Structural Analysis of Soluble Elastin in Dry and Hydrated States Using 13C Solid-State NMR
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Insoluble Elastin
2.2. Preparation of Soluble Elastin
2.3. Molecular Weight Analysis and Amino Acid Analyses
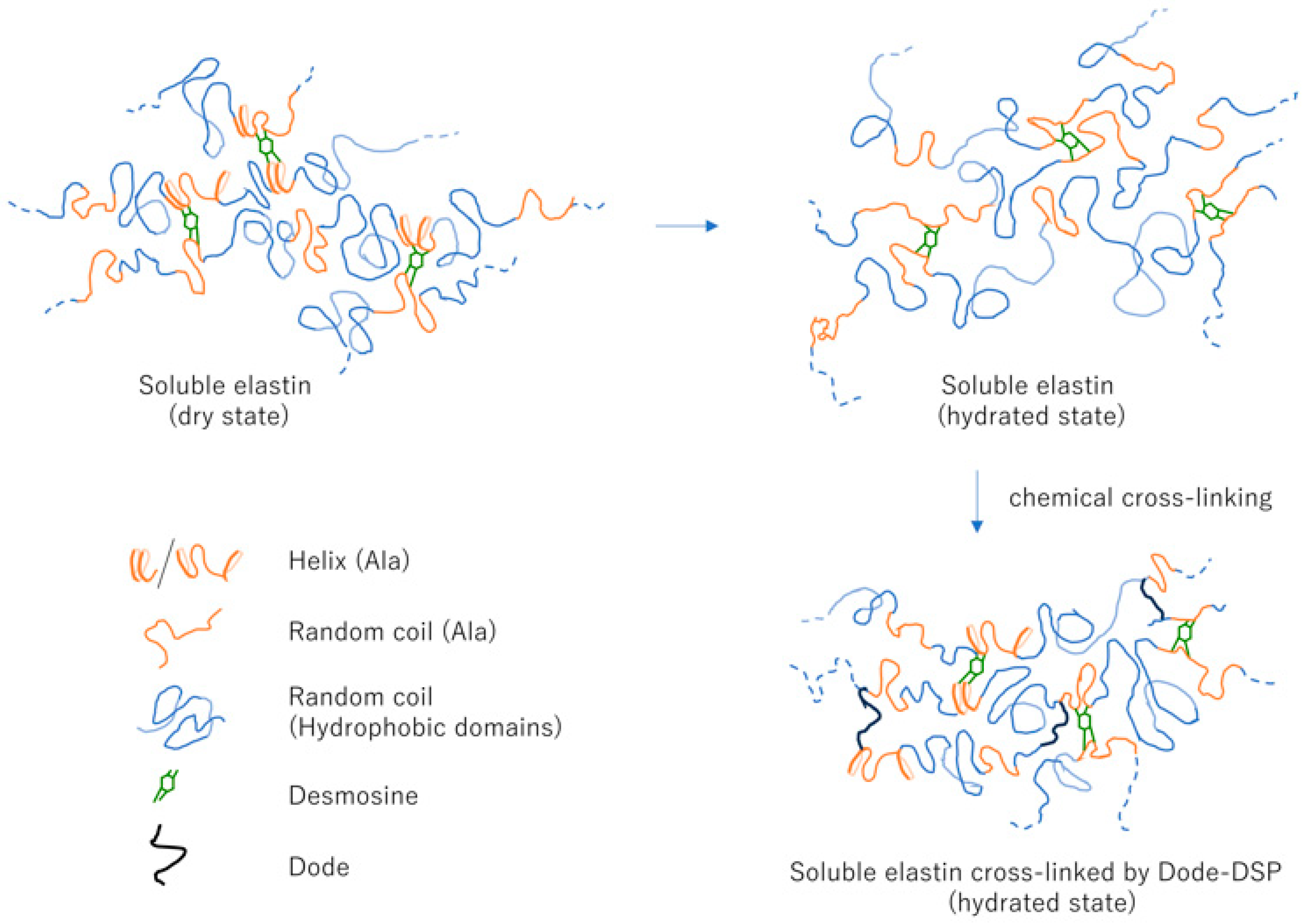
2.4. Cross-Linking of Soluble Elastin Using Dode-DSP
2.5. Preparation of (Val-Pro-Gly-Val-Gly)6
2.6. NMR Measurements
3. Results and Discussion
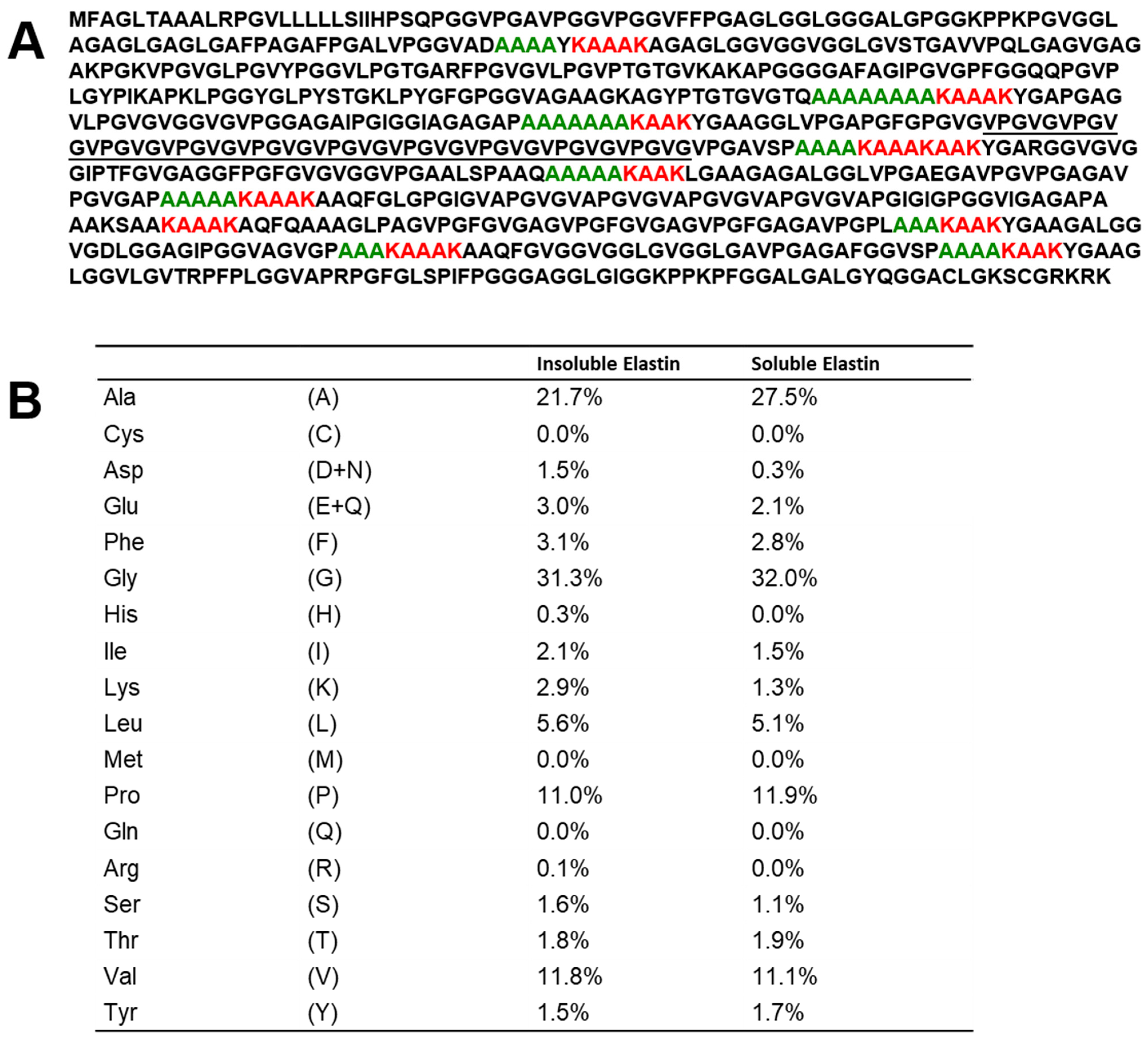
3.1. Amino Acid Sequence of Tropoelastin and Amino Acid Composition of Soluble Elastin
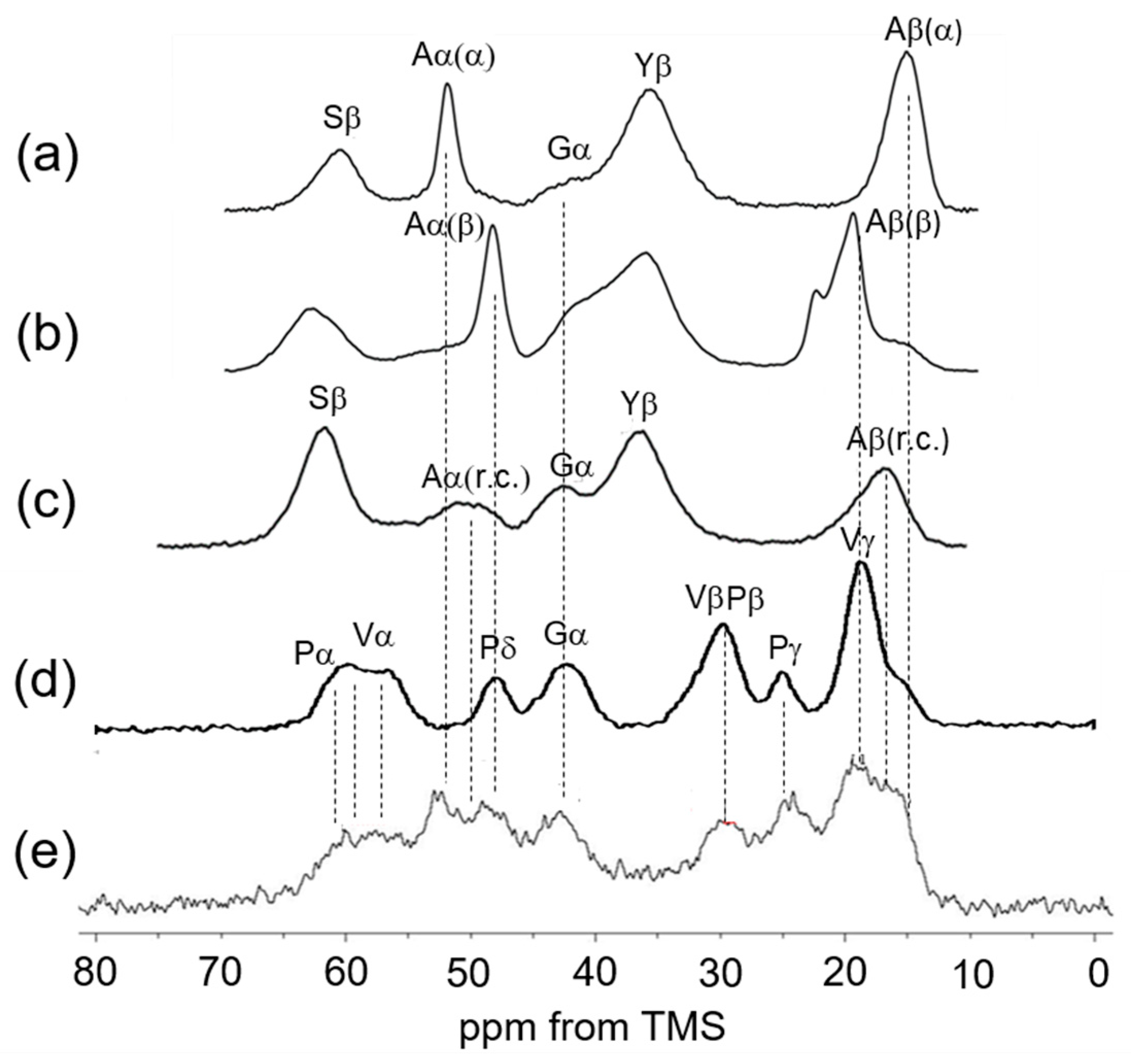
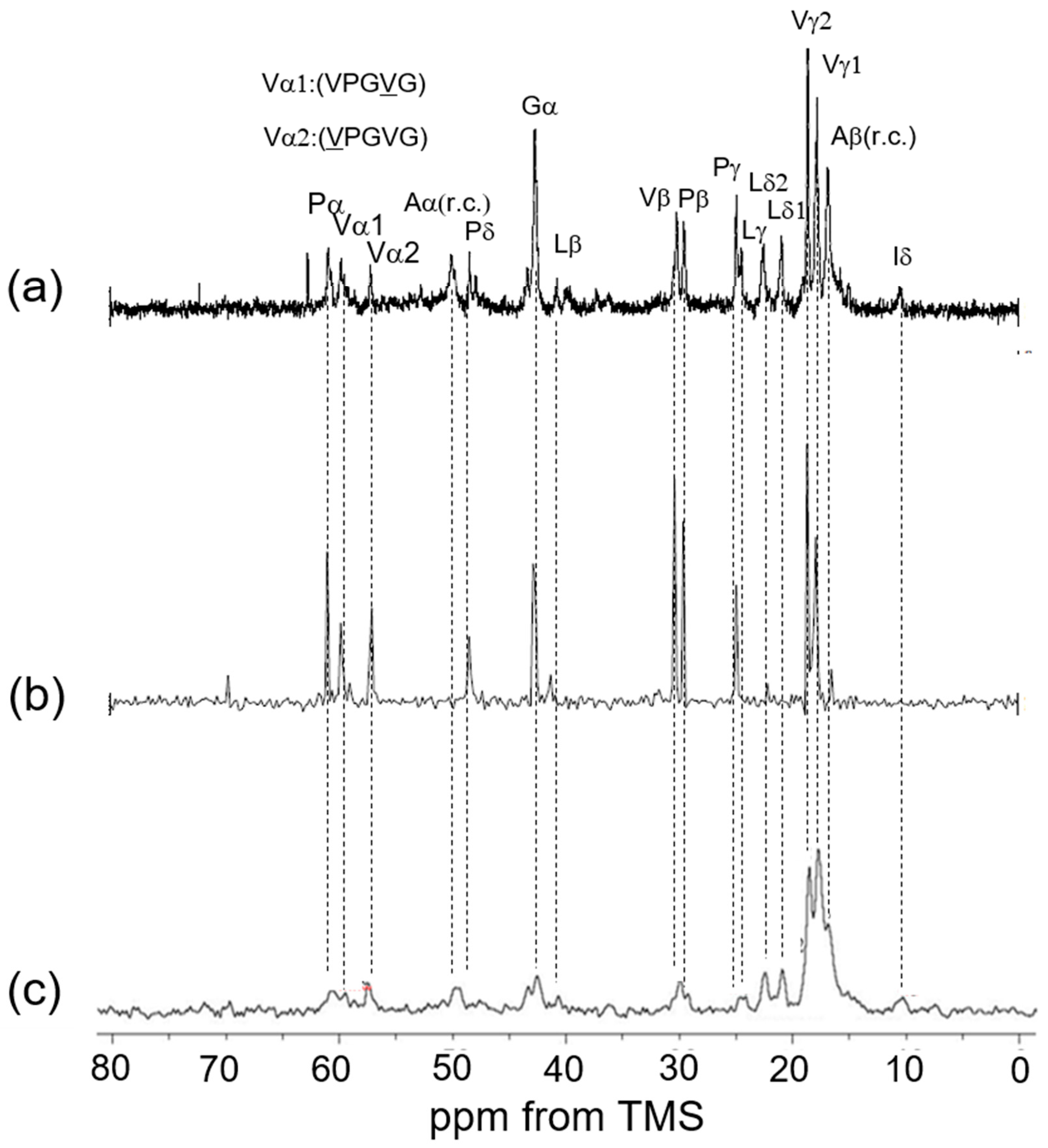
3.2. Peak Assignments of 13C Solid-State NMR Spectra of Soluble Elastin in the Dry and Hydrated States
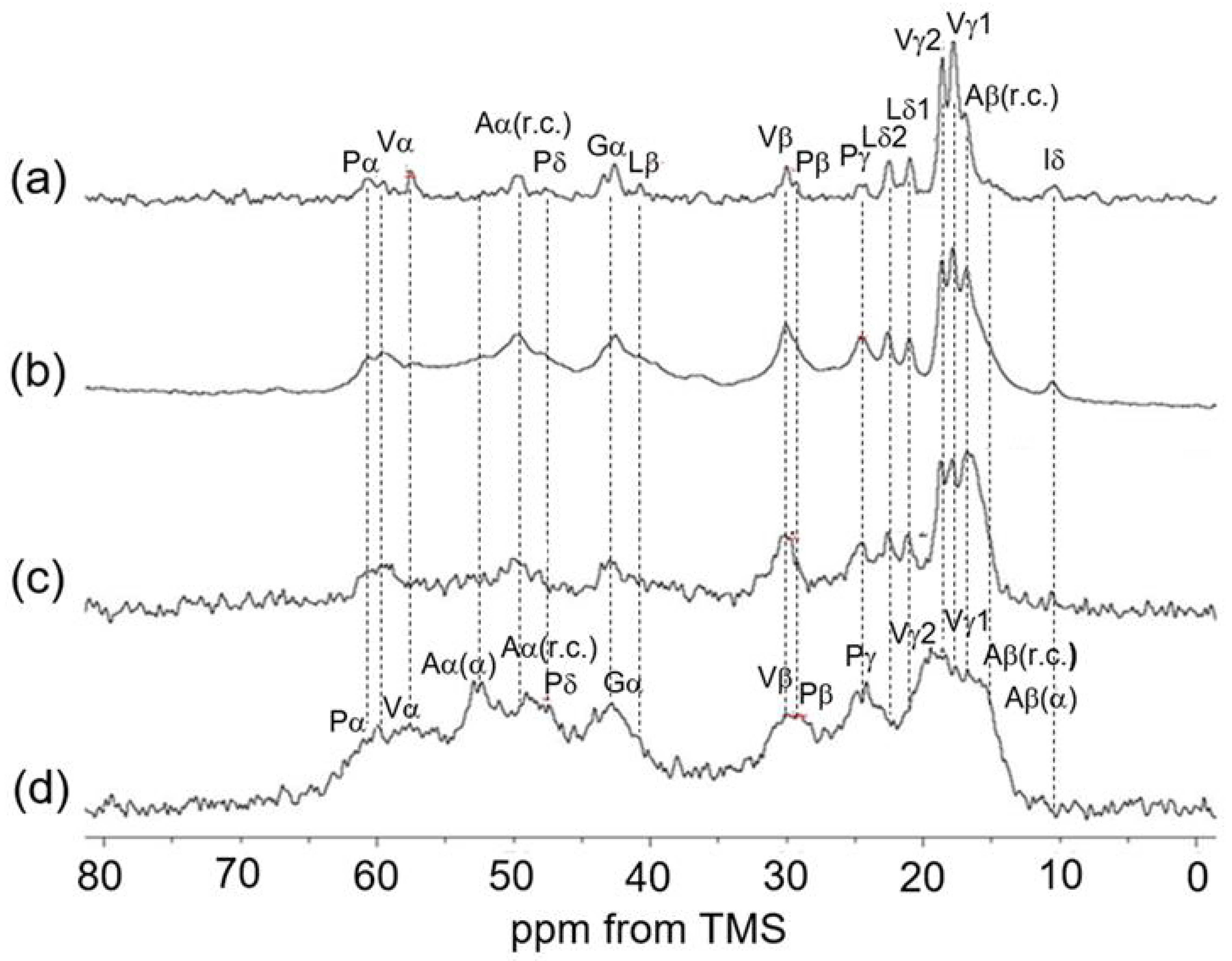
3.3. 13C Solid-State NMR Spectra of Soluble Elastin in the Dry and Hydrated States
3.4. 13C Solid-State NMR Spectra of Insoluble Elastin in the Dry and Hydrated States
3.5. Spectral Change of 13C Solid-State NMR Spectra of Soluble Elastin in the Dry and Hydrated States Cross-Linked Using Dode-DSP
3.6. Contextualizing the Findings in Biomaterial Design
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rosenbloom, J.; Abrams, W.R.; Mecham, R. Extracellular matrix. IV. The elastic fiber. FASEB J. 1993, 7, 1208–1218. [Google Scholar] [CrossRef]
- Wang, R.; de Kort Bente, J.; Smits, A.I.P.M.; Weiss, A.S. Elastinin Vascular Grafts, Tissue-Engineered Vascular Grafts, Reference Series in Biomedical Engineering; Walpothetal, B., Ed.; Springer Nature: Cham, Switzerland, 2019. [Google Scholar]
- Depenveiller, C.; Bochicchio, B.; Baud, S.; Dandurand, J.; Belloy, N.; Dauchez, M.; Pepe, A.; Pomès, R.; Samouillan, V.; Debelle, L. Structural and physical basis for the elasticity of elastin. Q. Rev. Biophys. 2024, 57, 1–23. [Google Scholar] [CrossRef]
- Kumashiro, K.K.; Kim, M.S.; Kaczmarek, S.E.; Sandberg, L.B.; Boyd, C.D. 13C Cross-Polarization/Magic Angle Spinning NMR Studies of α-Elastin Preparations Show Retention of Overall Structure and Reduction of Mobility with a Decreased Number of Cross-Links. Biopolymers 2001, 59, 266–275. [Google Scholar] [CrossRef]
- Toonkool, P.; Jensen, S.A.; Maxwell, A.L.; Weiss, A.S. Hydrophobic domains of human tropoelastin interact in a context-dependent manner. J. Biol. Chem. 2001, 276, 44575–44580. [Google Scholar] [CrossRef]
