A Novel Fractional Brownian Dynamics Method for Simulating the Dynamics of Confined Bottle-Brush Polymers in Viscoelastic Solution
Abstract
1. Introduction
2. Models and Simulation Methods
2.1. Force Fields
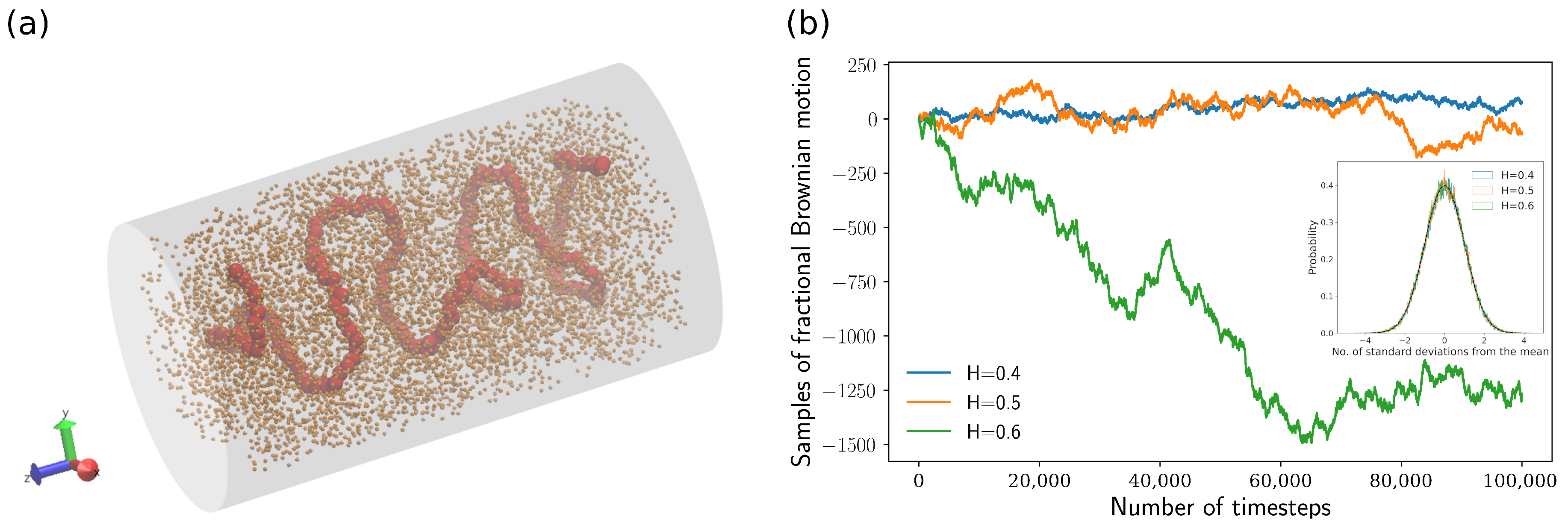
2.2. Fractional Brownian Dynamics Simulation
3. Results and Discussion
3.1. Potential Energy vs. Time for Standard and Fractional Brownian Dynamics Simulations
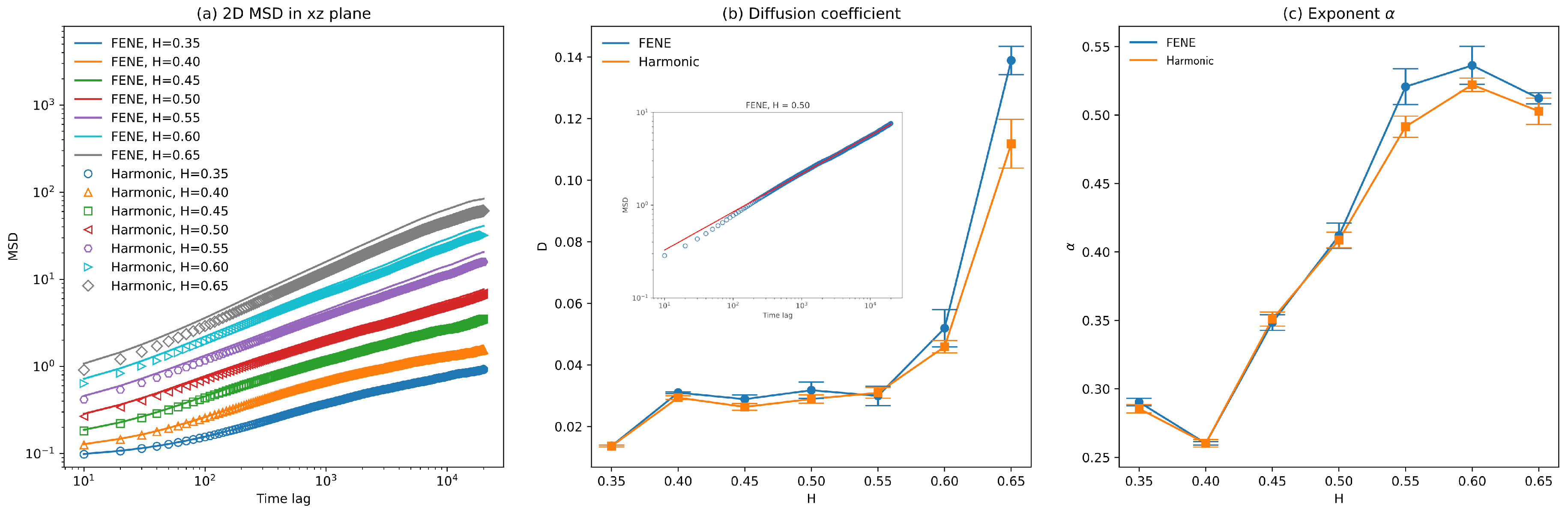
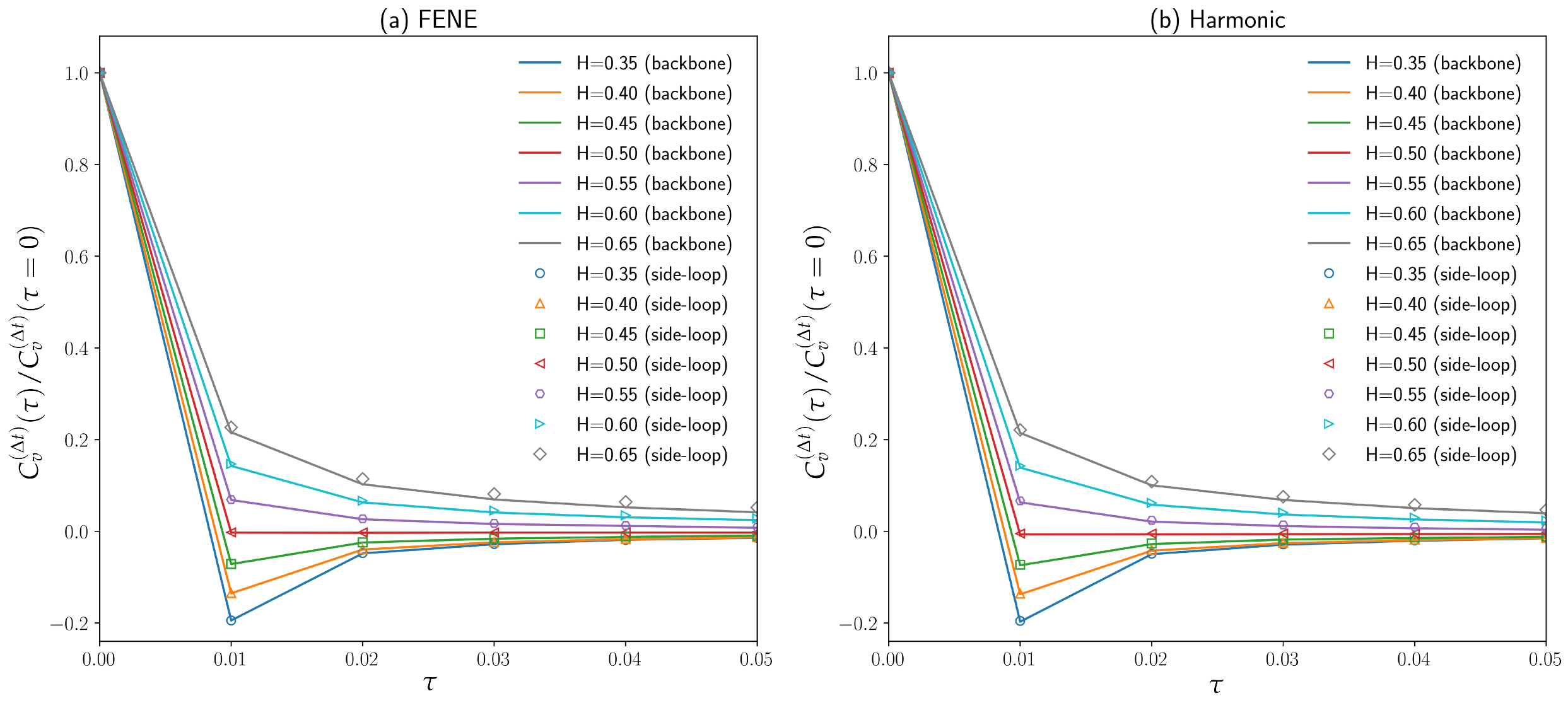
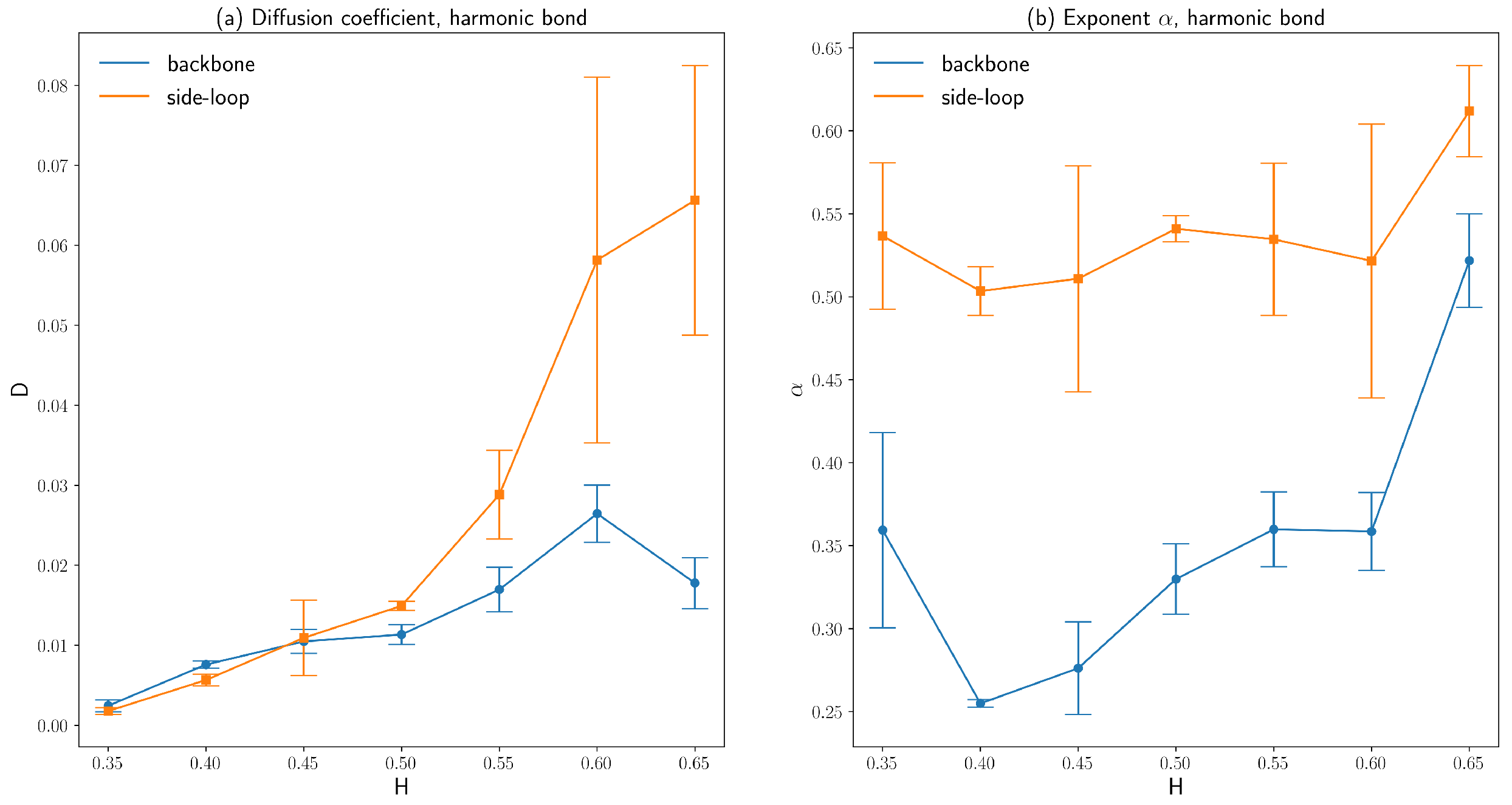
3.2. Diffusion Coefficient and Exponent
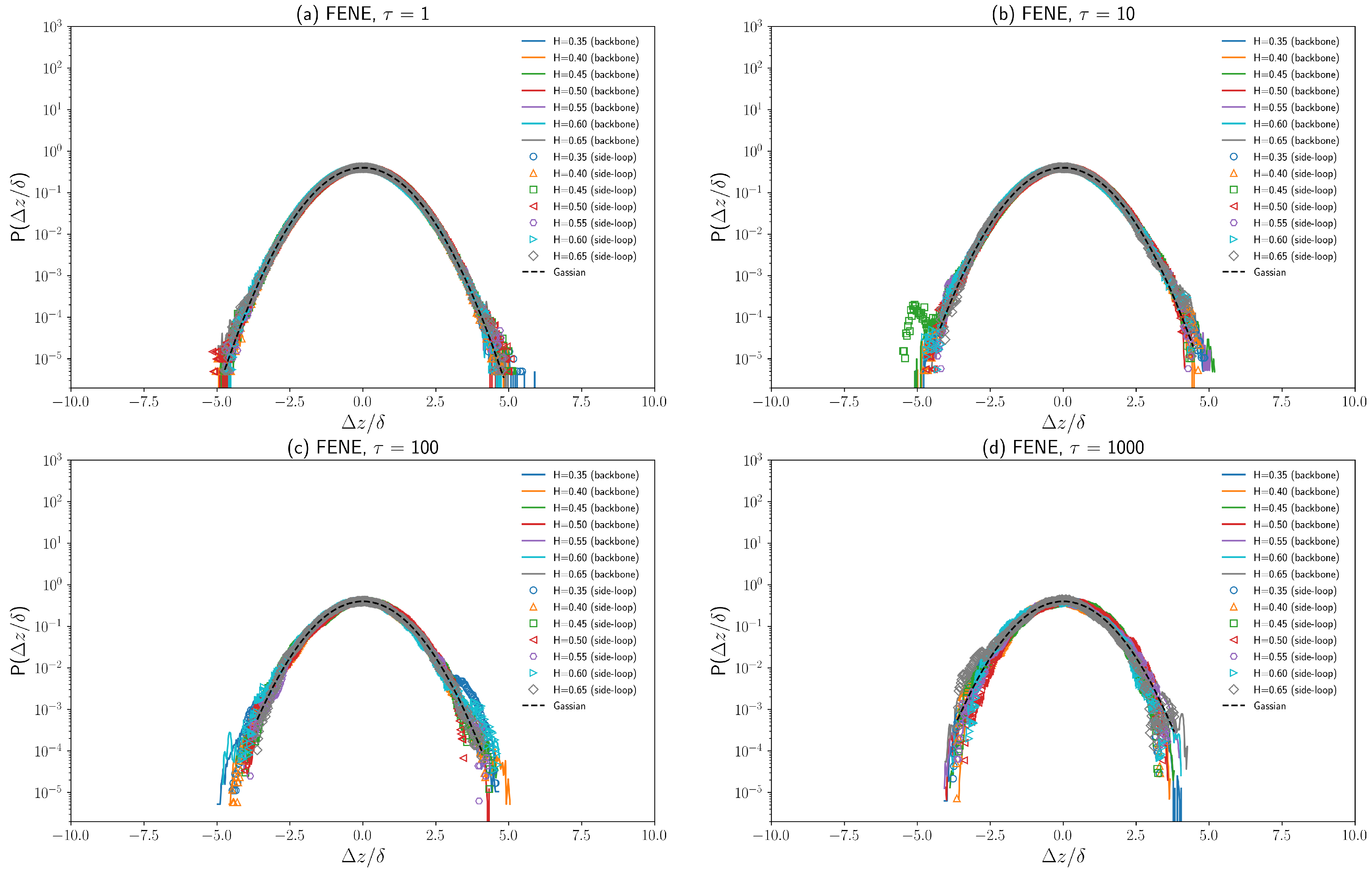
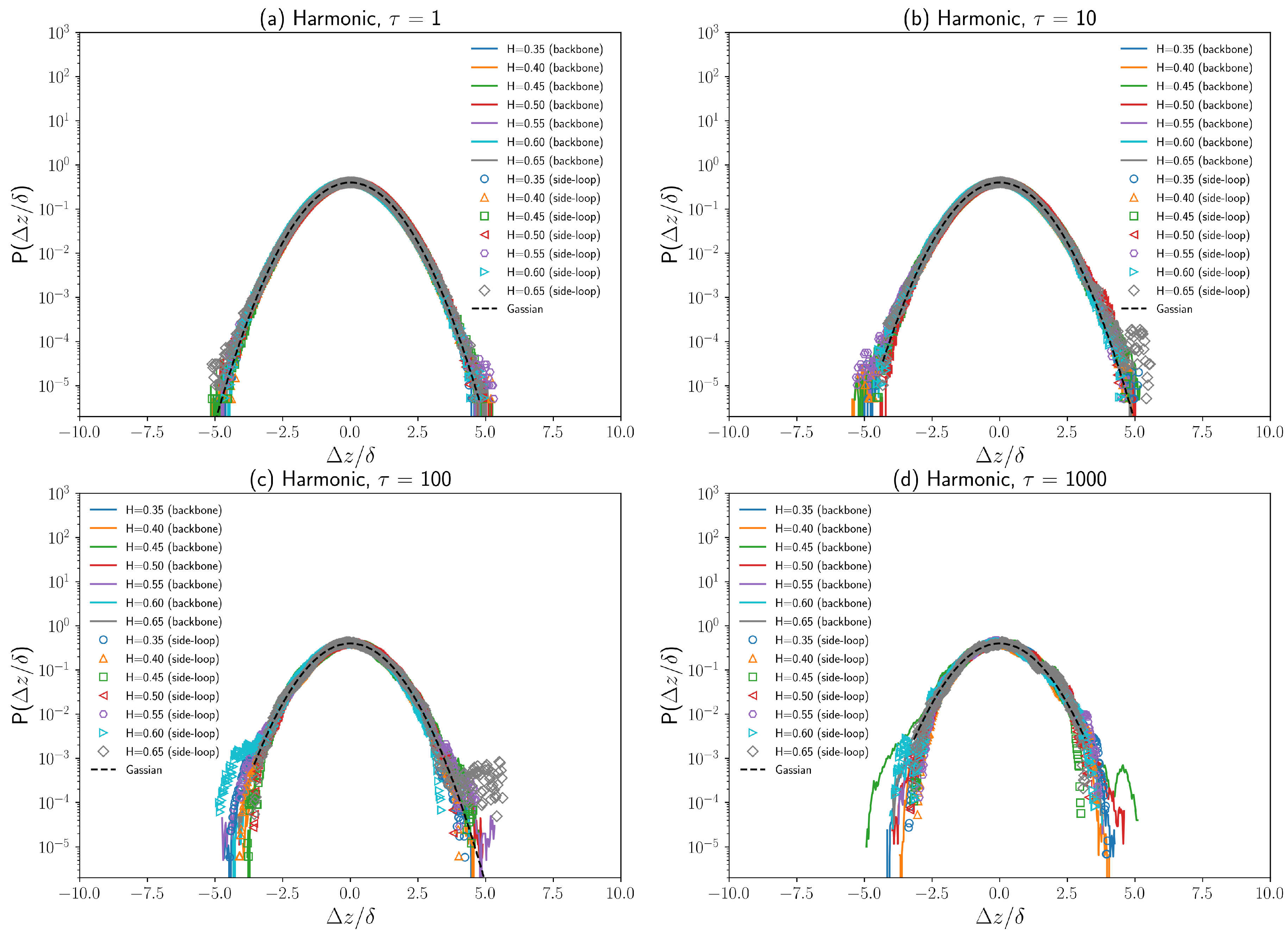
3.3. Displacement Distributions of Polymer Beads with Fractional Brownian Noises
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Soh, S.; Byrska, M.; Kandere-Grzybowska, K.; Grzybowski, B.A. Reaction-diffusion systems in intracellular molecular transport and control. Angew. Chem. Int. Ed. 2010, 49, 4170–4198. [Google Scholar] [CrossRef]
- Schavemaker, P.E.; Boersma, A.J.; Poolman, B. How important is protein diffusion in Prokaryotes? Front. Mol. Biosci. 2018, 5, 93. [Google Scholar] [CrossRef]
- Weber, S.C.; Spakowitz, A.J.; Theriot, J.A. Bacterial chromosomal loci move subdiffusively through a viscoelastic cytoplasm. Phys. Rev. Lett. 2010, 104, 27–30. [Google Scholar] [CrossRef]
