, 65, 373–386. [Google Scholar] [CrossRef]
- Ajaiyeoba, E. Phytochemical and antimicrobial studies of Gynandropsis gynandra and Buchholzia coriaceae extracts. Afr. J. Biomed. Res. 2000, 3, 161–165. [Google Scholar]
- Barker, M.S.; Vogel, H.; Schranz, M.E. Paleopolyploidy in the Brassicales: Analyses of the Cleome transcriptome elucidate the history of genome duplications in Arabidopsis and other Brassicales. Genome Biol. Evol. 2009, 1, 391–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marshall, D.M.; Muhaidat, R.; Brown, N.J.; Liu, Z.; Stanley, S.; Griffiths, H.; Sage, R.F.; Hibberd, J.M. Cleome, a genus closely related to Arabidopsis, contains species spanning a developmental progression from C3 to C4 photosynthesis. Plant J. 2007, 51, 886–896. [Google Scholar] [CrossRef]
- Van den Bergh, E.; Külahoglu, C.; Bräutigam, A.; Hibberd, J.M.; Weber, A.P.; Zhu, X.-G.; Schranz, M.E. Gene and genome duplications and the origin of C4 photosynthesis: Birth of a trait in the Cleomaceae. Curr. Plant Biol. 2014, 1, 2–9. [Google Scholar] [CrossRef] [Green Version]
- Amtmann, A. Learning from evolution: Thellungiella generates new knowledge on essential and critical components of abiotic stress tolerance in plants. Mol. Plant 2009, 2, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Mobility for Breeders in Africa. Available online: https://mobreed.com (accessed on 15 October 2019).
- Sorrells, M.E.; La Rota, M.; Bermudez-Kandianis, C.E.; Greene, R.A.; Kantety, R.; Munkvold, J.D.; Mahmoud, A.; Ma, X.; Gustafson, P.J.; Qi, L.L. Comparative DNA sequence analysis of wheat and rice genomes. Genome Res. 2003, 13, 1818–1827. [Google Scholar]
- Zhu, H.; Choi, H.-K.; Cook, D.R.; Shoemaker, R.C. Bridging model and crop legumes through comparative genomics. Plant Physiol. 2005, 137, 1189–1196. [Google Scholar] [CrossRef] [Green Version]
- Nadeem, M.A.; Nawaz, M.A.; Shahid, M.Q.; Doğan, Y.; Comertpay, G.; Yıldız, M.; Hatipoğlu, R.; Ahmad, F.; Alsaleh, A.; Labhane, N. DNA molecular markers in plant breeding: Current status and recent advancements in genomic selection and genome editing. Biotechnol. Biotechnol. Equip. 2018, 32, 261–285. [Google Scholar] [CrossRef] [Green Version]