- Gosline, J.; Lillie, M.; Carrington, E.; Guerette, P.; Ortlepp, C.; Savage, K. Elastic proteins: Biological roles and mechanical properties. Philosophical Transactions of the Royal Society of London. Proc. R. Soc. B. 2002, 357, 121–132. [Google Scholar]
- Tamburro, A.M.; Bochicchio, B.; Pepe, A. Dissection of human tropoelastin: Exon-by-exon chemical synthesis and related conforma tional studies. Biochemistry 2003, 42, 13347–13362. [Google Scholar] [CrossRef] [PubMed]
- Rauscher, S.; Baud, S.; Miao, M.; Keeley, F.W.; Pomès, R. Proline and glycine control protein self-organization into elastomeric or amyloid fibrils. Structure 2006, 14, 1667–1676. [Google Scholar] [CrossRef]
- Muiznieks, L.D.; Weiss, A.S. Flexibility in the solution structure of human tropoelastin. Biochemistry 2007, 46, 8196–8205. [Google Scholar] [CrossRef] [PubMed]
- Rauscher, S.; Pomès, R. Molecular simulations of protein dis order. Biochem. Cell Biol. 2010, 88, 269–290. [Google Scholar] [CrossRef]
- Muiznieks, L.D.; Weiss, A.S.; Keeley, F.W. Structuraldisorder and dynamics of elastin. Biochem. Cell Biol. 2010, 88, 239–250. [Google Scholar] [CrossRef]
- Rauscher, S.; Pomès, R. Structural disorder and protein elasticity. Adv. Exp. Med. Biol. 2012, 725, 159–183. [Google Scholar]
- Sandberg, L.B.; Weissman, N.; Gray, W.R. Structural features of tropoelastin related to the sites of cross-links in aortic elastin. Biochemistry 1971, 10, 52–56. [Google Scholar] [CrossRef]
- Foster, J.A.; Bruenger, E.; Rubin, L.; Imberman, M.; Kagan, H.; Mecham, R.; Franzblau, C. Circular dichroism studies of an elastin cross linked peptide. Biopolymers 1976, 15, 833–841. [Google Scholar] [CrossRef] [PubMed]
- Tamburro, A.M.; Pepe, A.; Bochicchio, B. Localizing-helices in human tropoelastin: Assembly of the elastin “puzzle”. Biochemistry 2006, 45, 9518–9530. [Google Scholar] [CrossRef] [PubMed]
- Miao, M.; Cirulis, J.T.; Lee, S.; Keeley, F.W. Structural determinants of cross-linking and hydrophobic domains for self-assembly of elastin-like polypeptides. Biochemistry 2005, 44, 14367–14375. [Google Scholar] [CrossRef]