- Javer, A.; Long, Z.; Nugent, E.; Grisi, M.; Siriwatwetchakul, K.; Dorfman, K.D.; Cicuta, P.; Cosentino Lagomarsino, M. Short-time movement of E. coli chromosomal loci depends on coordinate and subcellular localization. Nat. Commun. 2013, 4, 3003. [Google Scholar] [CrossRef] [PubMed]
- Javer, A.; Kuwada, N.J.; Long, Z.; Benza, V.G.; Dorfman, K.D.; Wiggins, P.A.; Cicuta, P.; Lagomarsino, M.C. Persistent super-diffusive motion of Escherichia coli chromosomal loci. Nat. Commun. 2014, 5, 3854. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lampo, T.J.; Stylianidou, S.; Backlund, M.P.; Wiggins, P.A.; Spakowitz, A.J. Cytoplasmic RNA-protein particles exhibit non-Gaussian subdiffusive behavior. Biophys. J. 2017, 112, 532–542. [Google Scholar] [CrossRef] [PubMed]
- Parry, B.R.; Surovtsev, I.V.; Cabeen, M.T.; O’Hern, C.S.; Dufresne, E.R.; Jacobs-Wagner, C. The bacterial cytoplasm has glass-like properties and is fluidized by metabolic activity. Cell 2014, 156, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Okumus, B.; Landgraf, D.; Lai, G.C.; Bakhsi, S.; Arias-Castro, J.C.; Yildiz, S.; Huh, D.; Fernandez-Lopez, R.; Peterson, C.N.; Toprak, E.; et al. Mechanical slowing-down of cytoplasmic diffusion allows in vivo counting of proteins in individual cells. Nat. Commun. 2016, 7, 11641. [Google Scholar] [CrossRef]