- Van den Bergh, E. Comparative Genomics and Trait Evolution in Cleomaceae, a Model Family for Ancient Polyploidy. Ph.D. Thesis, Wageningen University, Wageningen, The Netherlands, 2017. [Google Scholar]
- Mutwil, M.; Klie, S.; Tohge, T.; Giorgi, F.M.; Wilkins, O.; Campbell, M.M.; Fernie, A.R.; Usadel, B.; Nikoloski, Z.; Persson, S. PlaNet: Combined sequence and expression comparisons across plant networks derived from seven species. Plant Cell 2011, 23, 895–910. [Google Scholar] [CrossRef] [Green Version]
- Das, G.; Rao, G.J.; Varier, M.; Prakash, A.; Prasad, D. Improved Tapaswini having four BB resistance genes pyramided with six genes/QTLs, resistance/tolerance to biotic and abiotic stresses in rice. Sci. Rep. 2018, 8, 2413. [Google Scholar] [CrossRef] [PubMed]
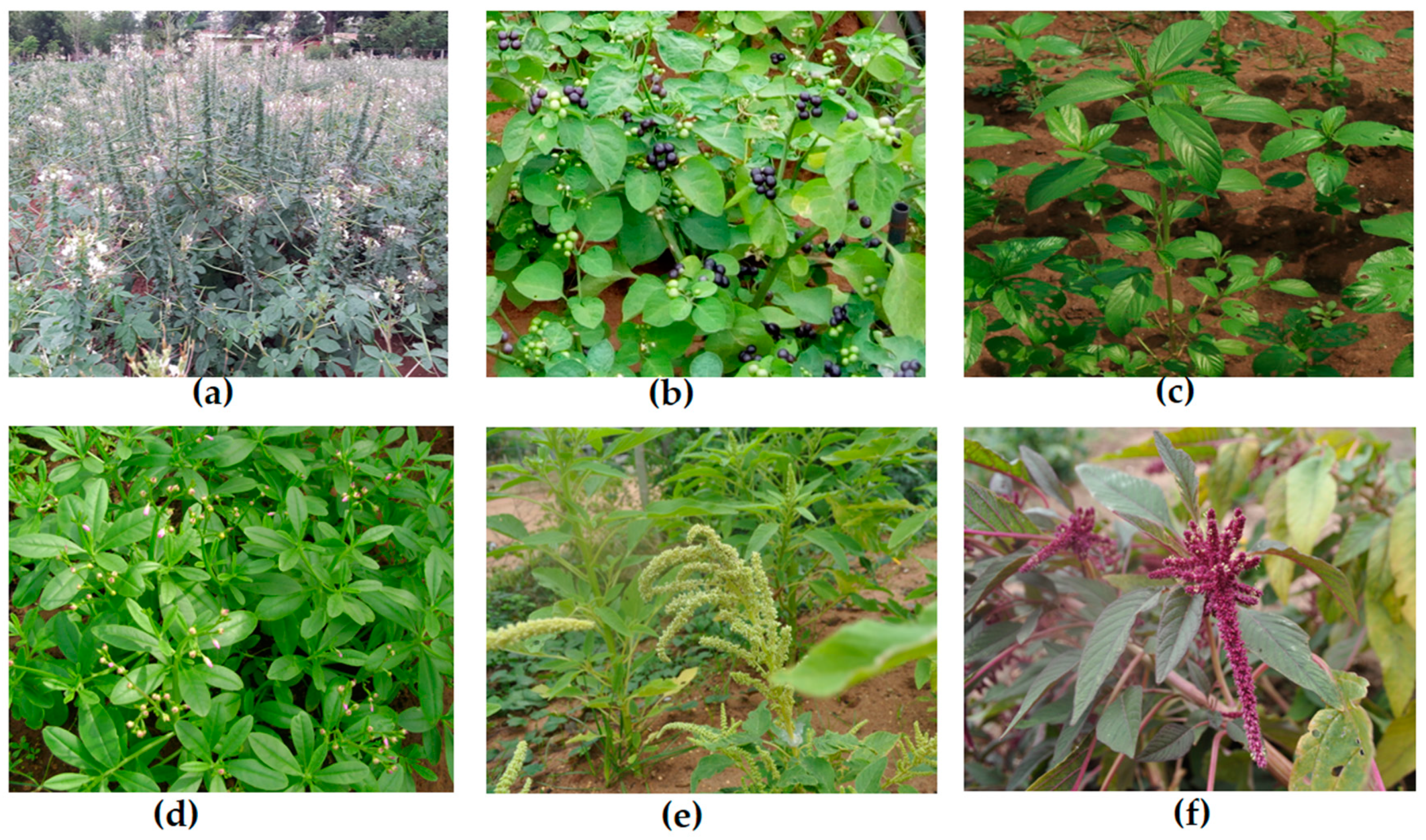
| Common Name | Scientific Name (Family) | Seed Constraints | References |
|---|---|---|---|
| Spider plant | Gynandropsis gynandra (Cleomaceae) | Primary non-deep physiological dormancy Physical dormancy and secondary dormancy Oxygen barrier between embryo and tissue Low germination of freshly harvested seeds Delayed, poor, and absence of germination Inaccessibility of quality seed for seed analysts and gene bank curators Low vigor and reduced number of viable seeds harvested by farmers Physiological dormancy | [31,32] |
| Jute mallow | Corchorus olitorius (Malvaceae) | Loss of viability and poor germination of fresh and old seeds Impermeable seed coat | [8,33] |
| African Nightshade | Solanum nigrum (Solanaceae) | Poor germination of seeds Improper seed extraction Deeper level of primary dormancy | [8,34] |
| Waterleaf | Talinum triangulare (Portulacaceae) | Dormancy due to the nature of the seed testa Undetermined physiological factors | [35] |
| Amaranths species | Amaranthus spp. (Amaranthaceae) | Primary dormancy and secondary dormancy occur among amaranths species | [36] |
| TLVs Species | Strategies for Seed Dormancy-Breaking |
|---|---|
| Gynandropsis gynandra |
|
| Amaranthus species |
|
| Corchorus olitorius |
|
| Talinum triangulare |
|
| Solanum nigrum |
|
| Mutants | Description/Action | References |
|---|---|---|
| nced6/nced9 and nced5 | Promote germination | [75,109,110] |
| aao3, aba1, and aba2 | Reduce dormancy | [73,111] |
