Valorization of Rice-Bran and Corn-Flour Hydrolysates for Optimized Polyhydroxybutyrate Biosynthesis: Statistical Process Design and Structural Verification
Abstract
1. Introduction
2. Material and Methods
2.1. Soil Sampling and Bacterial Extraction Techniques
2.2. Screening of PHB-Accumulating Bacterial Isolates Using Sudan Black B Staining
2.3. Morphology and Biochemical Characterization of the Isolates
2.4. Molecular Identification and GenBank Accession Submission
2.5. Production, Extraction, and Quantification of PHB
2.6. Polymer Characterization Using FTIR and NMR Analysis
2.7. Optimization of Culture Conditions for PHB Production by OVAT
2.8. Experimental Design for Medium Optimization Using Response Surface Methodology (RSM)
2.9. Statistical Analysis and Model Validation
3. Results
3.1. Screening Soil-Derived Bacteria for Polyhydroxybutyrate (PHB) Production
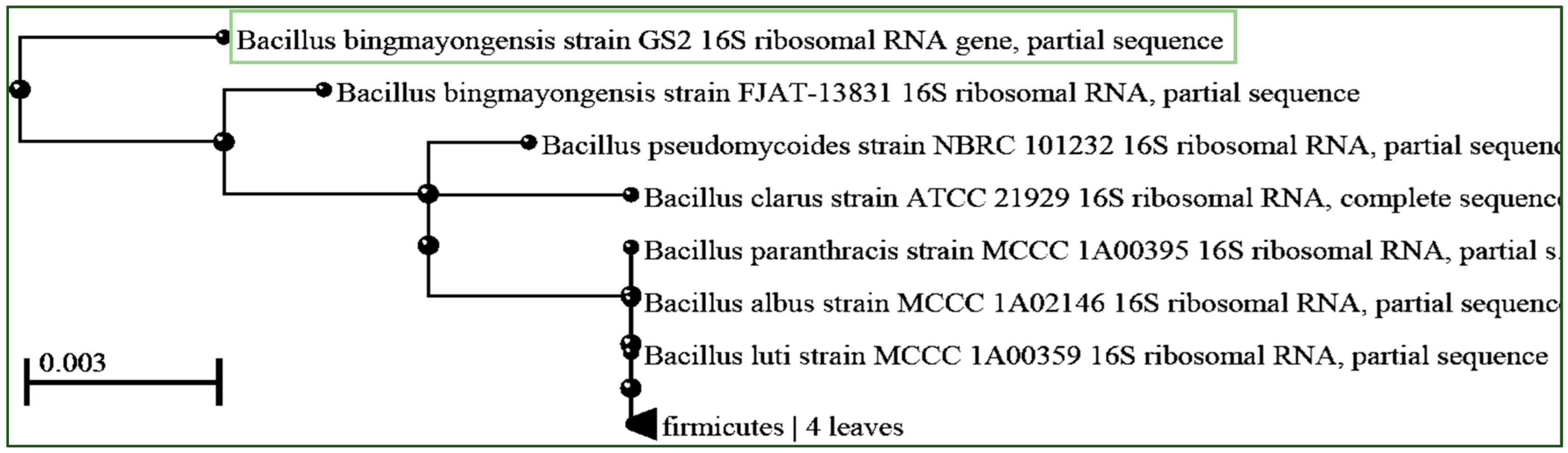
3.2. Molecular Identification of High PHB-Producing Isolate GS2 by 16S rDNA Gene Sequencing
3.3. Biochemical Profiling of Bacillus bingmayongensis GS2 Using VITEK-2: Insights into Metabolic and Resistance Traits
3.4. Production Optimization
3.4.1. Impact of Inoculum Age on PHB Production
3.4.2. Influence of Inoculum Size on PHB Production
3.4.3. Influence of Incubation Time on PHB Production
3.4.4. Influence of Incubation Temperature on PHB Production
3.4.5. Influence of pH of Media on PHB Production
3.4.6. Influence of Agitation Rate on PHB Production
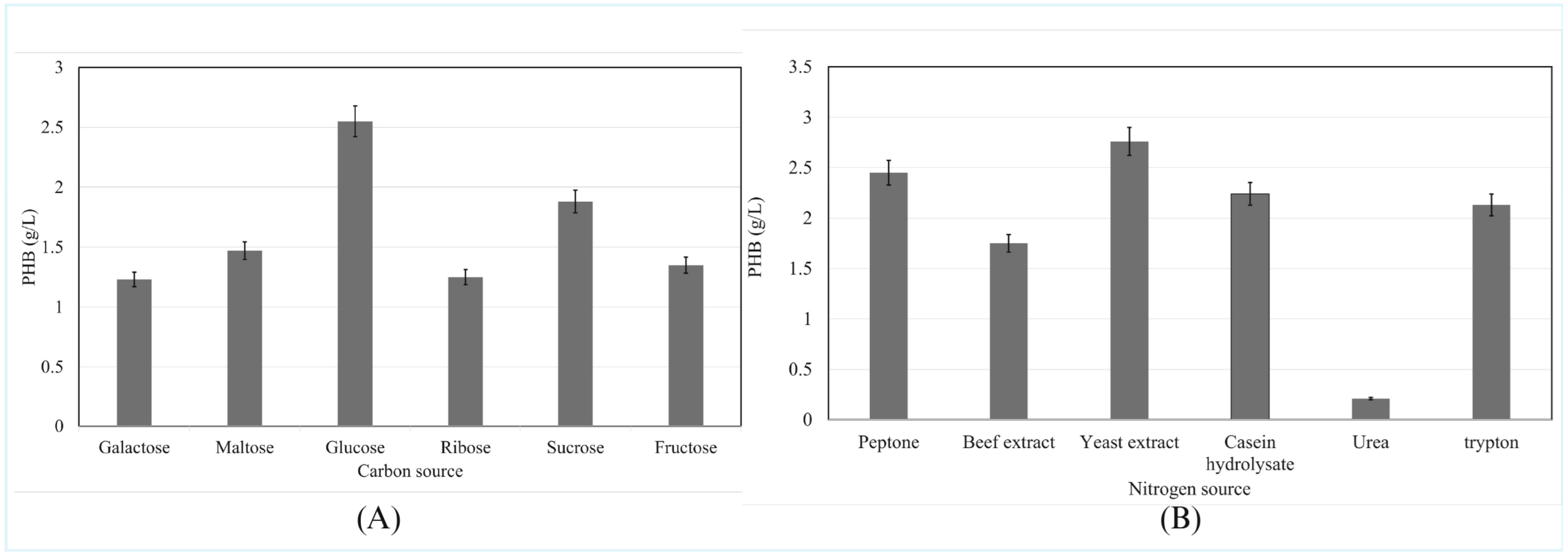
3.4.7. Influence of Carbon Sources on PHB Production
3.4.8. Influence of Nitrogen Source on PHB Production
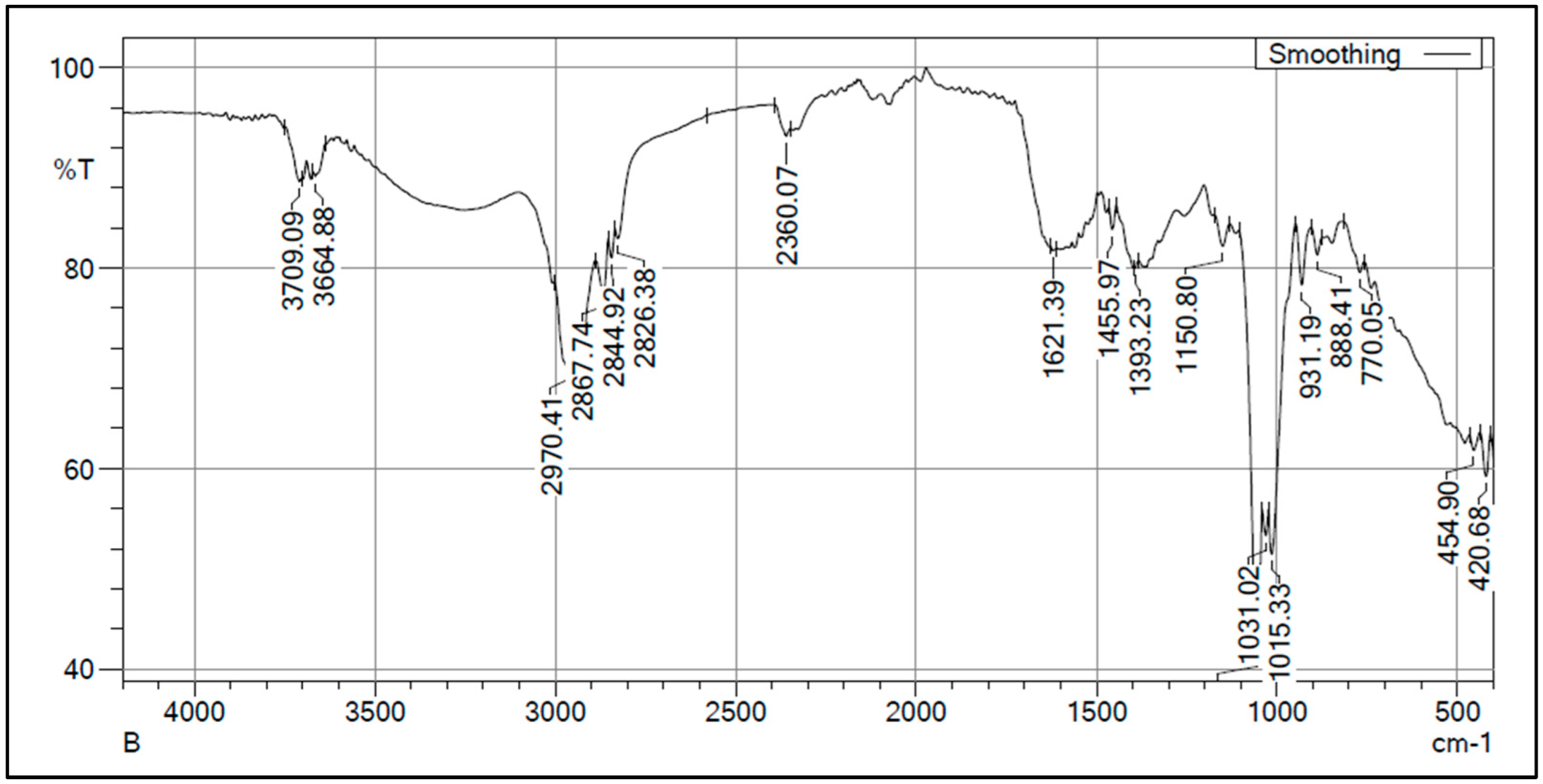
3.5. Characterization of PHB by FTIR and NMR Analysis
3.6. Production Optimization by Response Surface Methodology (RSM) Approach
(0.0438 × AC) + (0.4478 × AD) + (0.0510 × BC) + (0.2426 × BD) + (0.0203 × CD)
3.6.1. Statistical Evaluation of the Model
3.6.2. Effect of Nutrient Interactions on PHB Production
4. Discussion
4.1. Isolation and Identification of Bacillus bingmayongensis GS2 Demonstrate Specialized Metabolic Capabilities Suited for PHB Production
4.2. Optimization of Culture Conditions Significantly Enhances PHB Productivity in Bacillus bingmayongensis GS2
4.2.1. Temporal and Cultural Parameters
4.2.2. Environmental Parameters
4.2.3. Nutritional Requirements
4.3. Structural Characterization Validates the Successful Biosynthesis and Purity of PHB Produced by Bacillus bingmayongensis GS2
4.4. Response Surface Methodology Effectively Optimizes PHB Production, Substantially Improving Biopolymer Yields for Industrial Applications
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PHB | Polyhydroxybutyrate |