- Djajamuliadi, J.; Ohgo, K.; Kumashiro, K.K. Targeting Alanines in the Hydrophobic and Cross-Linking Domains of Native Elastin with Isotopic Enrichment and Solid-State NMR Spectroscopy. Macromolecules 2018, 51, 2157–2168. [Google Scholar] [CrossRef]
- Djajamuliadi, J.; Ohgo, K.; Kumashiro, K.K. A Two-State Model Describes the Temperature-Dependent Conformational Equilibrium in the Alanine-Rich Domains in Elastin. J. Phys. Chem. B. 2020, 124, 9017–9028. [Google Scholar] [CrossRef]
- Hoeve, C.A.J.; Flory, P.J. Elastic properties of elastin. Biopolymers 1974, 13, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Gray, W.R.; Sandberg, L.B.; Foster, J.A. Molecular model for elastin structure and function. Nature 1973, 246, 461–466. [Google Scholar] [CrossRef]
- Venkatachalam, C.M.; Urry, D.W. Development of a linear helical conformation from its cyclic correlate. β-Spiral model of the elastin poly(pentapeptide) (VPGVG)n. Macromolecules 1981, 14, 1225–1229. [Google Scholar] [CrossRef]
- Debelle, L.; Tamburro, A.M. Elastin: Molecular description and function. Int. J. Biochem. Cell Biol. 1999, 31, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.L.; Hong, M. Structure Distribution in an Elastin Mimetic Peptide (VPGVG)3 Investigated by Solid-State NMR. J. Am. Chem. Soc. 2004, 126, 4199–4210. [Google Scholar] [CrossRef] [PubMed]
- Ohgo, K.; Ashida, J.; Kumashiro, K.K.; Asakura, T. Structural Determination of an Elastin-Mimetic Model Peptide, (Val-Pro-Gly-Val-Gly)6, Studied by 13C CP/MAS NMR Chemical Shifts, Two Dimensional off Magic Angle Spinning Spin-Diffusion NMR, Rota tional Echo Double Resonance, and Statistical Distribution of Torsion Angles from Protein Data Bank. Macromolecules 2005, 38, 6038–6047. [Google Scholar]
- Kumashiro, K.K.; Ohgo, K.; Niemczura, W.P.; Onizuka, A.K.; Asakura, T. Structural insights into the elastin mimetic (LGGVG)6 using solid-state 13C NMR experiments and statistical analysis of the PDB. Biopolymers 2008, 89, 668–679. [Google Scholar] [CrossRef]
- Vrhovski, B.; Jensen, S.; Weiss, A.S. Coacervation characteristics of recombinant human tropoelastin. Eur. J. Biochem. 1997, 250, 92–98. [Google Scholar] [CrossRef]
- Reichheld, S.E.; Muiznieks, L.D.; Stahl, R.; Simonetti, K.; Sharpe, S.; Keeley, F.W. Conformational Transitions of the Cross linking Domains of Elastin during Self-assembly. J. Biol. Chem. 2014, 289, 10057–10068. [Google Scholar] [CrossRef]