| cyp707a | Enhance seed dormancy level | [17,77,112,113] |
| abi1 | Reduce dormancy through chilling and dry storage, reduce ABA sensitivity for germination and no precocious germination | [80,114] |
| abi3 | Leads to seed dormancy even in immature seeds | [115,116,117,118] |
| cts | Leads to the seed dormancy protection even after stratification and after-ripening | [80] |
| yuc1/yuc6 (Auxin) | Reduce seed dormancy | [89] |
| ein2 (Ethylene) | Leads to higher expression of NCED3 | [17,119,120] |
| etr1 | Induces lower activation of CYP707A2 genes | |
| snl1 and snl2 | Reduce seed dormancy together with the increased Ethylene content | [96] |
| hub1(rdo4) | Characterized by a reduced dormancy | [48] |
| tfiis | Reduces seed dormancy | [68] |
| dog1 and rdo4 | Reduce seed longevity phenotype | [55,69,70] |
| rdo1 and rdo2 | Not affected in their response to ABA |
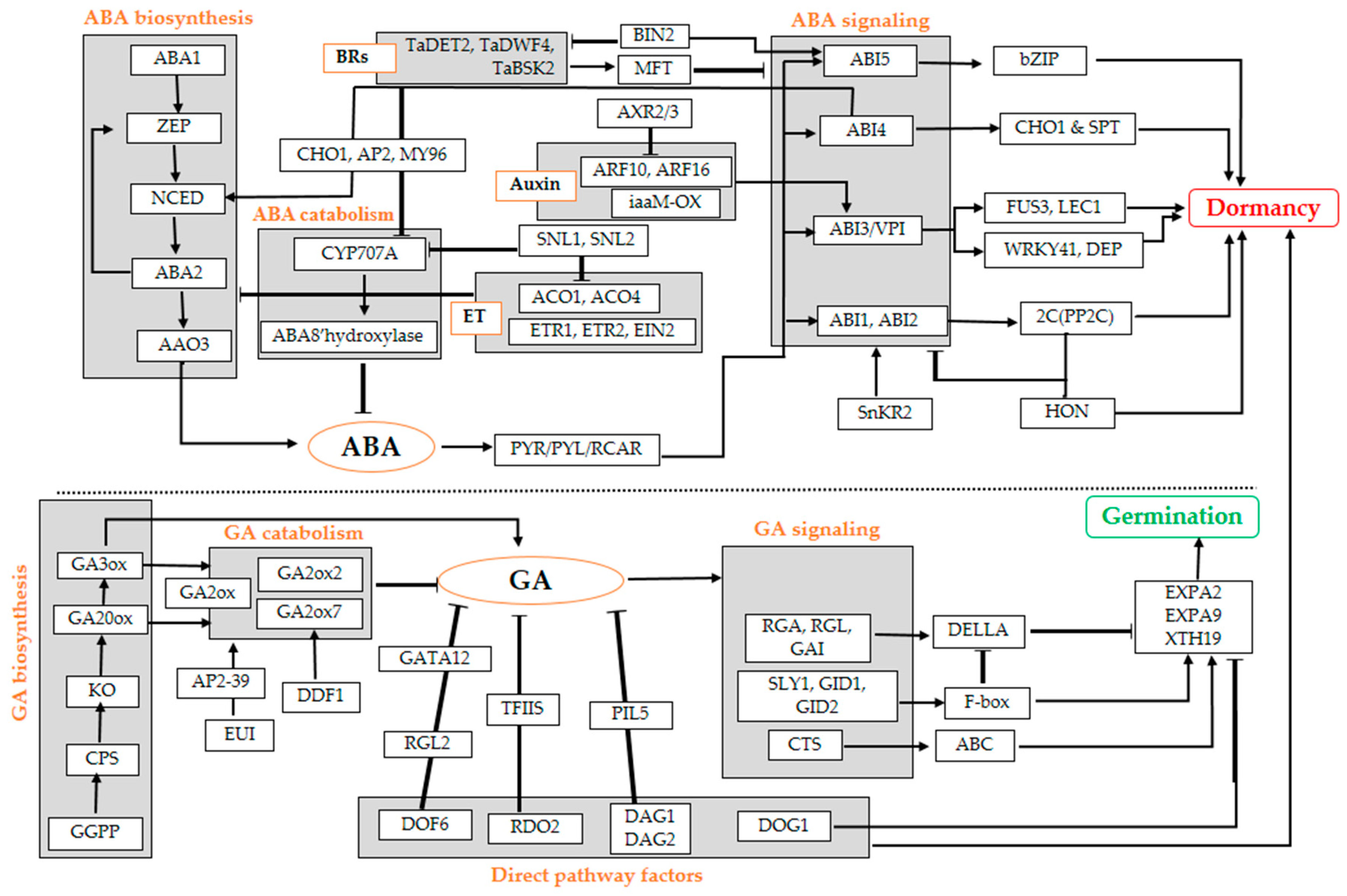
| Process | Genes | Description | Related Species | References |
|---|---|---|---|---|
| ABA biosynthesis | NCED5, NCED6, NCED9 | Induction of seed dormancy | Arabidopsis thaliana | [17,75,109,121,122] |
| NCED1, NCED2 | Induction of seed dormancy | Oryza sativa, Hordeum vulgare | [17] | |
| ABA1, ABA2 | Encode for zeaxanthine poxidase | Arabidopsis thaliana; Zea mays; Nicotiana plumbaginifolia | [80] | |
| AAO3 | Encodes final step of ABA biosynthesis | Arabidopsis thaliana | [80,123] | |
| ABA catabolism | CYP707A1, CYP707A2, CYP707A3 | Encode for ABA8′hydroxylase; loss of dormancy | Arabidopsis. thaliana, Hordeum vulgare | [17] |
| ABA signalling | ABI1, ABI2 | Encode for Serine/threonine phosphatase 2C (PP2C) inducing seed dormancy | Arabidopsis thaliana and monocot | [80,114] |
| ABI3/VP1 | Regulation of chlorophyll, anthocyanin, and storage proteins accumulation with FUS3 and LEC1 | Arabidopsis thaliana and monocot | [17,124,125] | |
| Regulated by WRKY41 and by DEP for primary seed dormancy establishment | Arabidopsis thaliana and monocot | [77,126,127] | ||
| ABI4 | Regulated by transcription factors CHO1 and SPT for dormancy establishment and maintenance through NCED2 and NCED3; Represses CYP707A1 and CYP707A2 | Arabidopsis thaliana and monocot | [77,128,129,130] | |
