Polypy: A Framework to Interpret Polymer Properties from Mass Spectrometry Data
Abstract
1. Introduction
2. Materials and Methods
2.1. Electrospray Ionization–Time of Flight–Mass Spectrometry (ESI-µTOF-MS)
2.2. Spectra Interpretation through Python Algorithm—Polypy
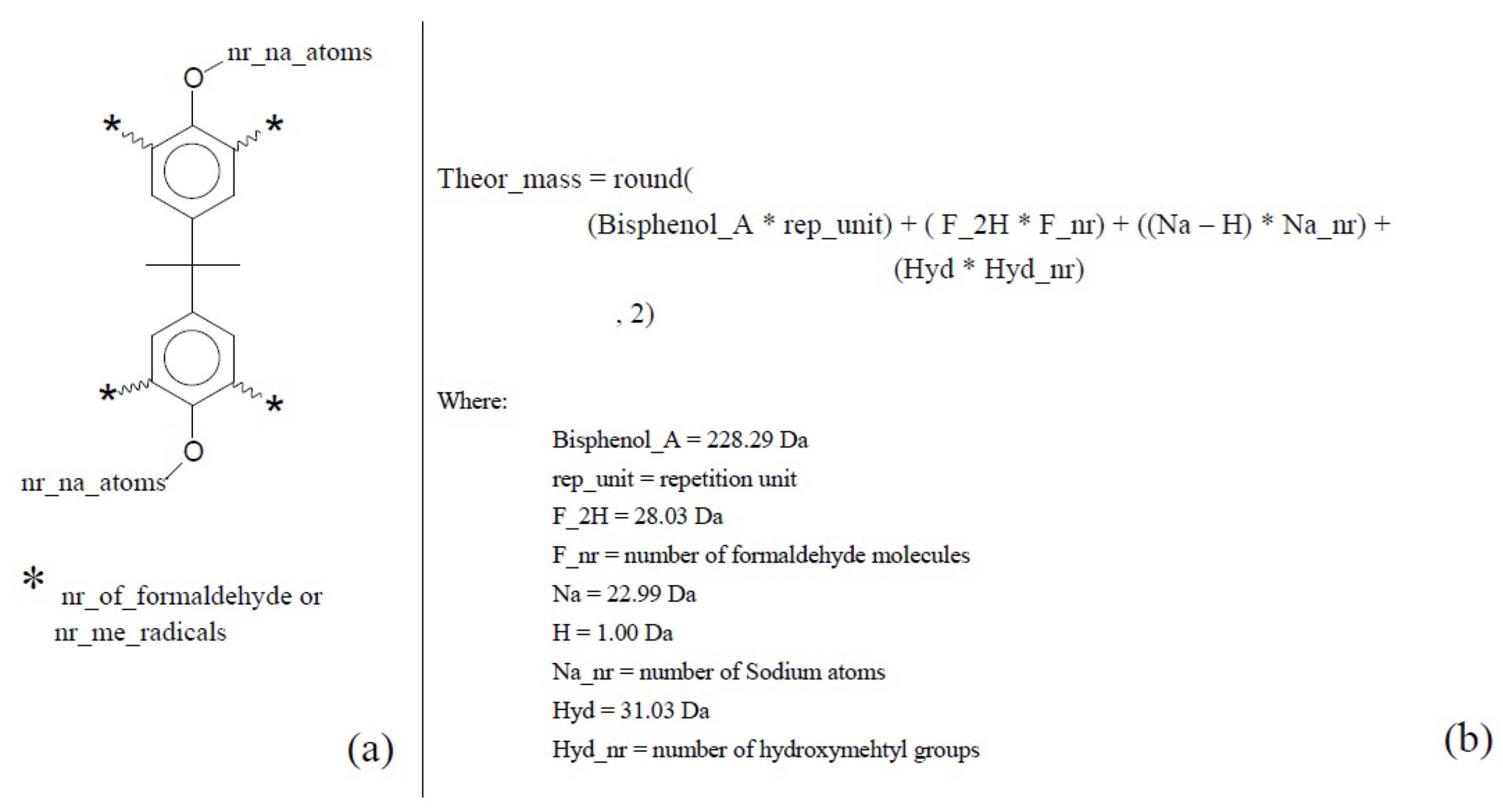
- (I)
- Theoretical masses calculation;
- (II)
- MS raw data reading and filtering;
- (III)
- Background signal noise filtering;
- (IV)
- Classification of neat, derivative, and side-product peaks;
- (V)
- Separation of repetitive unit regions and distribution percentage calculation;
- (VI)
- Polymer properties calculation;
- (VII)
- Polypy and MS caveats.