| RSM | Response Surface Methodology |
| CCD | Central Composite Design |
| OVAT | One-Variable-at-a-Time |
| O/129 | Vibriostatic agent |
| DCW | Dry Cell Weight |
| FTIR | Fourier Transform Infrared Spectroscopy |
| NMR | Nuclear Magnetic Resonance |
| PCR | Polymerase Chain Reaction |
| OD | Optical Density |
| rpm | Revolutions Per Minute |
| ANOVA | Analysis of Variance |
| LCA | Life Cycle Assessment |
| RBA | Rice Bran (Amylase-treated) |
| CFA | Corn Flour (Amylase-treated) |
| VITEK 2 | Automated Biochemical Identification System (bioMérieux) |
| ppm | Parts Per Million |
| δ | Chemical Shift (NMR) |
| CDCl3 | Deuterated Chloroform |
| w/v | Weight/Volume |
| °C | Degrees Celsius |
| NCBI | National Center for Biotechnology Information |
| BLAST | Basic Local Alignment Search Tool |
| PHA | Polyhydroxyalkanoate |
| phbA, phbB, phbC | Genes encoding enzymes involved in PHB biosynthesis |
References
- Chidambarampadmavathy, K.; Karthikeyan, O.P.; Heimann, K. Sustainable Bio-Plastic Production through Landfill Methane Recycling. Renew. Sustain. Energy Rev. 2017, 71, 555–562. [Google Scholar] [CrossRef]
- Adnan, M.; Siddiqui, A.J.; Ashraf, S.A.; Snoussi, M.; Badraoui, R.; Alreshidi, M.; Elasbali, A.M.; Al-Soud, W.A.; Alharethi, S.H.; Sachidanandan, M.; et al. Polyhydroxybutyrate (PHB)-Based Biodegradable Polymer from Agromyces Indicus: Enhanced Production, Characterization, and Optimization. Polymers 2022, 14, 3982. [Google Scholar] [CrossRef] [PubMed]
- Fayshal, M.A.; Tasnuva Dhara, F.; Mehedi Hasan, M.; Fairooz Adnan, H.M.; Mizan, T. Global plastic waste scenario: A review on production, fate and future prospects. In Proceedings of the 8th International Conference on Integrated Solid Waste and Faecal Sludge Managemen, Khulna, Bangladesh, 25–26 February 2023. [Google Scholar]
- Attapong, M.; Chatgasem, C.; Siripornadulsil, W.; Siripornadulsil, S. Ability of Converting Sugarcane Bagasse Hydrolysate into Polyhydroxybutyrate (PHB) by Bacteria Isolated from Stressed Environmental Soils. Biocatal. Agric. Biotechnol. 2023, 50, 102676. [Google Scholar] [CrossRef]
- Kourmentza, C.; Plácido, J.; Venetsaneas, N.; Burniol-Figols, A.; Varrone, C.; Gavala, H.N.; Reis, M.A.M. Recent Advances and Challenges towards Sustainable Polyhydroxyalkanoate (PHA) Production. Bioengineering 2017, 4, 55. [Google Scholar] [CrossRef] [PubMed]
- Anjum, A.; Zuber, M.; Zia, K.M.; Noreen, A.; Anjum, M.N.; Tabasum, S. Microbial Production of Polyhydroxyalkanoates (PHAs) and Its Copolymers: A Review of Recent Advancements. Int. J. Biol. Macromol. 2016, 89, 161–174. [Google Scholar] [CrossRef]