- Debelle, L.; Alix, A.J.P.; Jacob, M.; Huvenne, J.; Berjot, M.; Sombret, B.; Legrand, P. Bovine Elastin and κ-Elastin Secondary Structure Determination by Optical Spectroscopies. J. Biol. Chem. 1995, 270, 26099–26103. [Google Scholar] [CrossRef]
- Reichheld, S.E.; Muiznieks, L.D.; Keeley, F.W.; Sharpe, S. Direct observation of structure and dynamics during phase separation of an elastomeric protein. Proc. Natl. Acad. Sci. USA 2017, 114, E4408–E4415. [Google Scholar] [CrossRef]
- Tamburro, A.M.; Guantieri, V.; Pandolfo, L.; Scopa, A. Synthetic Fragments and Analogues of Elastin. II. Conformational Studies. Biopolymers 1990, 29, 855–870. [Google Scholar] [CrossRef]
- Urry, D.W.; Mitchell, L.W.; Ohnishi, T. Carbon-13 magnetic resonance assignments of the repeat peptides of elastin and solvent delineation of carbonyls. Biochemistry 1974, 13, 4083–4090. [Google Scholar] [CrossRef] [PubMed]
- Chang, D.K.; Venkatachalam, C.M.; Prasad, K.U.; Urry, D.W. Nuclear Overhauser effect and computational characterization of the beta-spiral of the polypentapeptide of elastin. J. Biomol. Struct. Dyn. 1989, 6, 851–858. [Google Scholar] [CrossRef]
- Perry, A.; Stypa, M.P.; Tenn, B.K.; Kumashiro, K.K. Solid-state 13C NMR reveals effects of temperature and hydration on elastin. Biophys. J. 2002, 82, 1086–1095. [Google Scholar] [CrossRef] [PubMed]
- Pometun, M.S.; Chekmenev, E.Y.; Wittebort, R.J. Quantitative Observation of Backbone Disorder in Native Elastin. J. Biol. Chem. 2004, 279, 7982–7987. [Google Scholar] [CrossRef]
- Ohgo, K.; Niemczura, W.P.; Muroi, T.; Onizuka, A.K.; Kumashiro, K.K. Wideline Separation (WISE) NMR of Native Elastin. Macromolecules 2009, 42, 8899–8906. [Google Scholar] [CrossRef]
- Ohgo, K.; Niemczura, W.P.; Kumashiro, K.K. Probing the Natural-Abundance 13C Populations of Insoluble Elastin Using 13C-1H Heteronuclear Correlation (HETCOR) NMR Spectroscopy. Macromolecules 2009, 42, 7024–7030. [Google Scholar] [CrossRef]
- Papaioannoua, A.; Louisb, M.; Dhitala, B.; Hob, H.P.; Changb, E.J.; Boutisa, G.S. Quantitative comparison of structure and dynamics of elastin following three isolation schemes by 13C solid state NMR and MALDI mass spectrometry. Biochim. Biophys. Acta 2015, 1854, 391–401. [Google Scholar] [CrossRef][Green Version]