3. Results and Discussion
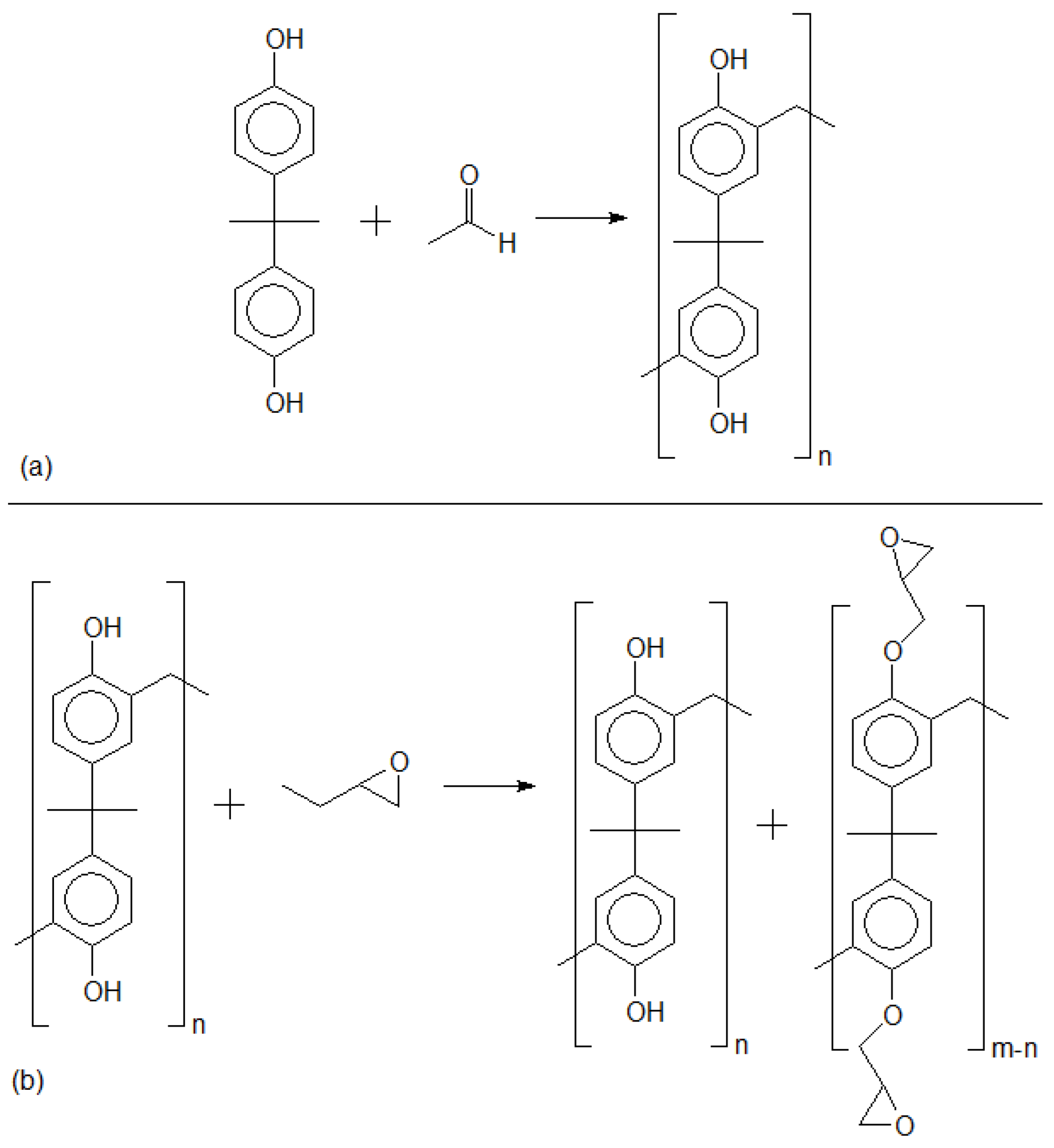
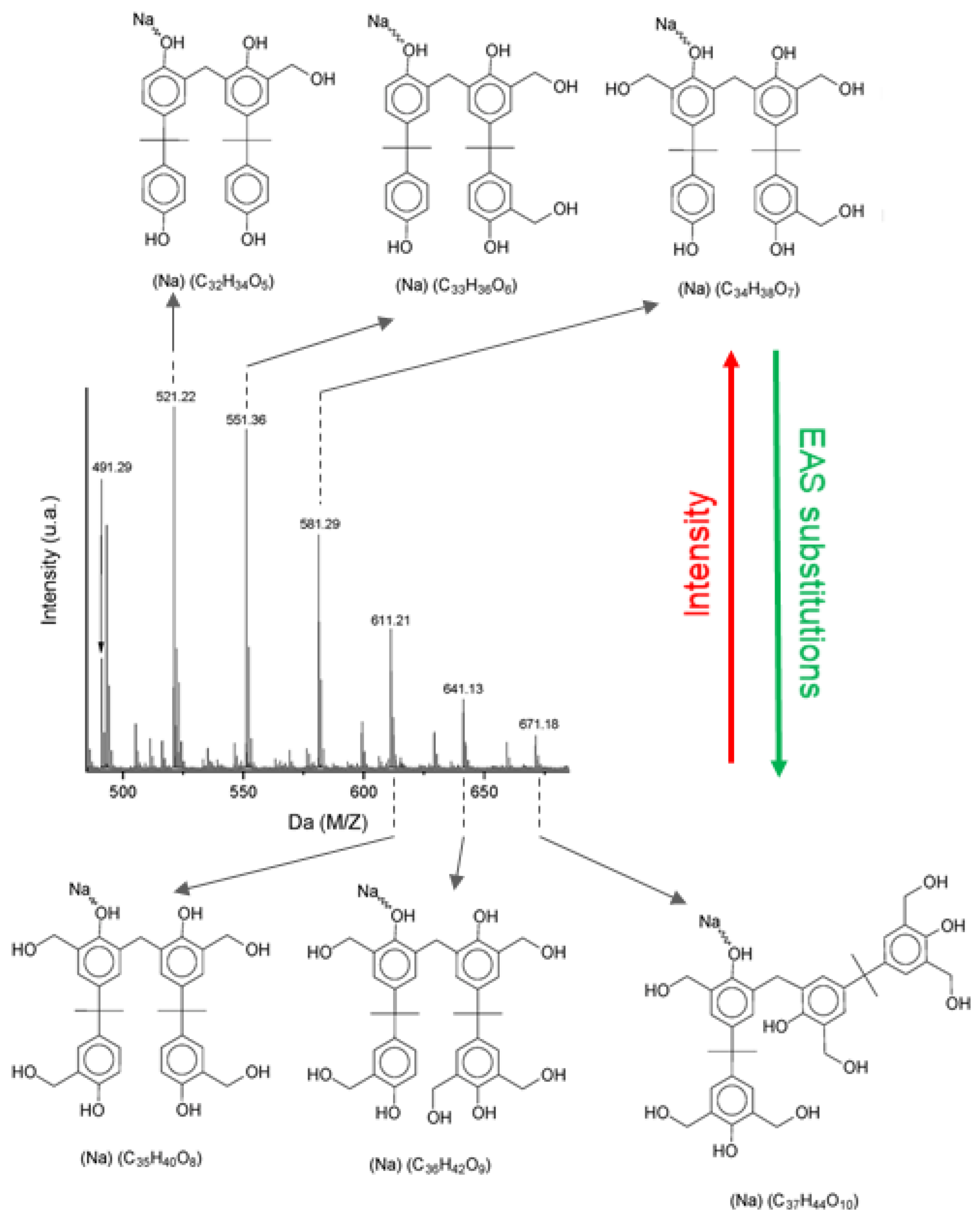
3.1. Aryl Resins—Neat, Derivative, and Adduct Product Definitions
- -
- Repetitive units, considered neat products from polymerization.