- McAdam, B.; Brennan Fournet, M.; McDonald, P.; Mojicevic, M. Production of Polyhydroxybutyrate (PHB) and Factors Impacting Its Chemical and Mechanical Characteristics. Polymers 2020, 12, 2908. [Google Scholar] [CrossRef]
- Bektas, K.İ.; Can, K.; Belduz, A.O. Isolation and Screening of Polyhydroxybutyrate (PHB) Producing Bacteria from Soils. Biol. Bull. Russ. Acad. Sci. 2023, 50, 319–328. [Google Scholar] [CrossRef]
- Sumaira; Afzal, S.; Islam, A. Optimization of Soil Bacteria for Bioplastic Production Using Organic Wastes as a Substrate Under Submerged Fermentation. Iran. J. Sci. 2023, 47, 375–387. [Google Scholar] [CrossRef]
- Yilmaz, M.; Beyatli, Y. Poly-β-Hydroxybutyrate (PHB) Production by a Bacillus Cereus M5 Strain in Sugarbeet Molasses. Zuckerindustrie 2005, 130, 109–112. [Google Scholar]
- Bhagowati, P.; Pradhan, S.; Dash, H.R.; Das, S. Production, Optimization and Characterization of Polyhydroxybutyrate, a Biodegradable Plastic by Bacillus Spp. Biosci. Biotechnol. Biochem. 2015, 79, 1454–1463. [Google Scholar] [CrossRef]
- Karolski, B.; Cardoso, L.O.B.; Gracioso, L.H.; Nascimento, C.A.O.; Perpetuo, E.A. MALDI-Biotyper as a Tool to Identify Polymer Producer Bacteria. J. Microbiol. Methods 2018, 153, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Aragosa, A.; Specchia, V.; Frigione, M. Isolation of Two Bacterial Species from Argan Soil in Morocco Associated with Polyhydroxybutyrate (PHB) Accumulation: Current Potential and Future Prospects for the Bio-Based Polymer Production. Polymers 2021, 13, 1870. [Google Scholar] [CrossRef]
- Basak, S.; Subramanian, B.; Thirumurugan, R.; Saleena, L.M. PHB Production by Bacillus Megaterium LSRB 0103 Using Cornstarch and Urea. Curr. Microbiol. 2024, 81, 139. [Google Scholar] [CrossRef] [PubMed]
- Thapa, C.; Shakya, P.; Shrestha, R.; Pal, S.; Manandhar, P. Isolation of Polyhydroxybutyrate (PHB) Producing Bacteria, Optimization of Culture Conditions for PHB Production, Extraction and Characterization of PHB. Nepal. J. Biotechnol. 2019, 6, 62–68. [Google Scholar] [CrossRef]
- Krieg, N.R.; Staley, J.T.; Brown, D.R.; Hedlund, B.P.; Paster, B.J.; Ward, N.L.; Ludwig, W.; Whitman, W.B. (Eds.) Bergey’s Manual® of Systematic Bacteriology: Volume Four The Bacteroidetes, Spirochaetes, Tenericutes (Mollicutes), Acidobacteria, Fibrobacteres, Fusobacteria, Dictyoglomi, Gemmatimonadetes, Lentisphaerae, Verrucomicrobia, Chlamydiae, and Planctomycetes; Springer New York: New York, NY, USA, 2010; ISBN 978-0-387-95042-6. [Google Scholar]