- Silverstein, M.C.; Bilici, K.; Morgan, S.W.; Wang, Y.; Zhang, Y.; Boutis, G.S. 13C, 2H NMR Studies of Structural and Dynamical Modifications of Glucose-Exposed Porcine Aortic Elastin. Biophs. J. 2015, 108, 1758–1772. [Google Scholar] [CrossRef][Green Version]
- Bilici, K.; Morgan, S.W.; Silverstein, M.C.; Wang, Y.; Hyung Jin Sun, H.J.; Zhang, K.; Boutis, G.S. Mechanical, Structural, and Dynamical Modifications of Cholesterol Exposed Porcine Aortic Elastin. Biophys. Chem. 2016, 218, 47–57. [Google Scholar] [CrossRef]
- Ohgo, K.; Dabalos, C.L.; Kumashiro, K.K. Solid-State NMR Spectroscopy and Isotopic Labeling Target Abundant Dipeptide Sequences in Elastin’s Hydrophobic Domains. Macromolecules 2018, 51, 2145–2156. [Google Scholar] [CrossRef]
- Eyre, D.R.; Paz, M.A.; Gallop, P.M. Cross-linking in collagen and elastin. Annu. Rev. Biochem. 1984, 53, 717–748. [Google Scholar] [CrossRef]
- Miyamoto, K.; Atarashi, M.; Kadozono, H.; Shibata, M.; Koyama, Y.; Okai, M.; Inakuma, A.; Kitazono, E.; Kaneko, H.; Takebayashi, T.; et al. Creation of cross-linked electrospun isotypic-elastin fibers controlled cell-differentiation with new cross-linker. Int. J. Biological. Macromol. 2008, 45, 33–41. [Google Scholar] [CrossRef]
- Partridge, S.M.; Davis, H.S.; Adair, G.S. The Chemistry of Connective Tissuee. 2. Soluble protein derived from partial hydrolysis of elastin. Biochem. J. 1955, 61, 11–21. [Google Scholar] [CrossRef]
- Partridge, S.M.; Elsden, D.F.; Thomas, J.; Dorfman, A.; Telser, A.; Ho, P.L. Biosynthesis of the desmosine and isodesmosine cross-bridges in elastin. Biochem. J. 1964, 93, 30–33. [Google Scholar]
- Asakura, T.; Endo, M.; Tasei, Y.; Ohkubo, T.; Hiraoki, T.; Boutis, G.S.; Li, J.L.; Wang, X.G. Hydration of Bombyx Mori Silk Cocoon, Silk Sericin and Silk Fibroin and Their Interactions with Water as Studied by 13C NMR and 2H NMR Relaxation. J. Mater. Chem. B 2017, 5, 1624–1632. [Google Scholar] [CrossRef] [PubMed]
- Asakura, T.; Isobe, K.; Kametani, S.; Ukpebor, O.T.; Silverstein, M.C.; Boutis, G.S. Characterization of Water in Hydrated Bombyx Mori Silk Fibroin Fiber and Films by 2H NMR Relaxation and 13C Solid State NMR. Acta Biomater. 2017, 50, 322–333. [Google Scholar] [CrossRef] [PubMed]