| ABI5 | Regulated by bZIP transcription factor for positive ABA signalling and repressing seed germination | Arabidopsis thaliana, Sorghum bicolor | [77,131] | |
| GA biosynthesis | GA3ox1, GA20ox3, KO1 | Inducing of hydrolytic enzymes that weaken the seed coat, inducing of mobilization of seed storage reserves, and stimulating of expansion of the embryo | Arabidopsis thaliana and monocot | [17,81,82] |
| CPS | Catalyzed geranylgeranyl pyrophosphate (GGDP) cyclization reaction in the provascular tissue | Arabidopsis thaliana | [80] | |
| GA signaling | CTS | Encodes a peroxisomal protein of the ATP-binding cassette (ABC) transporter class | Arabidopsis thaliana | [17,83] |
| RGA, RGL1, RGL2, GAI | Encode DELLA proteins as a repressor of GA signalling | Arabidopsis thaliana | [17,79,80,84] | |
| SLY1 | GA relieves DELLA repression of seed germination by F-box protein | Arabidopsis thaliana | [17,85] | |
| GID1 | Induce release of seed dormancy by promoting interaction of DELLA with the F-box protein | Arabidopsis thaliana, Oryza. sativa | [81,132,133] | |
| GID2 | Encodes for F-box subunits of an SCF E3 ubiquitin ligase that ubiquitinates DELLA proteins | Arabidopsis thaliana, Oryza sativa | [81,134,135,136,137] | |
| GA catabolism (GA2ox2) | DOG1 | Inhibition of genes encoding cell wall remodelling enzymes: EXPA2, EXPA9, XTH19 by regulates the expression of GA biosynthesis genes | Arabidopsis thaliana and monocot | [86] |
| DDF1 | Promotes transcription of the GA inactivation gene GA2ox7 | Arabidopsis thaliana | [77,138] | |
| EUI | Promoted by AP2 domain-containing transcription factor OsAP2-39 for GA inactivation | Oryza sativa | [77,139] | |
| Auxin | iaaM-OX | Strong seed dormancy | Triticum aestivum | [89] |
| ARF10 and ARF16 | Activates ABI3 by perceiving high level of IAA for dormancy maintenance | Arabidopsis thaliana | [17,77,91] | |
| AXR2/3 | Repress ARF10 and ARF16 | Arabidopsis thaliana | ||
| Ethylene | ACO1, ACO4 | Ethylene biosynthesis genes | Arabidopsis thaliana | [17,140] |
| ETR1, ETR2 EIN2 | Contrasting roles for ABA biosynthesis during seed germination under salt-stress conditions | Arabidopsis thaliana | [141] | |