- Wallet, F.; Loïez, C.; Renaux, E.; Lemaitre, N.; Courcol, R.J. Performances of VITEK 2 Colorimetric Cards for Identification of Gram-Positive and Gram-Negative Bacteria. J. Clin. Microbiol. 2005, 43, 4402–4406. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, C.; Ning, J.; Yu, T.; Yuan, K.; Liu, G.; Wang, Q.; Xu, X.; Liu, B.; Lu, X. Molecular Characteristics of IGF and IGF1R Genes and Their Potential Roles on Longevity in Two Scallops with Distinct Lifespans. Aquac. Rep. 2023, 33, 101812. [Google Scholar] [CrossRef]
- Bruno, W.J.; Socci, N.D.; Halpern, A.L. Weighted Neighbor Joining: A Likelihood-Based Approach to Distance-Based Phylogeny Reconstruction. Mol. Biol. Evol. 2000, 17, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Shrimali, G.; Gangawane, A.; Rami, E.; Shah, H.; Thummar, K.; Sahoo, D.K.; Patel, A.; Schmidt, J.E. Optimized Polyhydroxybutyrate Production by Neobacillus Niacini GS1 Utilizing Corn Flour, Wheat Bran, and Peptone: A Sustainable Approach. Biomass 2024, 4, 1164–1177. [Google Scholar] [CrossRef]
- Biglari, N.; Abdeshahian, P.; Orita, I.; Fukui, T.; Sudesh, K. Improving Bioplastic Production: Enhanced P3HB-Co-3HHx) Synthesis from Glucose by Using Mutant Cupriavidus Necator. Iran J. Biotech. 2024, 22, 38–53. [Google Scholar] [CrossRef]
- Sehgal, R.; Kumar, A.; Gupta, R. Bioconversion of Rice Husk as a Potential Feedstock for Fermentation by Priestia Megaterium POD1 for the Production of Polyhydroxyalkanoate. Waste Biomass Valor. 2023, 14, 3657–3670. [Google Scholar] [CrossRef]
- RamKumar Pandian, S.; Deepak, V.; Kalishwaralal, K.; Rameshkumar, N.; Jeyaraj, M.; Gurunathan, S. Optimization and Fed-Batch Production of PHB Utilizing Dairy Waste and Sea Water as Nutrient Sources by Bacillus Megaterium SRKP-3. Bioresour. Technol. 2010, 101, 705–711. [Google Scholar] [CrossRef]
- Gahlawat, G.; Srivastava, A. Development of a Mathematical Model for the Growth Associated Polyhydroxybutyrate Fermentation by Azohydromonas Australica and Its Use for the Design of Fed-Batch Cultivation Strategies. Bioresour. Technol. 2013, 137, 98–105. [Google Scholar] [CrossRef] [PubMed]
- De Rooy, S.L.; Wahyuni, E.T.; Wiratni, W.; Syamsiah, S.; Ismail, J. Purification and Characterization of Poly-Hydroxybutyrate (PHB) in Cupriavidus Necator. Indones. J. Chem. 2010, 7, 243–248. [Google Scholar] [CrossRef]
- Hassan, M.A.; Bakhiet, E.K.; Hussein, H.R.; Ali, S.G. Statistical Optimization Studies for Polyhydroxybutyrate (PHB) Production by Novel Bacillus Subtilis Using Agricultural and Industrial Wastes. Int. J. Environ. Sci. Technol. 2019, 16, 3497–3512. [Google Scholar] [CrossRef]
- Haaland, P.D. Experimental Design in Biotechnology; Taylor & Francis Group: Milton, UK, 1989; ISBN 978-0-8247-7881-1. [Google Scholar]
- Koller, M.; Maršálek, L.; Dias, M.M.; Braunegg, G. Producing Microbial Polyhydroxyalkanoate (PHA) Biopolyesters in a Sustainable Manner. New Biotechnol. 2017, 37, 24–38. [Google Scholar] [CrossRef]