- Asakura, T.; Endo, M.; Fukuhara, R.; Tasei, Y. 13C NMR Characterization of Hydrated 13C Labeled Bombyx Mori Silk Fibroin Sponges Prepared Using Glycerin, Poly(ethylene Glycol Diglycidyl Ether) and Poly(ethylene Glycol) as Porogens. J. Mater. Chem. B 2017, 5, 2152–2160. [Google Scholar] [CrossRef]
- Asakura, T.; Isobe, K.; Aoki, A.; Kametani, S. Conformation of Crystalline and Noncrystalline Domains of [3-13C]Ala-, [3-13C]Ser-, and [3-13C]Tyr-Bombyx Mori Silk Fibroin in a Hydrated State Studied with 13C DD/MAS NMR. Macromolecules 2015, 48, 8062–8069. [Google Scholar] [CrossRef]
- Miyamoto, K.; Tokita, M.; Komai, T. Novelcross-Linking Agents for Hydrogelation of Biological Polymers. Prot. Pept. Lett. 2001, 8, 231–236. [Google Scholar] [CrossRef]
- Urry, D.W.; Long, M.M. On the conformation, coacervation and function of polymeric models of elastin. In Elastin and Elastic Tissue, Advances in Experimental Medicine and Biology; Sandberg, L.B., Gray, W.R., Franzblau, C., Eds.; Springer: Boston, MA, USA, 1977; pp. 685–714. [Google Scholar]
- Urry, D.W. Physical Chemistry of Biological Free Energy Transduction as Demonstrated by Elastic Protein-Based Polymers. J. Phys. Chem. B. 1997, 101, 11007–11028. [Google Scholar] [CrossRef]
- Urry, D.W.; Trapane, T.L.; Sugano, H.; Prasad, K.U. Sequential polypeptides of elastin: Cyclic conformational correlates of the linear polypentapeptide. J. Am. Chem. Soc. 1981, 103, 2080–2089. [Google Scholar] [CrossRef]
- Asakura, T.; Ashida, J.; Ohgo, K. Conformational Characterization of (Val-Pro-Gly-Val-Gly)6 with 13C Solid State NMR. Polym. J. 2003, 35, 293–296. [Google Scholar] [CrossRef]
- Li, B.; Alonso, D.O.V.; Daggett, V. The molecular basis for the inverse temperature transition of elastin. J. Mol. Biol. 2001, 305, 581–592. [Google Scholar] [CrossRef]
- Asakura, T.; Nishimura, A.; Sato, Y. Quantitative Correlation between Primary Sequences and Conformations in 13C-Labeled Samia Cynthia Ricini Silk Fibroin during Strain-Induced Conformational Transition by 13C Solid State NMR. Macromolecules 2017, 50, 2871–2880. [Google Scholar] [CrossRef]
- Nishimura, A.; Matsuda, H.; Tasei, Y.; Asakura, T. Effect of Water on the Structure and Dynamics of Regenerated [3-13C] Ser, [3-13C], and [3-13C] Ala-Bombyx Mori Silk Fibroin Studied with 13C Solid-State Nuclear Magnetic Resonance. Biomacromolecules 2018, 19, 563–575. [Google Scholar] [CrossRef]