- -
- Derivatives, which are repetitive units added by an n numbers of hydroxymethyl groups.
- -
- Adducts, which are usually one of the categories above, with sodium atoms added at phenolic positions.
3.2. Polypy Interpreter
- (I)
- MS theoretical masses calculation
- (II)
- Raw data reading
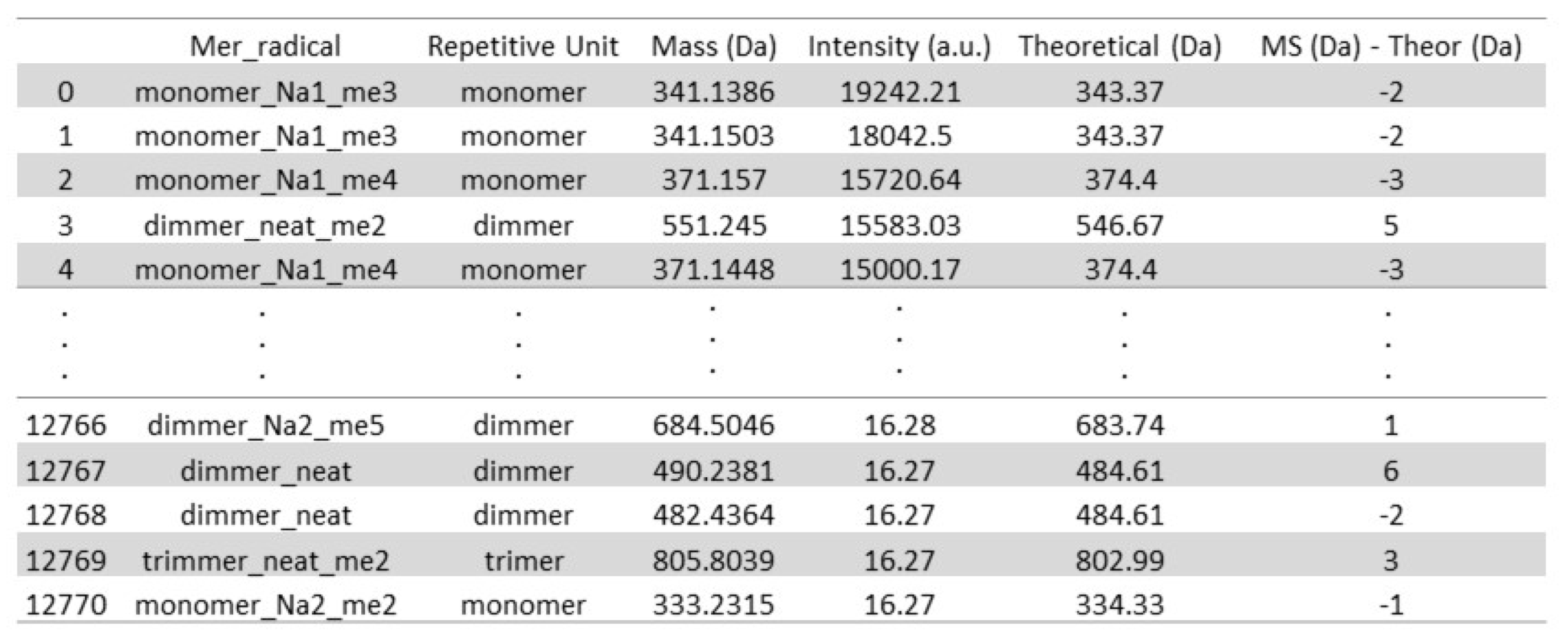
- (III)
- Background signal noise filtering
- (IV)
- Separation of neat, derivative, and adduct peaks
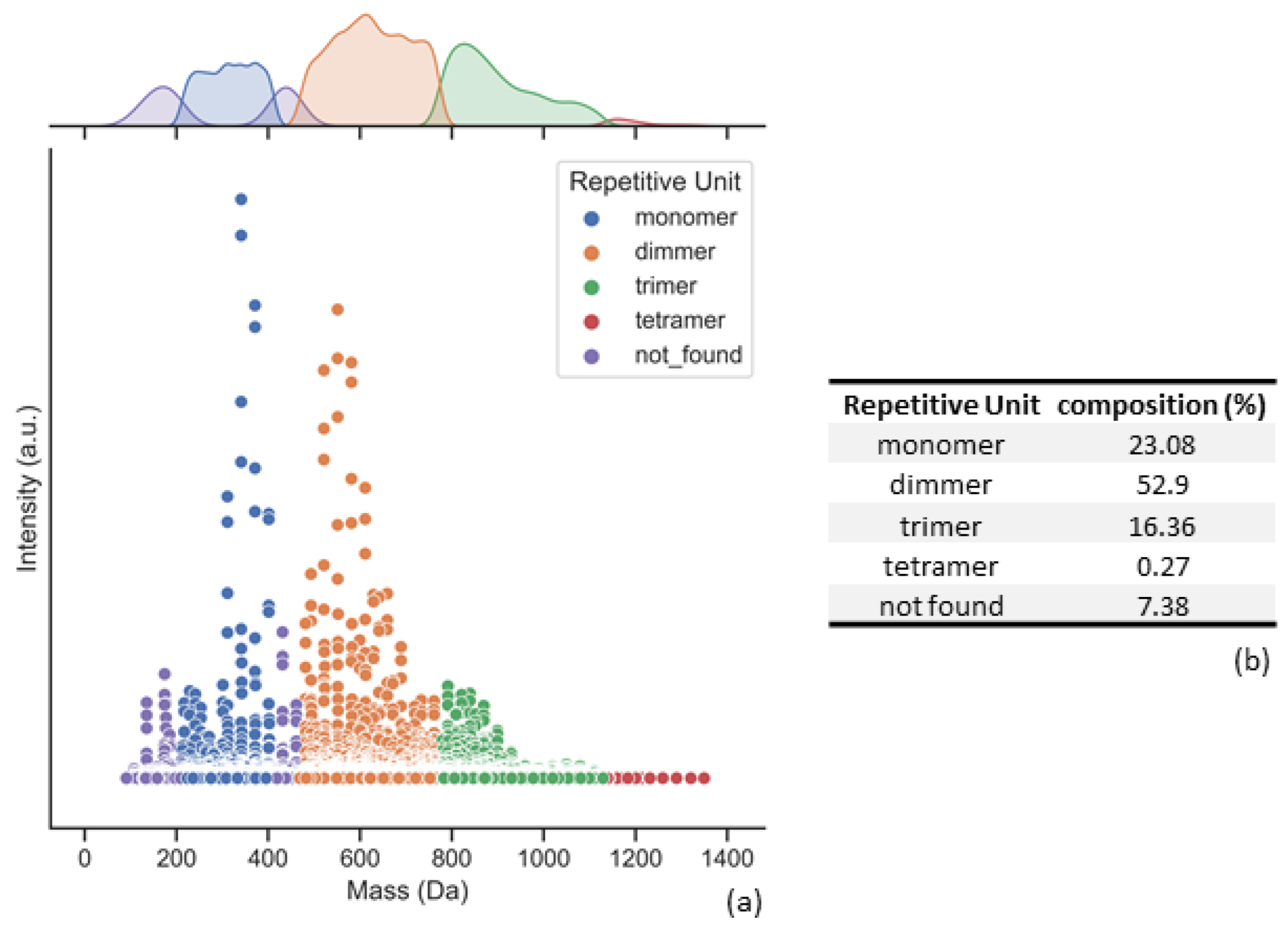
- (V)
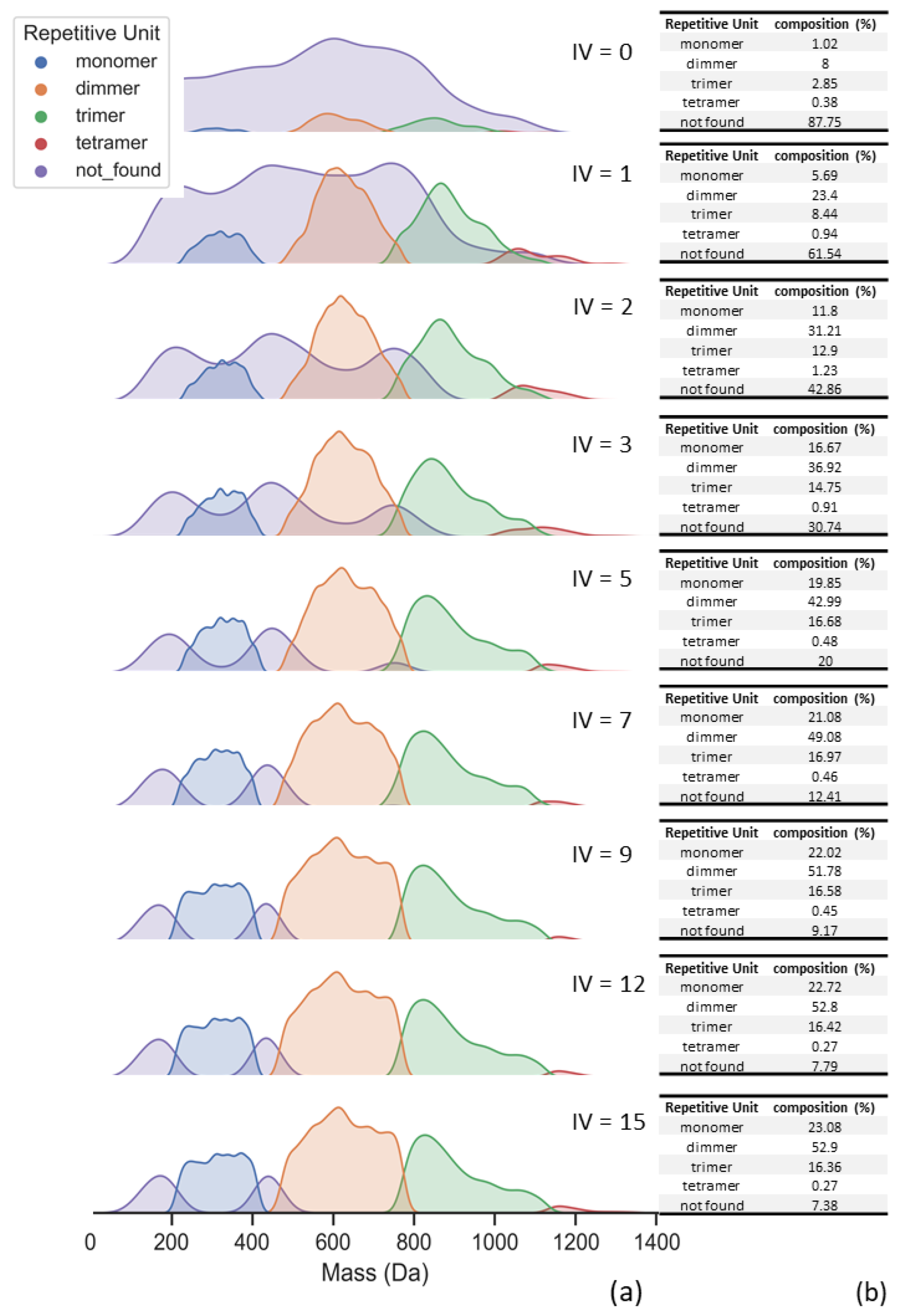
- Separation of repetitive unit regions and distribution percentage calculation
- (VI)
- Polymer property calculations
- (VII)
- Polypy and Mass Spectrometry caveats
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
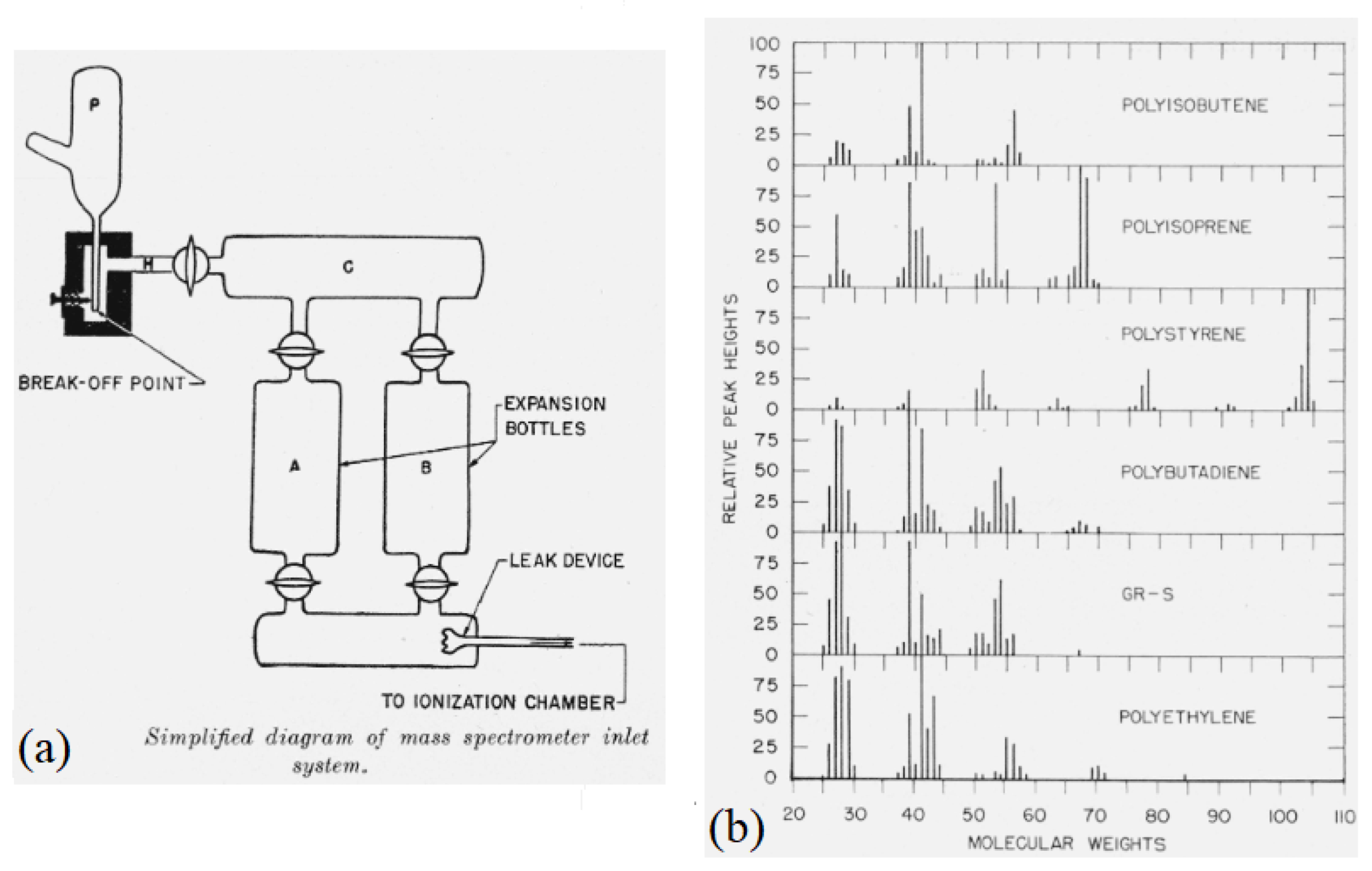
- Gruendling, T.; Weidner, S.; Falkenhagen, J.; Barner-Kowollik, C. Mass spectrometry in polymer chemistry: A state-of-the-art up-date. Polym. Chem. 2010, 1, 599–617. [Google Scholar] [CrossRef]
- Gies, A.P. Ionization Techniques for Polymer Mass Spectrometry. In Mass Spectrometry in Polymer Chemistry; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2011; Chapter 2; pp. 33–56. [Google Scholar] [CrossRef]
- De Bruycker, K.; Welle, A.; Hirth, S.; Blanksby, S.J.; Barner-Kowollik, C. Mass spectrometry as a tool to advance polymer science. Nat. Rev. Chem. 2020, 4, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Lietz, C.B.; Richards, A.L.; Marshall, D.D.; Ren, Y.; Trimpin, S. Matrix-Assisted Inlet Ionization and Solvent-Free Gas-Phase Separation Using Ion Mobility Spectrometry for Imaging and Electron Transfer Dissociation Mass Spectrometry of Polymers. In Mass Spectrometry in Polymer Chemistry; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2011; Chapter 4; pp. 85–118. [Google Scholar] [CrossRef]