- Madison, L.L.; Huisman, G.W. Metabolic Engineering of Poly(3-Hydroxyalkanoates): From DNA to Plastic. Microbiol. Mol. Biol. Rev. 1999, 63, 21–53. [Google Scholar] [CrossRef]
- Palmeiro-Sánchez, T.; O’Flaherty, V.; Lens, P.N.L. Polyhydroxyalkanoate Bio-Production and Its Rise as Biomaterial of the Future. J. Biotechnol. 2022, 348, 10–25. [Google Scholar] [CrossRef]
- Upadhayay, V.; Verma, S.; Kuila, A. Production of Poly Hydroxy Butyrate (PHB) from Eichhornia Crassipes through Microbial Fermentation Process. Plant Sci. Today 2019, 6, 541–550. [Google Scholar] [CrossRef]
- Sharma, P.; Bajaj, B.K. Cost-Effective-Substrates for Production of Poly-β-Hydroxybutyrate by a Newly Isolated Bacillus Cereus PS-10. J. Environ. Biol. 2015, 36, 1297–1304. [Google Scholar]
- Adnan, M.; Siddiqui, A.J.; Ashraf, S.A.; Snoussi, M.; Badraoui, R.; Ibrahim, A.M.M.; Alreshidi, M.; Sachidanandan, M.; Patel, M. Characterization and Process Optimization for Enhanced Production of Polyhydroxybutyrate (PHB)-Based Biodegradable Polymer from Bacillus Flexus Isolated from Municipal Solid Waste Landfill Site. Polymers 2023, 15, 1407. [Google Scholar] [CrossRef] [PubMed]
- Soni, S.; Chhokar, V.; Beniwal, V.; Kumar, R.; Badgujjar, H.; Chauhan, R.; Dudeja, S.; Kumar, A. Cost Effective Media Optimization for PHB Production by Bacillus Badius MTCC 13004 Using the Statistical Approach. Int. J. Biol. Macromol. 2023, 233, 123575. [Google Scholar] [CrossRef] [PubMed]
- Sathiyanarayanan, G.; Kiran, G.S.; Selvin, J.; Saibaba, G. Optimization of Polyhydroxybutyrate Production by Marine Bacillus Megaterium MSBN04 under Solid State Culture. Int. J. Biol. Macromol. 2013, 60, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Fukala, I.; Kučera, I. Natural Polyhydroxyalkanoates—An Overview of Bacterial Production Methods. Molecules 2024, 29, 2293. [Google Scholar] [CrossRef]
- San Miguel-González, G.D.J.; Alemán-Huerta, M.E.; Martínez-Herrera, R.E.; Quintero-Zapata, I.; De La Torre-Zavala, S.; Avilés-Arnaut, H.; Gandarilla-Pacheco, F.L.; De Luna-Santillana, E.D.J. Alkaline-Tolerant Bacillus Cereus 12GS: A Promising Polyhydroxybutyrate (PHB) Producer Isolated from the North of Mexico. Microorganisms 2024, 12, 863. [Google Scholar] [CrossRef]
- Dalton, B.; Bhagabati, P.; De Micco, J.; Padamati, R.B.; O’Connor, K. A Review on Biological Synthesis of the Biodegradable Polymers Polyhydroxyalkanoates and the Development of Multiple Applications. Catalysts 2022, 12, 319. [Google Scholar] [CrossRef]
- Ronďošová, S.; Legerská, B.; Chmelová, D.; Ondrejovič, M.; Miertuš, S. Optimization of Growth Conditions to Enhance PHA Production by Cupriavidus Necator. Fermentation 2022, 8, 451. [Google Scholar] [CrossRef]