- Wlodarski, M.; Raciti, B.; Kotar, J.; Lagomarsino, M.C.; Fraser, G.M.; Cicuta, P. Both genome and cytosol dynamics change in E. coli challenged with sublethal rifampicin. Phys. Biol. 2017, 14, 015005. [Google Scholar] [CrossRef]
- Wlodarski, M.; Mancini, L.; Raciti, B.; Sclavi, B.; Lagomarsino, M.C.; Cicuta, P. Cytosolic crowding drives the dynamics of both genome and cytosol in Escherichia coli challenged with sub-lethal antibiotic treatments. iScience 2020, 23, 101560. [Google Scholar] [CrossRef]
- Yu, I.; Mori, T.; Ando, T.; Harada, R.; Jung, J.; Sugita, Y.; Feig, M. Biomolecular interactions modulate macromolecular structure and dynamics in atomistic model of a bacterial cytoplasm. eLife 2016, 5, e19274. [Google Scholar] [CrossRef]
- Yoo, J.; Winogradoff, D.; Aksimentiev, A. Molecular dynamics simulations of DNA–DNA and DNA–protein interactions. Curr. Opin. Struct. Biol. 2020, 64, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Marrink, S.J.; Corradi, V.; Souza, P.C.T.; Ingólfsson, H.I.; Tieleman, D.P.; Sansom, M.S.P. Computational modeling of realistic cell membranes. Chem. Rev. 2019, 119, 6184–6226. [Google Scholar] [CrossRef] [PubMed]
- Uusitalo, J.J.; Ingólfsson, H.I.; Akhshi, P.; Tieleman, D.P.; Marrink, S.J. Martini coarse-grained force field: Extension to DNA. J. Chem. Theory Comput. 2015, 11, 3932–3945. [Google Scholar] [CrossRef]