- Lark-Horovitz, K.; Johnson, V.A. (Eds.) 2. Preparation and Purification of Materials. In Solid State Physics: Preparation, Structure, Mechanical and Thermal Properties; Methods in Experimental Physics; Academic Press: Cambridge, MA, USA, 1959; Volume 6, pp. 21–186. [Google Scholar] [CrossRef]
- Anufriev, G.; Pozdnyakov, O.; Regel, V. Mass-spectrometry method of studying the thermal degradation of polymers. Polym. Sci. U.S.S.R. 1966, 8, 916–923. [Google Scholar] [CrossRef]
- Zemany, P.D. Thermal Degradation of Polystyrene. Nature 1953, 171, 391–392. [Google Scholar] [CrossRef]
- Nakagawa, K. Mass Spectrometric Study on the Evaporation of Volatile Components in Polyethylene. J. Phys. Soc. Jpn. 1961, 16, 741–745. [Google Scholar] [CrossRef]
- Charlesby, A.; Callaghan, L. Crystal distribution in various polyethylenes. J. Phys. Chem. Solids 1958, 4, 227–238. [Google Scholar] [CrossRef]
- Waldron, J. (Ed.) Bibliography on Mass Spectrometry 1938–1957 inclusive. In Advances in Mass Spectrometry; Pergamon: Oxford, UK, 1959; pp. 592–689. [Google Scholar] [CrossRef]
- Madorsky, S.L.; Straus, J. Pyrolytic fractionation of polystyrene in a high vacuum and mass spectrometer analysis of some of the fractions. J. Res. Natl. Bur. Stand. 1948, 40, 417–425. [Google Scholar] [CrossRef]
- Ciapetta, F.G.; Macuga, S.J.; Leum, L.N. Depolymerization of Butylene Polymers. Anal. Chem. 1948, 20, 699–704. [Google Scholar] [CrossRef]
- Wall, L.A. Mass spectrometric investigation of the thermal decomposition of polymers. J. Res. Od Natl. Bur. Stand. 1948, 41, 315–322. [Google Scholar] [CrossRef]
