Towards Electrochemical Sensor Based on Molecularly Imprinted Polypyrrole for the Detection of Bacteria—Listeria monocytogenes
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Electrochemical Measurements
2.2. Pre-Treatment of Working Electrodes
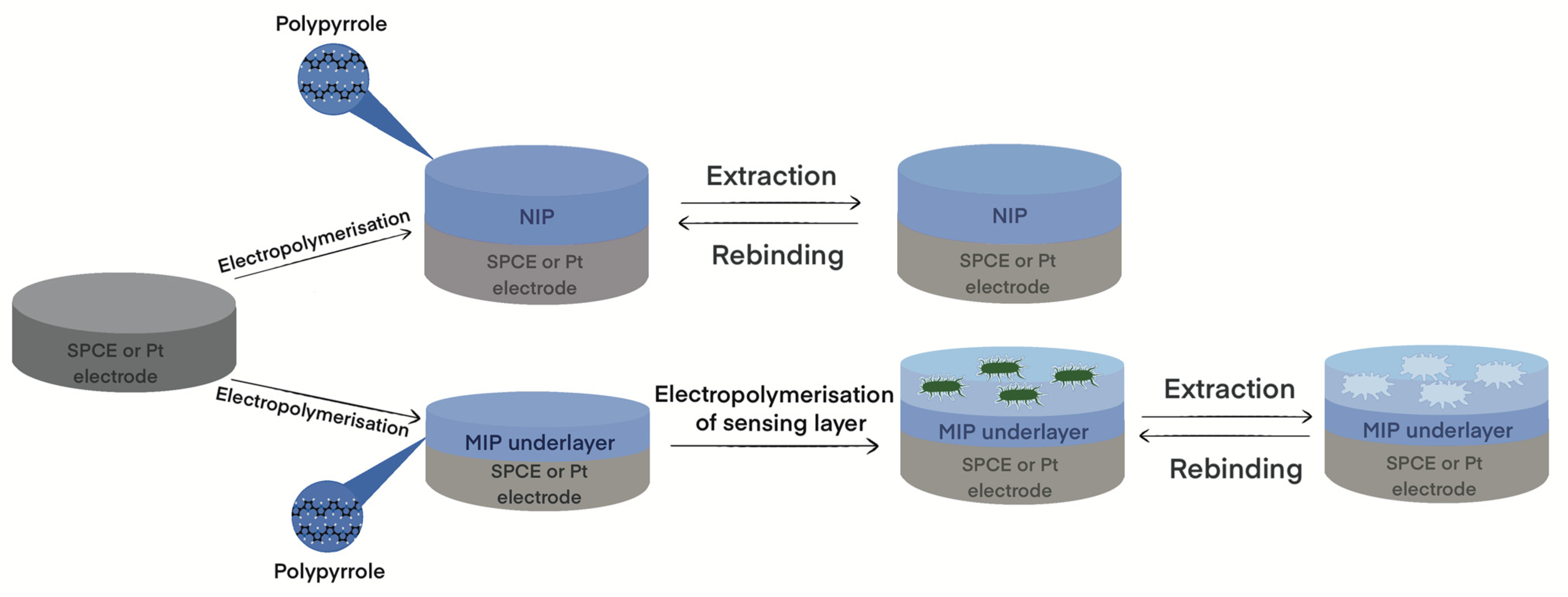
2.3. Electrochemical Modification of Electrodes by NIP-Ppy and MIP-Ppy Layers