- Sengar, A.; Ouldridge, T.E.; Henrich, O.; Rovigatti, L.; Šulc, P. A primer on the oxDNA model of DNA: When to use it, how to simulate it and how to interpret the results. Front. Mol. Biosci. 2021, 8, 693710. [Google Scholar] [CrossRef]
- Shinoda, W.; Discher, D.E.; Klein, M.L.; Loverde, S.M. Probing the structure of PEGylated-lipid assemblies by coarse-grained molecular dynamics. Soft Matter 2013, 9, 11549–11556. [Google Scholar] [CrossRef]
- Frembgen-Kesner, T.; Elcock, A.H. Computer simulations of the bacterial cytoplasm. Biophys. Rev. 2013, 5, 109–119. [Google Scholar] [CrossRef]
- Ando, T.; Skolnick, J. Crowding and hydrodynamic interactions likely dominate in vivo macromolecular motion. Proc. Natl. Acad. Sci. USA 2010, 107, 18457–18462. [Google Scholar] [CrossRef] [PubMed]
- Hasnain, S.; McClendon, C.L.; Hsu, M.T.; Jacobson, M.P.; Bandyopadhyay, P. A new coarse-grained model for E. coli cytoplasm: Accurate calculation of the diffusion coefficient of proteins and observation of anomalous diffusion. PLoS ONE 2014, 9, e106466. [Google Scholar] [CrossRef]
- Słyk, E.; Skóra, T.; Kondrat, S. How macromolecules softness affects diffusion under crowding. Soft Matter 2022, 18, 5366–5370. [Google Scholar] [CrossRef]
- Swain, P.; Mulder, B.M.; Chaudhuri, D. Confinement and crowding control the morphology and dynamics of a model bacterial chromosome. Soft Matter 2019, 15, 2677–2687. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, D.; Mulder, B.M. Spontaneous helicity of a polymer with side loops confined to a cylinder. Phys. Rev. Lett. 2012, 108, 268305. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, D.; Mulder, B.M. Molecular dynamics simulation of a feather-boa model of a bacterial chromosome. Methods Mol. Biol. 2018, 1837, 403–415. [Google Scholar] [PubMed]
- Berlatzky, I.A.; Rouvinski, A.; Ben-Yehuda, S. Spatial organization of a replicating bacterial chromosome. Proc. Natl. Acad. Sci. USA 2008, 105, 14136–14140. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.; Murray, S.M. Subdiffusive movement of chromosomal loci in bacteria explained by DNA bridging. Phys. Rev. Res. 2023, 5, 023034. [Google Scholar] [CrossRef]
- Ernst, D.; Hellmann, M.; Köhler, J.; Weiss, M. Fractional Brownian motion in crowded fluids. Soft Matter 2012, 8, 4886–4889. [Google Scholar] [CrossRef]
- Mardoukhi, Y.; Jeon, J.-H.; Metzler, R. Geometry controlled anomalous diffusion in random fractal geometries: Looking beyond the infinite cluster. Phys. Chem. Chem. Phys. 2015, 17, 30134–30147. [Google Scholar] [CrossRef]
- Magdziarz, M.; Weron, A.; Burnecki, K.; Klafter, J. Fractional Brownian motion versus the continuous-time random walk: A simple test for subdiffusive dynamics. Phys. Rev. Lett. 2009, 103, 180602. [Google Scholar] [CrossRef]