- Samuel L., M.; Sidney, S.; Dorothy, T.; Laura, W. Pyrolysis of polyisobutene (Vistanex) polyisoprene, polybutadiene GR-S and polyethylene in a high vacuum. J. Res. Natl. Bur. Stand. 1949, 42, 499–514. [Google Scholar]
- Roboz, J. Mass Spectrometry in Clinical Chemistry; Advances in Clinical Chemistry; Elsevier: Amsterdam, The Netherlands, 1975; Volume 17, pp. 109–191. [Google Scholar] [CrossRef]
- Glish, G.; Vachet, R. The basics of mass spectrometry in the twenty-first century. Nat. Rev. Drug Discov. 2003, 1, 140–150. [Google Scholar] [CrossRef]
- Kenndler, E.; Schmid, E.R. Chapter 10 Combination of Liquid Chromatography and Mass Spectrometry. In Instrumentation for High-Performance Liquid Chromatography; Huber, J., Ed.; Journal of Chromatography Library; Elsevier: Amsterdam, The Netherlands, 1978; Volume 13, pp. 163–177. [Google Scholar] [CrossRef]
- Hillenkamp, F.; Karas, M. Matrix-assisted laser desorption/ionisation, an experience. Int. J. Mass Spectrom. 2000, 200, 71–77. [Google Scholar] [CrossRef]
- Posthumus, M.A.; Kistemaker, P.G.; Meuzelaar, H.L.C.; Ten Noever de Brauw, M.C. Laser desorption-mass spectrometry of polar nonvolatile bio-organic molecules. Anal. Chem. 1978, 50, 985–991. [Google Scholar] [CrossRef]
- Fréchet, J.M.; Hawker, C.J. 3—Synthesis and Properties of Dendrimers and Hyperbranched Polymers. In Comprehensive Polymer Science and Supplements; Allen, G., Bevington, J.C., Eds.; Pergamon: Amsterdam, The Netherlands, 1989; pp. 71–132. [Google Scholar] [CrossRef]
- Weidner, S.M.; Falkenhagen, J. LC-MALDI MS for Polymer Characterization. In Maldi Mass Spectrometry for Synthetic Polymer Analysis; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2009; Chapter 11; pp. 247–265. [Google Scholar] [CrossRef]