- Andler, R.; Valdés, C.; Urtuvia, V.; Andreeßen, C.; Díaz-Barrera, A. Fruit Residues as a Sustainable Feedstock for the Production of Bacterial Polyhydroxyalkanoates. J. Clean. Prod. 2021, 307, 127236. [Google Scholar] [CrossRef]
- Muneer, F.; Rasul, I.; Qasim, M.; Sajid, A.; Nadeem, H. Optimization, Production and Characterization of Polyhydroxyalkanoate (PHA) from Indigenously Isolated Novel Bacteria. J. Polym. Environ. 2022, 30, 3523–3533. [Google Scholar] [CrossRef]
- Martínez-Avila, O.; Llenas, L.; Ponsá, S. Sustainable Polyhydroxyalkanoates Production via Solid-State Fermentation: Influence of the Operational Parameters and Scaling up of the Process. Food Bioprod. Process. 2022, 132, 13–22. [Google Scholar] [CrossRef]
- Bonthrone, K.M.; Clauss, J.; Horowitz, D.M.; Hunter, B.K.; Sanders, J.K.M. The Biological and Physical Chemistry of Polyhydroxyalkanoates as Seen by NMR Spectroscopy. FEMS Microbiol. Lett. 1992, 103, 269–277. [Google Scholar] [CrossRef]
- Rizzarelli, P.; Leanza, M.; Rapisarda, M. Investigations into the Characterization, Degradation, and Applications of Biodegradable Polymers by Mass Spectrometry. Mass. Spectrom. Rev. 2023, mas.21869. [Google Scholar] [CrossRef] [PubMed]
- Myers, R.H. Response Surface Methodology: Process and Product Optimization Using Designed Experiments, 1st ed.; New York Academy of Sciences Series; John Wiley & Sons, Incorporated: Newark, NJ, USA, 2016; ISBN 978-1-118-91601-8. [Google Scholar]
- Chen, J.; Li, W.; Zhang, Z.-Z.; Tan, T.-W.; Li, Z.-J. Metabolic Engineering of Escherichia coli for the Synthesis of Polyhydroxyalkanoates Using Acetate as a Main Carbon Source. Microb. Cell Fact. 2018, 17, 102. [Google Scholar] [CrossRef] [PubMed]
- Hauf, W.; Schlebusch, M.; Hüge, J.; Kopka, J.; Hagemann, M.; Forchhammer, K. Metabolic Changes in Synechocystis PCC6803 upon Nitrogen-Starvation: Excess NADPH Sustains Polyhydroxybutyrate Accumulation. Metabolites 2013, 3, 101–118. [Google Scholar] [CrossRef]
- Meng, D.; Wang, S.; Zhao, K.; Luo, Y.; Li, X.; Wang, Y. Improvement of Acetate Tolerance of Escherichia coli by Introducing the PHB Mobilization Pathway. Appl. Environ. Microbiol. 2025, 91, e02454-24. [Google Scholar] [CrossRef]
- Hamdy, S.M.; Danial, A.W.; Gad El-Rab, S.M.F.; Shoreit, A.A.M.; Hesham, A.E.-L. Production and Optimization of Bioplastic (Polyhydroxybutyrate) from Bacillus Cereus Strain SH-02 Using Response Surface Methodology. BMC Microbiol. 2022, 22, 183. [Google Scholar] [CrossRef]



| Independent Variables | Symbols | Code Levels | ||||
|---|---|---|---|---|---|---|
| −α | −1 | 0 | +1 | +α | ||
| RBA | A | 7.25 | 8 | 8.75 | 9.5 | 10.25 |