- Weber, S.C.; Theriot, J.A.; Spakowitz, A.J. Subdiffusive motion of a polymer composed of subdiffusive monomers. Phys. Rev. E 2010, 82, 011913. [Google Scholar] [CrossRef]
- Weiss, M. Single-particle tracking data reveal anticorrelated fractional Brownian motion in crowded fluids. Phys. Rev. E 2013, 88, 010101. [Google Scholar] [CrossRef] [PubMed]
- Metzler, R.; Klafter, J. The random walk’s guide to anomalous diffusion: A fractional dynamics approach. Phys. Rep. 2000, 339, 1–77. [Google Scholar] [CrossRef]
- Chechkin, A.V.; Seno, F.; Metzler, R.; Sokolov, I.M. Brownian yet non-Gaussian diffusion: From superstatistics to subordination of diffusing diffusivities. Phys. Rev. X 2017, 7, 021002. [Google Scholar] [CrossRef]
- Chubynsky, M.V.; Slater, G.W. Diffusing diffusivity: A model for anomalous, yet Brownian, diffusion. Phys. Rev. Lett. 2014, 113, 098302. [Google Scholar] [CrossRef]
- dos Santos, M.A.F.; Junior, L.M. Random diffusivity models for scaled Brownian motion. Chaos Solit. Fractals 2021, 144, 110634. [Google Scholar] [CrossRef]
- Spakowitz, A.J. Transient anomalous diffusion in a heterogeneous environment. Front. Phys. 2019, 7, 119. [Google Scholar] [CrossRef]
- Baldovin, F.; Orlandini, E.; Seno, F. Polymerization induces non-Gaussian diffusion. Front. Phys. 2019, 7, 124. [Google Scholar] [CrossRef]
- Fodor, É.; Guo, M.; Gov, N.S.; Visco, P.; Weitz, D.A.; van Wijland, F. Activity-driven fluctuations in living cells. EPL 2015, 110, 48005. [Google Scholar] [CrossRef]