| SNL1 and SNL2 | Reduce acetylation level of histone 3 lysine 9/18 and histone 3 lysine 14 repressing ABA accumulation at high level of ET | Arabidopsis thaliana | [96] | |
| SNL1 and SNL2 | Promote seed dormancy through simultaneous modulation of ACO1, ACO4 and CYP707A1, CYP707A2 | Arabidopsis thaliana | [96] | |
| Brassinosteroid biosynthesis | TaDE-etiolated 2 (TaDET2) and TaDWARF 4 | Ensure BR production in plant | Triticum aestivum | [17,99] |
| Brassinosteroid signaling | TaBR signalling kinase 2 (TaBSK2) | Promote BR signalling | Triticum aestivum | [17,99] |
| MFT | Forming a negative feedback loop to modulate ABA signalling | Arabidopsis thaliana | [142,143] | |
| BIN2 | Key repressor of the BR signalling | Arabidopsis thaliana | [17,98] | |
| Jasmonic acid | OPDA | Promote effect of ABA1, ABI5, and RGL2 and its regulatory action on MFT gene for seed dormancy maintenance | Arabidopsis thaliana | [17,77,100,144]. |
| Other genes | DOG1 | Shows strong dormancy | Arabidopsis thaliana, Hordeum vulgare, Triticum aestivum | [29,50,51,52,53,55] |
| DAG1 and DAG2 | Inhibiting germination by mediating PIL5 activity as well as directly affecting gibberellin biosynthesis | Arabidopsis thaliana | [63,64] | |
| DOF6 | Negatively regulates germination by affecting abscisic acid signalling in seeds | Arabidopsis thaliana | [60] | |
| RDO2 | Encodes TFIIS for strong dormancy | Arabidopsis thaliana | [67] | |
| GATA12 | Encodes a GATA-type zinc finger transcription factor for novel RGL2–DOF6 complex enforcing primary seed dormancy via GA signalling repression | Arabidopsis thaliana | [59,61,62] | |
| NR (Nitrate reductase) | Promotes dormancy release | Arabidopsis thaliana | [81,145] |
| Environment Factors | Situations | Role in Seed Dormancy Regulation | Description | Species |
|---|---|---|---|---|
| After-ripening | Seed dry storage period at room temperature | Reduced dormancy | Positive relationship with CYP707A2 Induces GA insensitive dwarf1 GID1b | Arabidopsis thaliana |
| Promotes expression of JA biosynthesis genes: Allene oxide synthase (AOS), 3-ketoacyl coenzyme A (KAT3) and Lipoxygenase 5 (LOX5); Induces GA20ox1 and GA3ox2 | Triticum aestivum | |||
| Increases the expression of ABA8′OH-1 | Hordeum vulgare, Brachypodium distachyon | |||