| CFA | B | 6.25 | 7.5 | 8.75 | 10 | 11.25 |
| Peptone | C | 0 | 3.2 | 6.4 | 9.6 | 12.8 |
| pH | D | 6 | 6.5 | 7 | 7.5 | 8 |
| Sr No. | Test | Result (+/−) | Sr No | Test | Result (+/−) |
|---|---|---|---|---|---|
| 1 | Alpha-Amylase | − | 22 | d-Galactose Fermentation | + |
| 2 | Phosphatidylinositol Phospholipase C (PIPLC) | − | 23 | d-Ribose Fermentation | + |
| 3 | Arginine Dihydrolase 1 | + | 24 | Lactose Fermentation | − |
| 4 | Beta-Galactosidase | − | 25 | N-Acetyl-Glucosamine Fermentation | + |
| 5 | Alpha-Glucosidase | + | 26 | d-Maltose Fermentation | + |
| 6 | Alkaline Phosphatase | − | 27 | d-Mannose Fermentation | − |
| 7 | L-Aspartate Arylamidase | − | 28 | d-Mannitol Fermentation | − |
| 8 | Beta-Galactosidase | − | 29 | Methyl-Beta-D-Glucopyranoside Fermentation | − |
| 9 | Alpha-Mannosidase | − | 30 | Pullulan Fermentation | − |
| 10 | Phosphatase | − | 31 | d-Raffinose Fermentation | − |
| 11 | Leucine Arylamidase | + | 32 | Salicin Fermentation | − |
| 12 | Proline Arylamidase | − | 33 | Saccharose Fermentation | + |
| 13 | Beta-Glucuronidase | − | 34 | d-Trehalose Fermentation | + |
| 14 | Alpha-Galactosidase | − | 35 | Urease | − |
| 15 | Pyroglutamyl Aminopeptidase | − | 36 | Polymyxin B Resistance | + |
| 16 | Beta-Glucuronidase | − | 37 | Bacitracin Resistance | + |
| 17 | Alanine Arylamidase | − | 38 | Novobiocin Resistance | − |
| 18 | Tyrosine Arylamidase | − | 39 | O/129 (2,4-diamino-6,7-diisopropylteridine) Resistance | + |
| 19 | Alcohol Dehydrogenase 2s | − | 40 | Optochin Resistance | + |
| 20 | d-Xylose Fermentation | − | 41 | L-Lactate Alkalinization | − |
| 21 | d-Sorbitol Fermentation | − | - | - | − |
| Std | Run | RBA (g/L) | Peptone (g/L) | CFA(g/L) | pH | PHB Yield (g/L) |
|---|---|---|---|---|---|---|
| 27 | 1 | 8.75 | 8.75 | 6.4 | 7 | 3.188 |
| 13 | 2 | 8 | 7.5 | 9.6 | 7.5 | 1.758 |
| 2 | 3 | 9.5 | 7.5 | 3.2 | 6.5 | 0.563 |
| 15 | 4 | 8 | 10 | 9.6 | 7.5 | 0.496 |
| 1 | 5 | 8 | 7.5 | 3.2 | 6.5 | 0.306 |
| 7 | 6 | 8 | 10 | 9.6 | 6.5 | 0.577 |
| 17 | 7 | 7.25 | 8.75 | 6.4 | 7 | 0.098 |
| 8 | 8 | 9.5 | 10 | 9.6 | 6.5 | 0.758 |
| 30 | 9 | 8.75 | 8.75 | 6.4 | 7 | 3.188 |
| 3 | 10 | 8 | 10 | 3.2 | 6.5 | 0.468 |
| 16 | 11 | 9.5 | 10 | 9.6 | 7.5 | 0.479 |
| 25 | 12 | 8.75 | 8.75 | 6.4 | 7 | 3.188 |
| 10 | 13 | 9.5 | 7.5 | 3.2 | 7.5 | 0.91 |
| 28 | 14 | 8.75 | 8.75 | 6.4 | 7 | 3.188 |
| 9 | 15 | 8 | 7.5 | 3.2 | 7.5 | 0.879 |
| 11 | 16 | 8 | 10 | 3.2 | 7.5 | 1.181 |
| 18 | 17 | 10.25 | 8.75 | 6.4 | 7 | 0.374 |
| 23 | 18 | 8.75 | 8.75 | 6.4 | 6 | 0.353 |
| 5 | 19 | 8 | 7.5 | 9.6 | 6.5 | 0.569 |
| 6 | 20 | 9.5 | 7.5 | 9.6 | 6.5 | 0.493 |
| 26 | 21 | 8.75 | 8.75 | 6.4 | 7 | 3.188 |
| 22 | 22 | 8.75 | 8.75 | 12.8 | 7 | 0.198 |