- Metzler, R.; Klafter, J. Accelerating Brownian motion: A fractional dynamics approach to fast diffusion. Europhys. Lett. 2000, 51, 492–498. [Google Scholar] [CrossRef]
- Florescu, A.-M.; Joyeux, M. Description of nonspecific DNA-protein interaction and facilitated diffusion with a dynamical model. J. Chem. Phys. 2009, 130, 015103. [Google Scholar] [CrossRef]
- Mandelbrot, B.B.; van Ness, J.W. Fractional Brownian motions, fractional noises and applications. SIAM Rev. 1968, 10, 422–437. [Google Scholar] [CrossRef]
- Dieker, A.B.; Mandjes, M. On spectral simulation of fractional Brownian motion. Probab. Eng. Inf. Sci. 2003, 17, 417–434. [Google Scholar] [CrossRef]
- Davies, R.B.; Harte, D.S. Tests for Hurst Effect. Biometrika 1987, 74, 95–101. [Google Scholar] [CrossRef]
- Ha, B.-Y.; Jung, Y. Polymers under confinement: Single polymers, how they interact, and as model chromosomes. Soft Matter 2015, 11, 2333–2352. [Google Scholar] [CrossRef]
- Yu, S.; Dorfman, K.D.; Sheats, J.; Sclavi, B.; Cosentino Lagomarsino, M.; Cicuta, P. Subdiffusion of loci and cytoplasmic particles are different in compressed Escherichia coli cells. Commun. Biol. 2018, 1, 176. [Google Scholar] [CrossRef]
- Jeon, C.; Jung, Y.; Ha, B.-Y. A ring-polymer model shows how macromolecular crowding controls chromosome-arm organization in Escherichia coli. Sci. Rep. 2017, 7, 11896. [Google Scholar] [CrossRef]