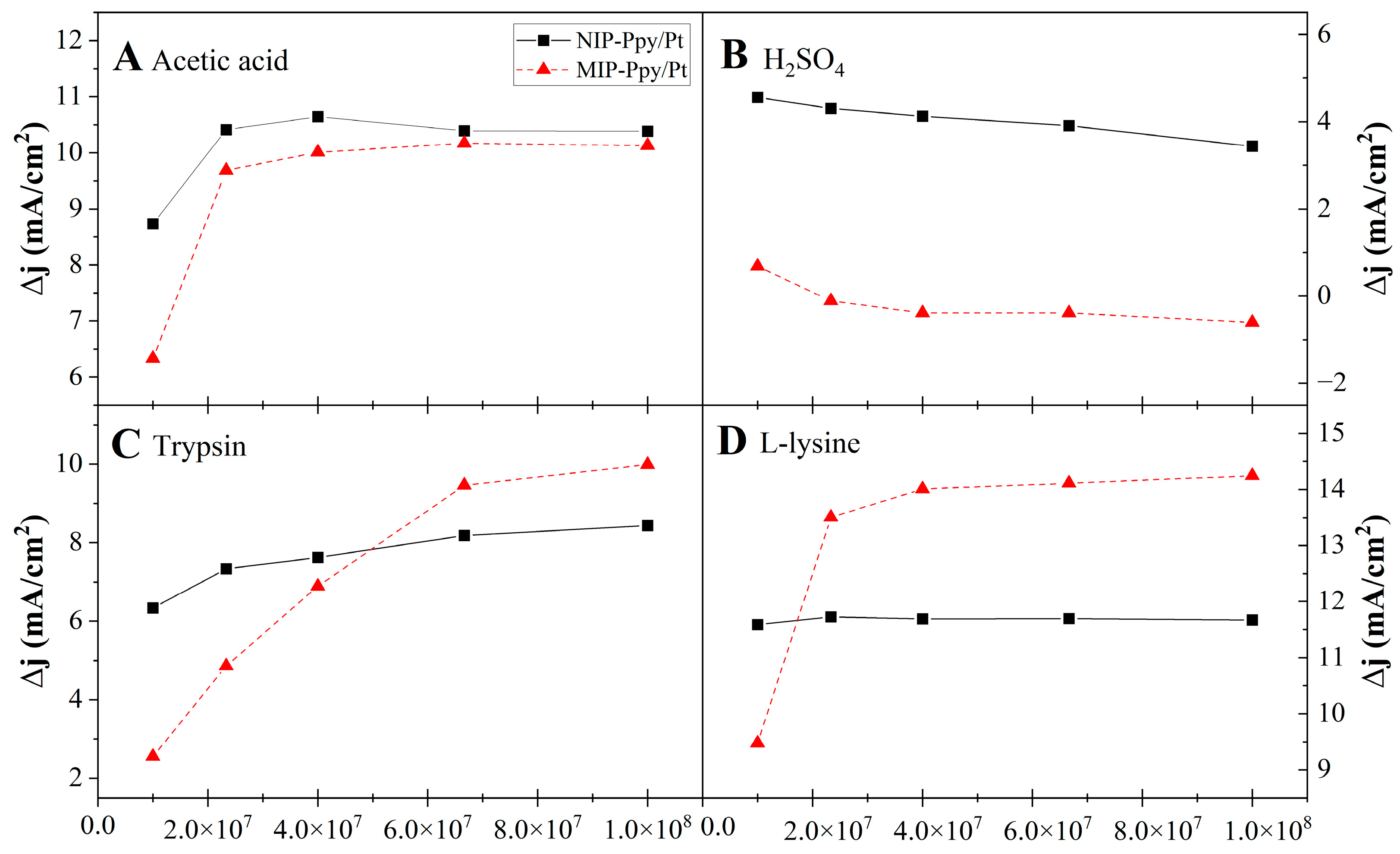
3. Results
3.1. Electrodeposition of Molecularly Imprinted Polypyrrole
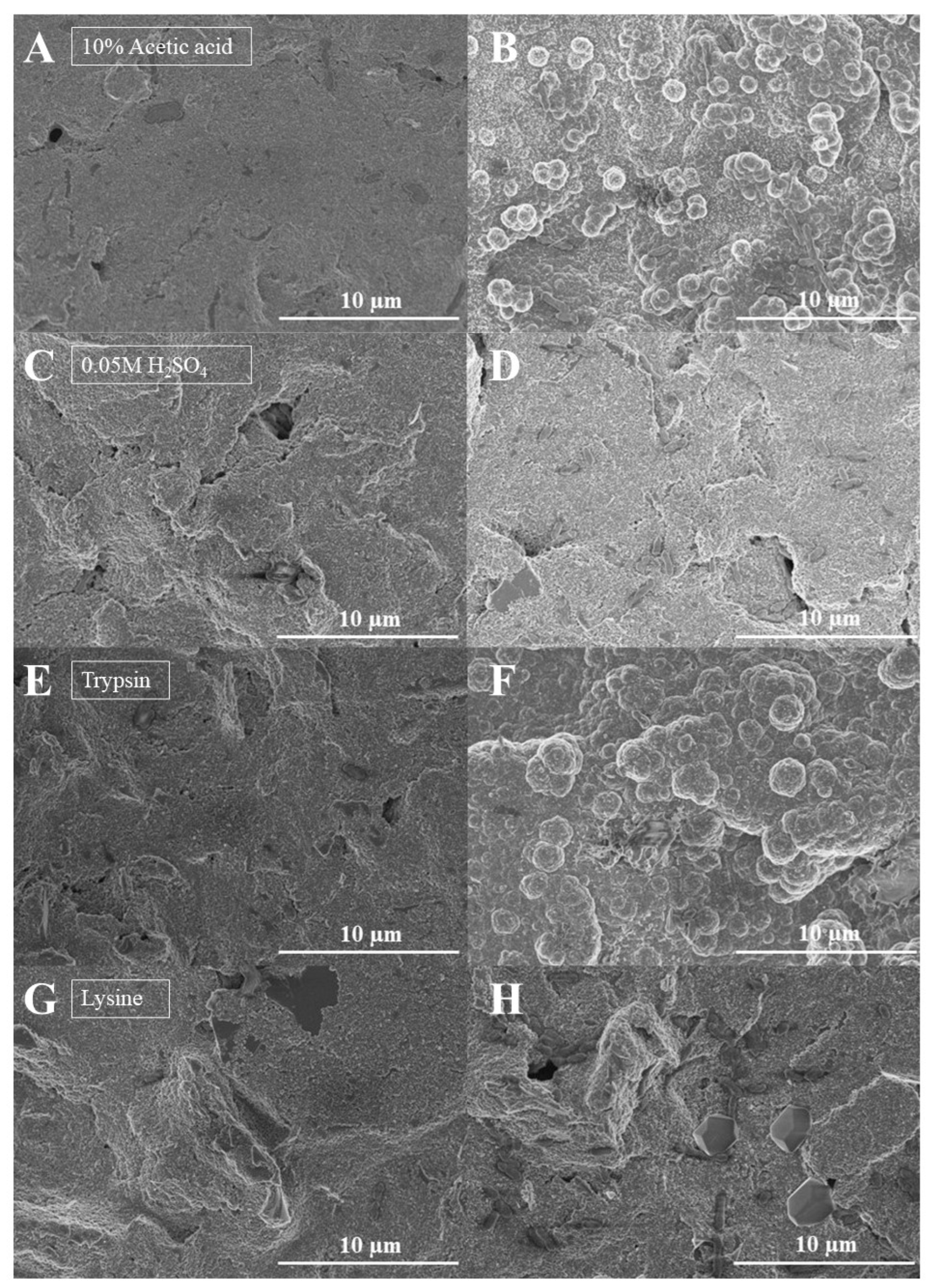