- MacArthur, M.W.; Thornton, J.M. Influence of proline residues on protein conformation. J. Mol. Biol. 1991, 218, 397–412. [Google Scholar] [CrossRef]
- Sandberg, L.B.; Leslie, J.G.; Leach, C.T.; Alvarez, V.L.; Torres, A.R.; Smith, D.W. Elastin covalent structure as determined pubs.acs.org/JPCB Article by solid phase amino acid sequencing. Pathol. Biol. 1985, 33, 266–274. [Google Scholar]
- Vrhovski, B.; Weiss, A.S. Biochemistry of tropoelastin. Eur. J. Biochem. 1998, 258, 1–18. [Google Scholar] [CrossRef]
- Wise, S.G.; Yeo, G.C.; Hiob, M.A.; Rnjak-Kovacina, J.; Kaplan, D.L.; Ng, M.K.C.; Weiss, A.S. Tropoelastin: A versatile, bioactive assembly module. Acta Biomater. 2014, 10, 1532–1541. [Google Scholar] [CrossRef]
- Kumashiro, K.K.; Ho, J.P.; Niemczura, W.P.; Keeley, F.W. Cooperativity between the hydrophobic and cross-linking domains of elastin. J. Biol. Chem. 2006, 281, 23757–23765. [Google Scholar] [CrossRef] [PubMed]
- Muiznieks, L.D.; Jensen, S.A.; Weiss, A.S. Structural changes and facilitated association of tropoelastin. Arch. Biochem. Biophys. 2003, 410, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Keeley, F.W. The evolution of elastin. In Evolution of Extracellular Matrix, Biology of Extracellular Matrix; Keeley, F.W., Mecham, R.P., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 73–119. [Google Scholar]
- Shewry, P.R.; Tatham, A.S.; Bailey, A.J. (Eds.) Elastomeric Proteins: Structures, Biomechanical Properties, and Biological Roles, 1st ed.; Cambridge University Press: Cambridge, UK, 2003. [Google Scholar]
- Muiznieks, L.D.; Keeley, F.W. Biomechanical design of elastic protein biomaterials: A balance of protein structure and conformational disorder. ACS Biomater. Sci. Eng. 2017, 3, 661–679. [Google Scholar] [CrossRef] [PubMed]
- Reichheld, S.E.; Muiznieks, L.D.; Huynh, Q.; Wang, N.; Ing, C.; Miao, M.; Sitarz, E.E.; Pomès, R.; Sharpe, S.; Keeley, F.W. Thee volutionary background and functional consequences of the rs2071307 polymorphism in human tropoelastin. Biopolymers 2021, 112, e23414. [Google Scholar]
- Tanaka, T.; Abe, Y.; Cheng, C.-J.; Tanaka, R.; Naito, A.; Asakura, T. Development of Small-Diameter Elastin-Silk Fibroin Vascular Grafts. Front. Bioeng. Biotechnol. 2021, 8, 1531. [Google Scholar] [CrossRef] [PubMed]