| Temperature | Low temperature | Reduced dormancy | Promotes GA3ox1 expression; Represses GA2ox2 gene | Arabidopsis thaliana |
| Higher level of dormancy during seed development | Activates MFT gene | Triticum aestivum | ||
| High temperature | Increased dormancy during seed imbibition | Represses GA20ox1, GA20ox2, GA20ox3, GA3ox1, and GA3ox2 genes; Promotes the expression of ABA biosynthesis genes | Arabidopsis thaliana | |
| Light | Red (R) light | Reduced dormancy | Inhibits the expression of NCED6 | Arabidopsis thaliana |
| Fared (FR) light | Increase dormancy | Inhibits the expression of CYP707A2 | ||
| Blue light | Increased dormancy | Promotes NCED1, NCED2, GA2ox3 and GA2ox5 genes; Represses GA3ox2 | Hordeum vulgare |
| Species | Part | Flavonoid (mg/100 g) |
|---|---|---|
| Gynandropsis gynandra | Shoot | 64.3 |
| Corchorus olitorius | Shoot | 63.9 |
| Solanum nigrum | Seed | 1.01 |
| Amaranthus cruentus | Seed | 667 |
| Chenopodium quinoa Willd. | Seed | 2238 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sohindji, F.S.; Sogbohossou, D.E.O.; Zohoungbogbo, H.P.F.; Houdegbe, C.A.; Achigan-Dako, E.G. Understanding Molecular Mechanisms of Seed Dormancy for Improved Germination in Traditional Leafy Vegetables: An Overview. Agronomy 2020, 10, 57. https://doi.org/10.3390/agronomy10010057
Sohindji FS, Sogbohossou DEO, Zohoungbogbo HPF, Houdegbe CA, Achigan-Dako EG. Understanding Molecular Mechanisms of Seed Dormancy for Improved Germination in Traditional Leafy Vegetables: An Overview. Agronomy. 2020; 10(1):57. https://doi.org/10.3390/agronomy10010057
Chicago/Turabian StyleSohindji, Fernand S., Dêêdi E. O. Sogbohossou, Herbaud P. F. Zohoungbogbo, Carlos A. Houdegbe, and Enoch G. Achigan-Dako. 2020. "Understanding Molecular Mechanisms of Seed Dormancy for Improved Germination in Traditional Leafy Vegetables: An Overview" Agronomy 10, no. 1: 57. https://doi.org/10.3390/agronomy10010057
APA StyleSohindji, F. S., Sogbohossou, D. E. O., Zohoungbogbo, H. P. F., Houdegbe, C. A., & Achigan-Dako, E. G. (2020). Understanding Molecular Mechanisms of Seed Dormancy for Improved Germination in Traditional Leafy Vegetables: An Overview. Agronomy, 10(1), 57. https://doi.org/10.3390/agronomy10010057