- Chauhan, G.; Simpson, M.L.; Abel, S.M. Crowding-induced interactions of ring polymers. Soft Matter 2021, 17, 16–23. [Google Scholar] [CrossRef]
- Si, F.; Li, B.; Margolin, W.; Sun, S.X. Bacterial growth and form under mechanical compression. Sci. Rep. 2015, 5, 11367. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, S.; Chu, R.; Wu, G.; Meng, X. A Novel Fractional Brownian Dynamics Method for Simulating the Dynamics of Confined Bottle-Brush Polymers in Viscoelastic Solution. Polymers 2024, 16, 524. https://doi.org/10.3390/polym16040524
Yu S, Chu R, Wu G, Meng X. A Novel Fractional Brownian Dynamics Method for Simulating the Dynamics of Confined Bottle-Brush Polymers in Viscoelastic Solution. Polymers. 2024; 16(4):524. https://doi.org/10.3390/polym16040524
Chicago/Turabian StyleYu, Shi, Ruizhi Chu, Guoguang Wu, and Xianliang Meng. 2024. "A Novel Fractional Brownian Dynamics Method for Simulating the Dynamics of Confined Bottle-Brush Polymers in Viscoelastic Solution" Polymers 16, no. 4: 524. https://doi.org/10.3390/polym16040524
APA StyleYu, S., Chu, R., Wu, G., & Meng, X. (2024). A Novel Fractional Brownian Dynamics Method for Simulating the Dynamics of Confined Bottle-Brush Polymers in Viscoelastic Solution. Polymers, 16(4), 524. https://doi.org/10.3390/polym16040524