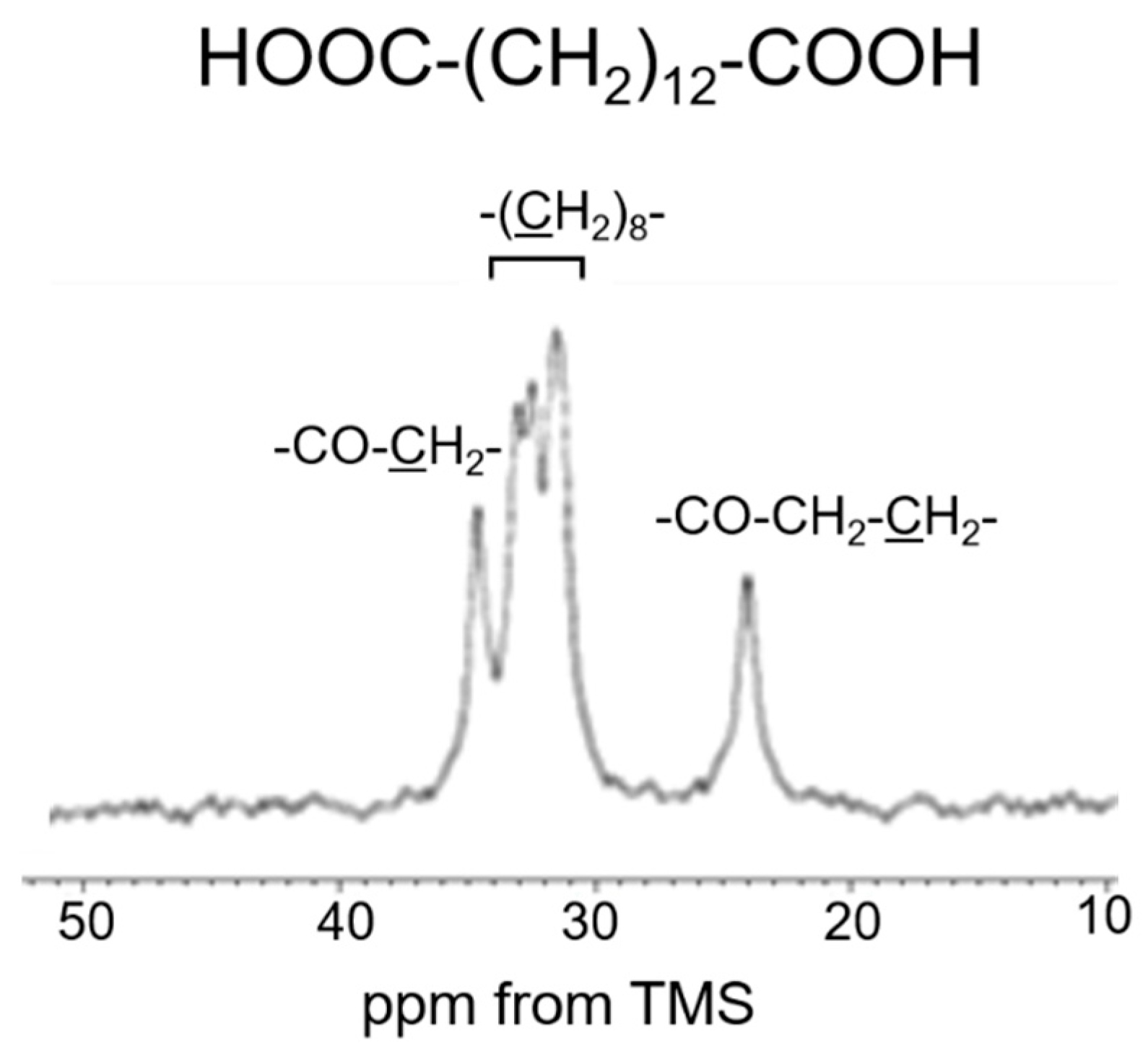


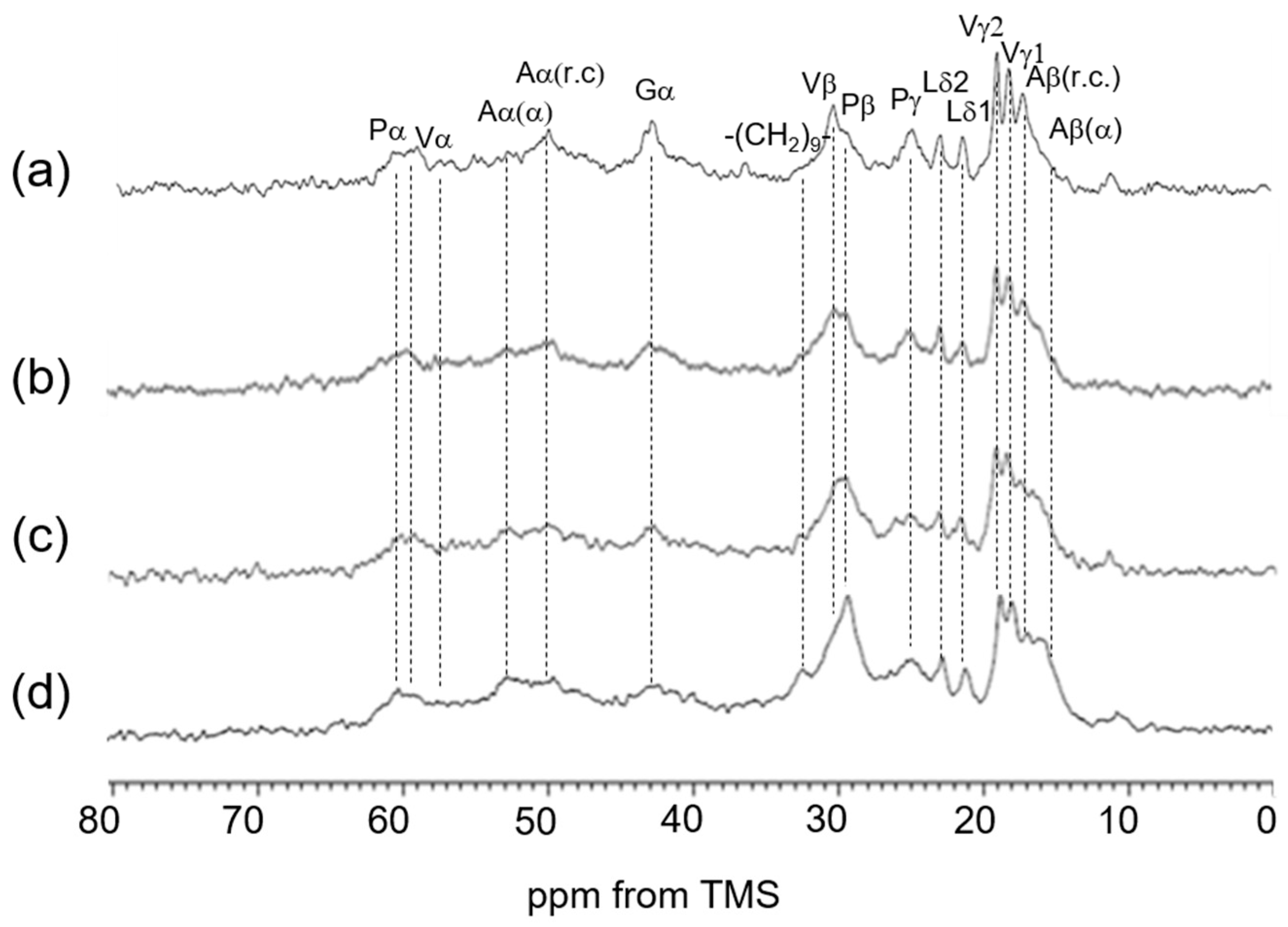
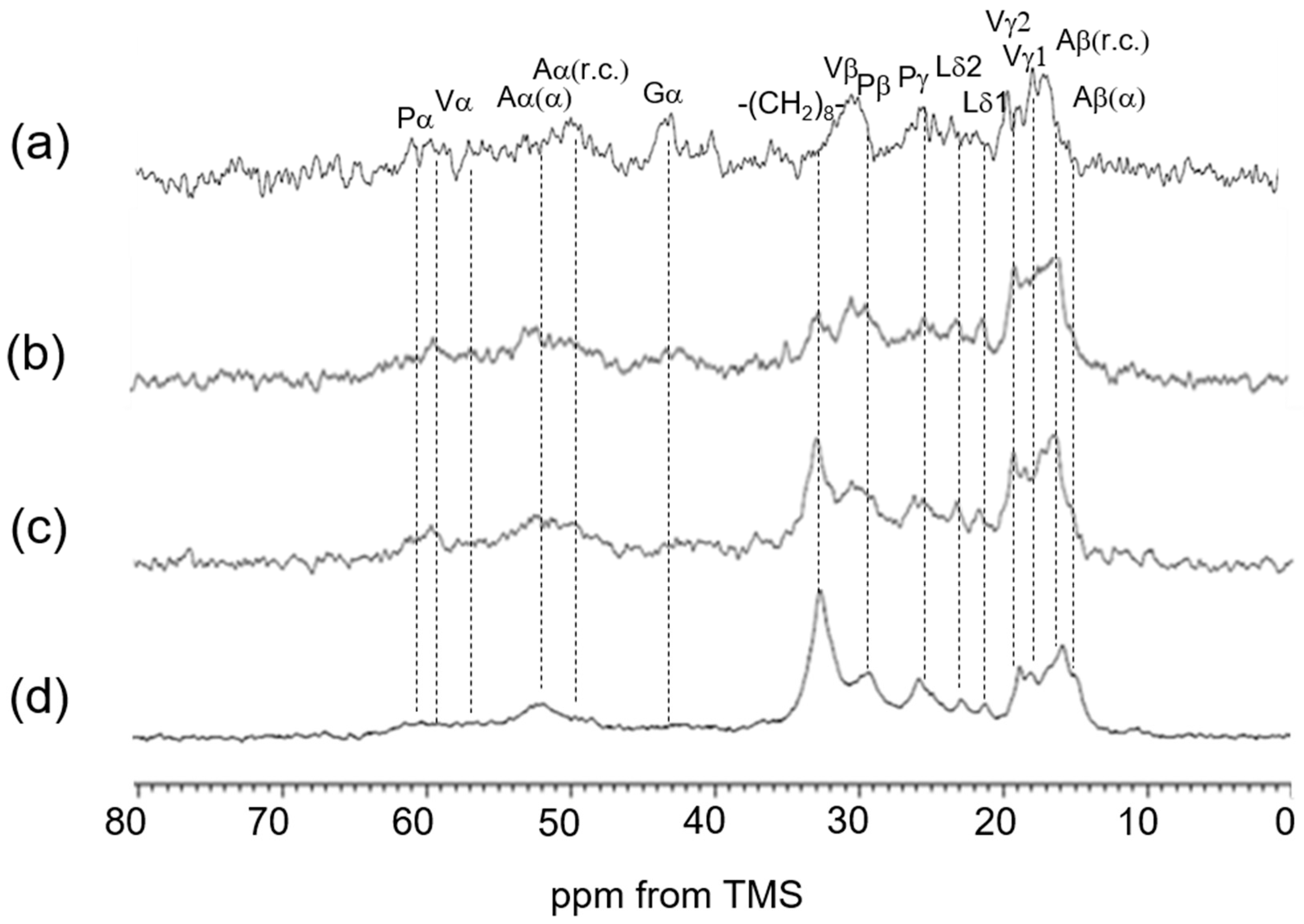
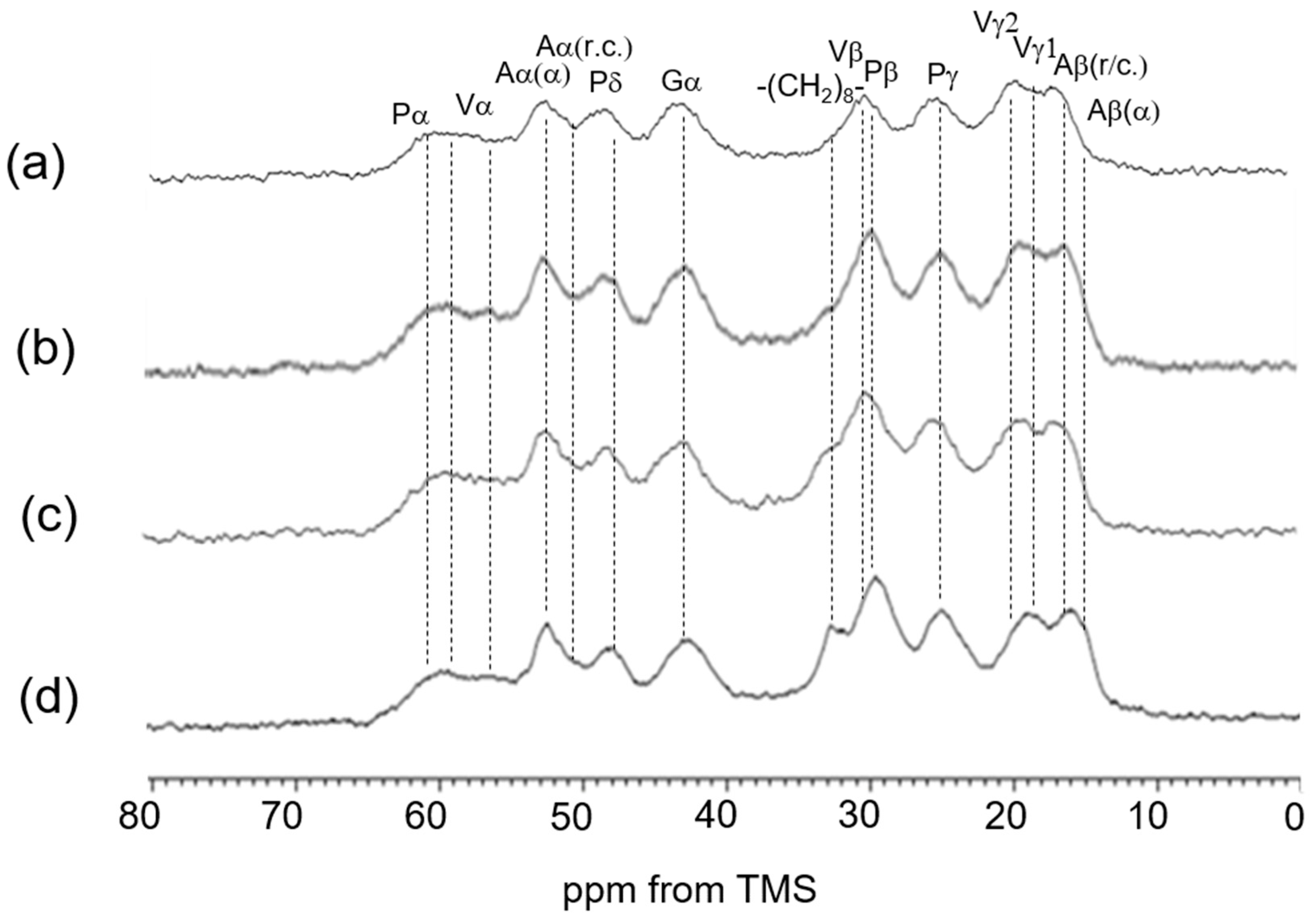
| 13C Solid-State NMR | Chemical Shifts (ppm) | Assignment | Chemical Shifts (ppm) | Assignment |
|---|---|---|---|---|
| (a) INEPT (hydrated) | 10.5 | Iδ | 30.2 | Vβ |
| 16.6 | Aβ (r.c.) | 40.5 | Lβ | |
| 17.7 | Vγ2 | 42.6 | Gα | |
| 18.5 | Vγ1 | 48.4 | Pδ | |
| 21.0 | Lδ2 | 50.0 | Aα (r.c.) | |
| 22.6 | Lδ1 | 57.0 | Vα2 | |
| 24.3 | Lγ | 59.7 | Vα1 | |
| 24.7 | Pγ | 60.9 | Pα | |
| 29.5 | Pβ | |||
| (b) DD/MAS (hydrated) | 10.5 | Iδ | 42.6 | Gα |
| 16.6 | Aβ (r.c.) | 48.4 | Pδ | |
| 17.8 | Vγ2 | 50.0 | Aα (r.c.) | |
| 18.7 | Vγ1 | 57.0 | Vα2 | |
| 21.1 | Lδ2 | 59.4 | Vα1 | |
| 22.5 | Lδ1 | 60.8 | Pα | |
| 24.6 | Pγ + Lγ | 129.2 | Yγ,δ + Fδ,ε,ζ | |
| 30.1 | Pβ + Vβ | |||
| (c) CP/MAS (hydrated) | 16.6 | Aβ (r.c.) | 42.6 | Gα |
| 17.8 | Vγ2 | 48.4 | Pδ | |
| 18.9 | Vγ1 | 50.0 | Aα (r.c.) | |
| 21.1 | Lδ2 | 57–61 | Vα + Pα | |
| 22.7 | Lδ1 | 114.9 | Yε | |
| 24.6 | Pγ | 128.6 | Yγ,δ + Fδ,ε,ζ | |
| 30.0 | Pβ + Vβ | 155.0 | Yζ | |
| (d) CP/MAS (dried) | 15.0 | Aβ (α) | 50.0 | Aα (r.c.) |
| 16.6 | Aβ (r.c.) | 52.5 | Aα (α) | |
| 18–19 | Vγ | 57–61 | Vα + Pα | |
| 24.6 | Pγ | 114.9 | Yε | |
| 30.0 | Pβ + Vβ | 128.7 | Yγ,δ + Fδ,ε,ζ | |
| 42.6 | Gα | 155.2 | Yζ | |
| 48.4 | Pδ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asakura, T.; Naito, A.; Miyamoto, K. Structural Analysis of Soluble Elastin in Dry and Hydrated States Using 13C Solid-State NMR. Polymers 2025, 17, 2638. https://doi.org/10.3390/polym17192638
Asakura T, Naito A, Miyamoto K. Structural Analysis of Soluble Elastin in Dry and Hydrated States Using 13C Solid-State NMR. Polymers. 2025; 17(19):2638. https://doi.org/10.3390/polym17192638
Chicago/Turabian StyleAsakura, Tetsuo, Akira Naito, and Keiichi Miyamoto. 2025. "Structural Analysis of Soluble Elastin in Dry and Hydrated States Using 13C Solid-State NMR" Polymers 17, no. 19: 2638. https://doi.org/10.3390/polym17192638
APA StyleAsakura, T., Naito, A., & Miyamoto, K. (2025). Structural Analysis of Soluble Elastin in Dry and Hydrated States Using 13C Solid-State NMR. Polymers, 17(19), 2638. https://doi.org/10.3390/polym17192638