- Hanton, S.D. Mass Spectrometry of Polymers and Polymer Surfaces. Chem. Rev. 2001, 101, 527–570. [Google Scholar] [CrossRef] [PubMed]
- Polce, M.J.; Wesdemiotis, C. Tandem Mass Spectrometry and Polymer Ion Dissociation. In Maldi Mass Spectrometry for Synthetic Polymer Analysis; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2009; Chapter 5; pp. 85–127. [Google Scholar] [CrossRef]
- Falkenhagen, J.; Weidner, S. Hyphenated Techniques. In Mass Spectrometry in Polymer Chemistry; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2011; Chapter 7; pp. 209–235. [Google Scholar] [CrossRef]
- Hu, X.; Mar, D.; Suzuki, N.; Zhang, B.; Peter, K.T.; Edward, P.; Kolodziej, D.A.C.B. Mass-Suite: A novel open-source python package for high-resolution mass spectrometry data analysis. J. Cheminform. 2023, 15, 87. [Google Scholar] [CrossRef] [PubMed]
- Dávila-Santiago, E.; Shi, C.; Mahadwar, G.; Medeghini, B.; Insinga, L.; Hutchinson, R.; Good, S.; Jones, G.D. Machine Learning Applications for Chemical Fingerprinting and Environmental Source Tracking Using Non-target Chemical Data. Environ. Sci. Technol. 2022, 56, 4080–4090. [Google Scholar] [CrossRef] [PubMed]
- Nikolopoulou, V.; Aalizadeh, R.; Nika, M.C.; Thomaidis, N.S. TrendProbe: Time profile analysis of emerging contaminants by LC-HRMS non-target screening and deep learning convolutional neural network. J. Hazard. Mater. 2022, 428, 128194. [Google Scholar] [CrossRef] [PubMed]
- Helmus, R.; ter Laak, T.L.; van Wezel, A.P.; de Voogt, P.; Schymanski, E.L. patRoon: Open source software platform for environmental mass spectrometry based non-target screening. J. Cheminform. 2021, 13, 1. [Google Scholar] [CrossRef] [PubMed]