| 24 | 23 | 8.75 | 8.75 | 6.4 | 8 | 0.718 |
| 4 | 24 | 9.5 | 10 | 3.2 | 6.5 | 0.627 |
| 29 | 25 | 8.75 | 8.75 | 6.4 | 7 | 3.188 |
| 21 | 26 | 8.75 | 8.75 | 0 | 7 | 0.103 |
| 19 | 27 | 8.75 | 6.25 | 6.4 | 7 | 0.335 |
| 12 | 28 | 9.5 | 10 | 3.2 | 7.5 | 0.337 |
| 14 | 29 | 9.5 | 7.5 | 9.6 | 7.5 | 0.427 |
| 20 | 30 | 8.75 | 11.25 | 6.4 | 7 | 0.293 |
| Source | Sum of Squares | df | Mean Square | F-Value | p-Value |
|---|---|---|---|---|---|
| Model | 34.55 | 14 | 2.47 | 23.32 | <0.0001 significant |
| A-RBA | 0.0489 | 1 | 0.0489 | 0.4621 | 0.5070 |
| B-CFA | 0.0465 | 1 | 0.0465 | 0.4397 | 0.5173 |
| C-Peptone | 0.0092 | 1 | 0.0092 | 0.0870 | 0.7721 |
| D-pH | 0.3321 | 1 | 0.3321 | 3.14 | 0.0968 |
| AB | 0.0228 | 1 | 0.0228 | 0.2150 | 0.6495 |
| AC | 0.0438 | 1 | 0.0438 | 0.4135 | 0.5299 |
| AD | 0.4478 | 1 | 0.4478 | 4.23 | 0.0575 |
| BC | 0.0510 | 1 | 0.0510 | 0.4820 | 0.4981 |
| BD | 0.2426 | 1 | 0.2426 | 2.29 | 0.1508 |
| CD | 0.0203 | 1 | 0.0203 | 0.1919 | 0.6676 |
| A2 | 12.49 | 1 | 12.49 | 118.07 | <0.0001 |
| B2 | 11.79 | 1 | 11.79 | 111.46 | <0.0001 |
| C2 | 13.29 | 1 | 13.29 | 125.62 | <0.0001 |
| D2 | 9.87 | 1 | 9.87 | 93.29 | <0.0001 |
| Residual | 1.59 | 15 | 0.1058 | ||
| Lack of Fit | 1.59 | 10 | 0.1587 | ||
| Pure Error | 0.0000 | 5 | 0.0000 | ||
| Cor Total | 36.13 | 29 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shrimali, G.; Shah, H.; Thummar, K.; Rami, E.; Chaudhari, R.; Schmidt, J.E.; Gangawane, A. Valorization of Rice-Bran and Corn-Flour Hydrolysates for Optimized Polyhydroxybutyrate Biosynthesis: Statistical Process Design and Structural Verification. Polymers 2025, 17, 1904. https://doi.org/10.3390/polym17141904
Shrimali G, Shah H, Thummar K, Rami E, Chaudhari R, Schmidt JE, Gangawane A. Valorization of Rice-Bran and Corn-Flour Hydrolysates for Optimized Polyhydroxybutyrate Biosynthesis: Statistical Process Design and Structural Verification. Polymers. 2025; 17(14):1904. https://doi.org/10.3390/polym17141904
Chicago/Turabian StyleShrimali, Gaurav, Hardik Shah, Kashyap Thummar, Esha Rami, Rajeshkumar Chaudhari, Jens Ejbye Schmidt, and Ajit Gangawane. 2025. "Valorization of Rice-Bran and Corn-Flour Hydrolysates for Optimized Polyhydroxybutyrate Biosynthesis: Statistical Process Design and Structural Verification" Polymers 17, no. 14: 1904. https://doi.org/10.3390/polym17141904
APA StyleShrimali, G., Shah, H., Thummar, K., Rami, E., Chaudhari, R., Schmidt, J. E., & Gangawane, A. (2025). Valorization of Rice-Bran and Corn-Flour Hydrolysates for Optimized Polyhydroxybutyrate Biosynthesis: Statistical Process Design and Structural Verification. Polymers, 17(14), 1904. https://doi.org/10.3390/polym17141904