- Liebal, U.W.; Phan, A.N.T.; Sudhakar, M.; Raman, K.; Blank, L.M. Machine Learning Applications for Mass Spectrometry-Based Metabolomics. Metabolites 2020, 10, 243. [Google Scholar] [CrossRef]
- Melnikov, A.D.; Tsentalovich, Y.P.; Yanshole, V.V. Deep Learning for the Precise Peak Detection in High-Resolution LC–MS Data. Anal. Chem. 2020, 92, 588–592. [Google Scholar] [CrossRef]
- Riquelme, G.; Zabalegui, N.; Pablo Marchi, C.M.J.; Monge, M.E. A Python-Based Pipeline for Preprocessing LC–MS Data for Untargeted Metabolomics Workflows. Metabolites 2020, 10, 416. [Google Scholar] [CrossRef] [PubMed]
- Levitsky, L.I.; Klein, J.A.; Ivanov, M.V.; Gorshkov, M.V. Pyteomics 4.0: Five Years of Development of a Python Proteomics Framework. J. Proteome Res. 2019, 18, 709–714. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef] [PubMed]
- Röst, H.L.; Sachsenberg, T.; Aiche, S.; Bielow, C.; Weisser, H.; Aicheler, F.; Andreotti, S.; Ehrlich, H.C.; Gutenbrunner, P.; Kenar, E.; et al. OpenMS: A flexible open-source software platform for mass spectrometry data analysis. Nat. Methods 2016, 13, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Uppal, K.; Soltow, Q.A.; Strobel, F.H.; Pittard, W.S.; Gernert, K.M.; Yu, T.; Jones, D.P. xMSanalyzer: Automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinform. 2013, 14, 15. [Google Scholar] [CrossRef] [PubMed]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed]
- Vlnieska, V.; Mikhaylov, A.; Zakharova, M.; Blasco, E.; Kunka, D. Epoxy Resins for Negative Tone Photoresists. Polymers 2019, 11, 1457. [Google Scholar] [CrossRef] [PubMed]
- Achilias, D.S. A Review of Modeling of Diffusion Controlled Polymerization Reactions. Macromol. Theory Simul. 2007, 16, 319–347. [Google Scholar] [CrossRef]
- Kruve, A.; Kaupmees, K. Adduct Formation in ESI/MS by Mobile Phase Additives. J. Am. Soc. Mass Spectrom. 2017, 28, 887–894. [Google Scholar] [CrossRef] [PubMed]
- Vlnieska, V.; Zakharova, M.; Börner, M.; Bade, K.; Mohr, J.; Kunka, D. Chemical and Molecular Variations in Commercial Epoxide Photoresists for X-ray Lithography. Appl. Sci. 2018, 8, 528. [Google Scholar] [CrossRef]
- Robert, J.; Young, P.A.L. Introduction to Polymers, 3rd ed.; CRC Press: Boca Raton, FL, USA, 2011; pp. 262–265. [Google Scholar]
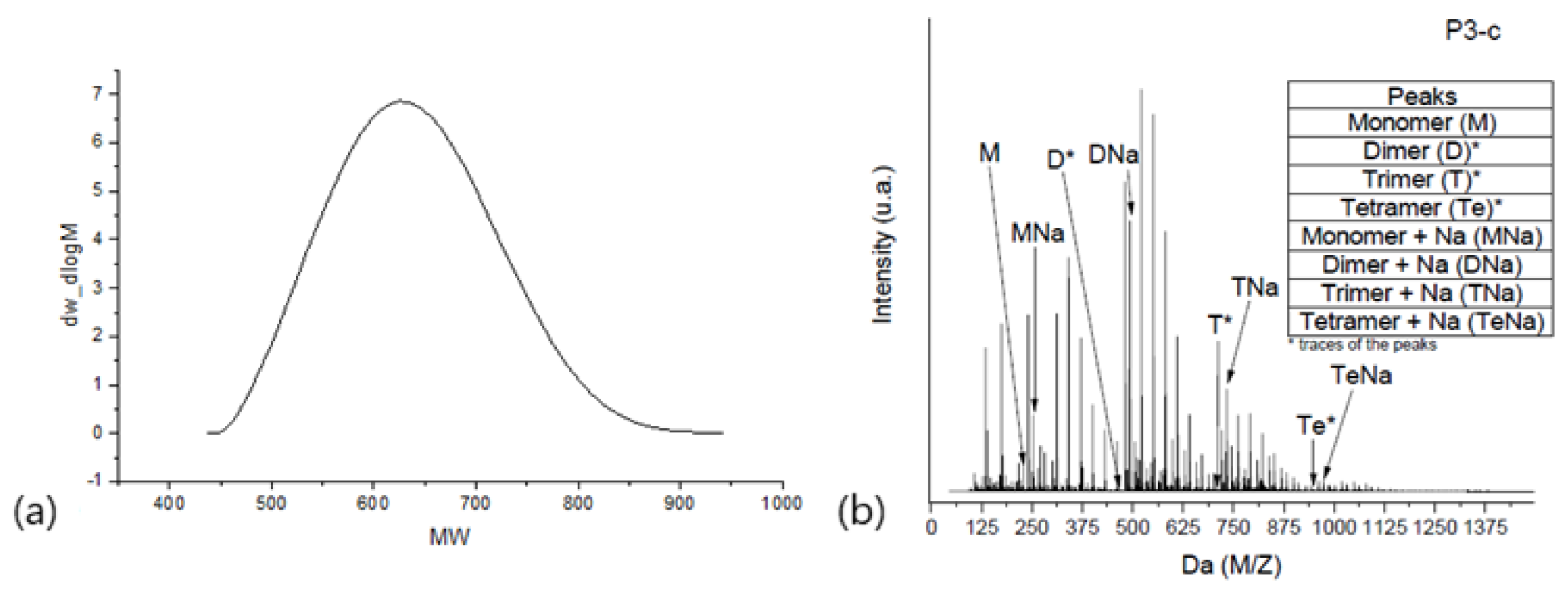





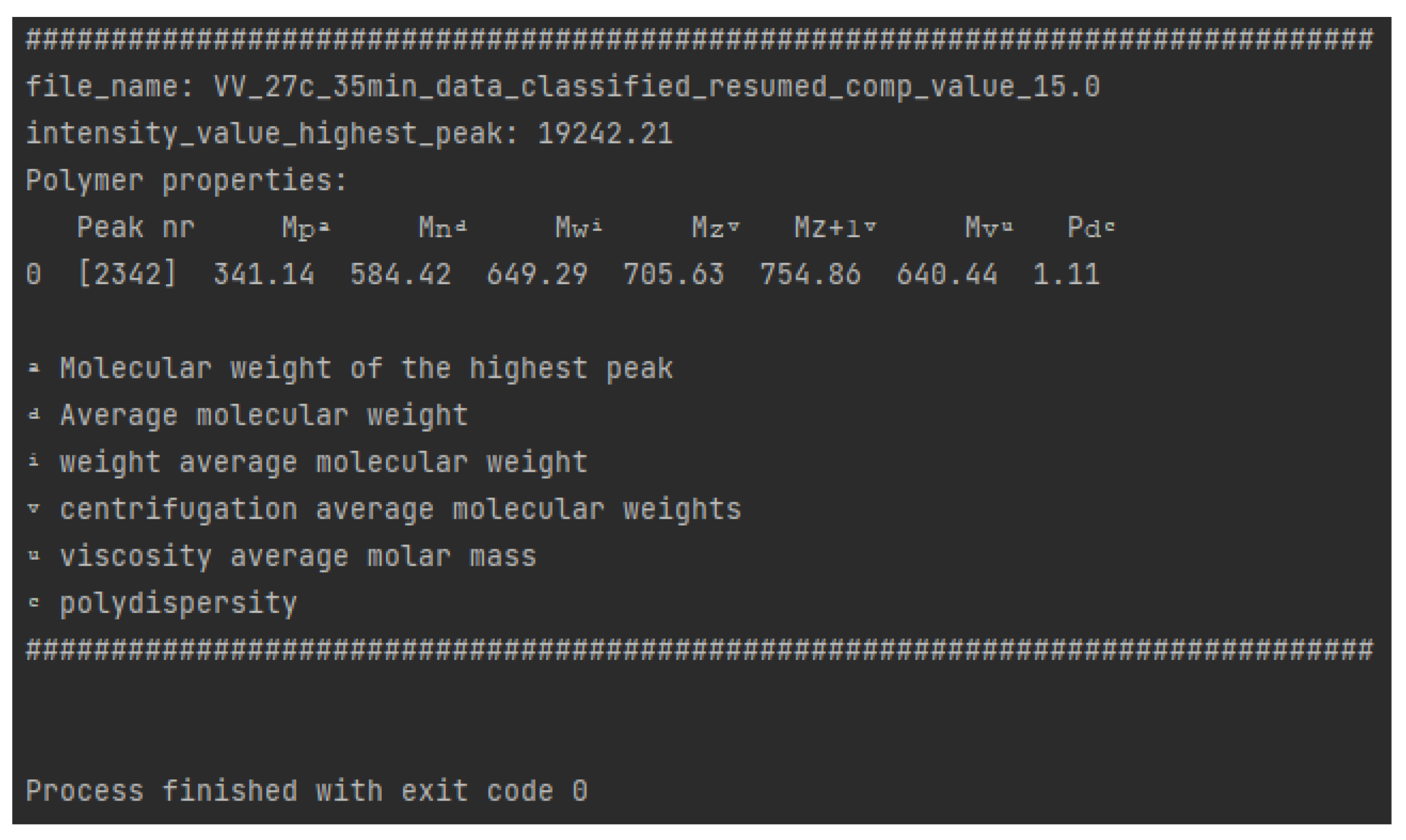
| Peak nr | Mp a | Mn d | Mw i | Mz v | Mz+1 v | Mv u | Pd c |
|---|---|---|---|---|---|---|---|
| 1 | 625 | 622 | 631 | 641 | 651 | 630 | 1.01447 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vlnieska, V.; Khanda, A.; Gilshtein, E.; Beltrán, J.L.; Heier, J.; Kunka, D. Polypy: A Framework to Interpret Polymer Properties from Mass Spectrometry Data. Polymers 2024, 16, 1771. https://doi.org/10.3390/polym16131771
Vlnieska V, Khanda A, Gilshtein E, Beltrán JL, Heier J, Kunka D. Polypy: A Framework to Interpret Polymer Properties from Mass Spectrometry Data. Polymers. 2024; 16(13):1771. https://doi.org/10.3390/polym16131771
Chicago/Turabian StyleVlnieska, Vitor, Ankita Khanda, Evgeniia Gilshtein, Jorge Luis Beltrán, Jakob Heier, and Danays Kunka. 2024. "Polypy: A Framework to Interpret Polymer Properties from Mass Spectrometry Data" Polymers 16, no. 13: 1771. https://doi.org/10.3390/polym16131771
APA StyleVlnieska, V., Khanda, A., Gilshtein, E., Beltrán, J. L., Heier, J., & Kunka, D. (2024). Polypy: A Framework to Interpret Polymer Properties from Mass Spectrometry Data. Polymers, 16(13), 1771. https://doi.org/10.3390/polym16131771