3.2. Extraction of Imprinted Bacteria from the MIP-Ppy Layer
3.3. Electrochemical Characterisation of Bacteria-Imprinted MIP-Ppy Layer
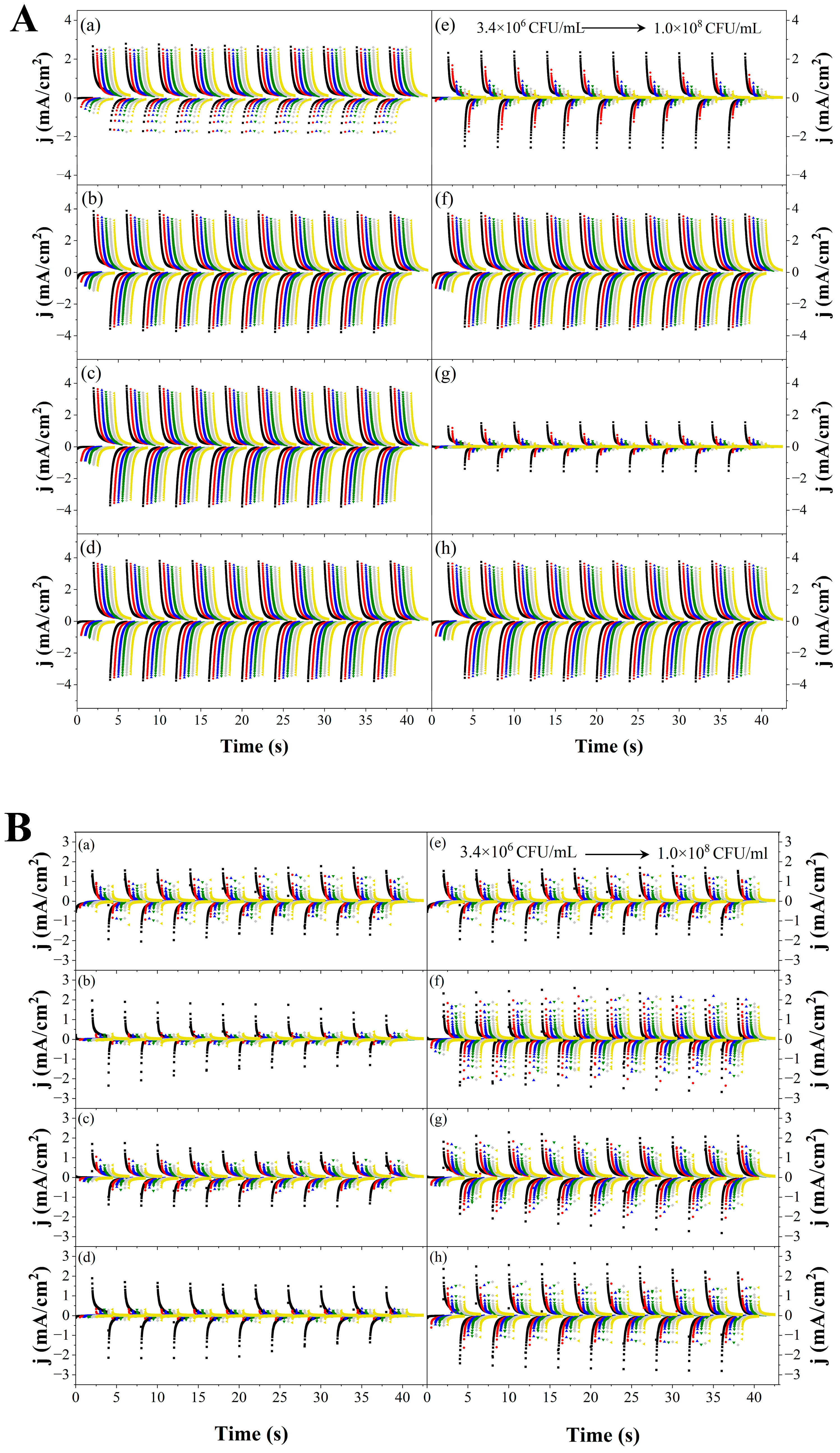
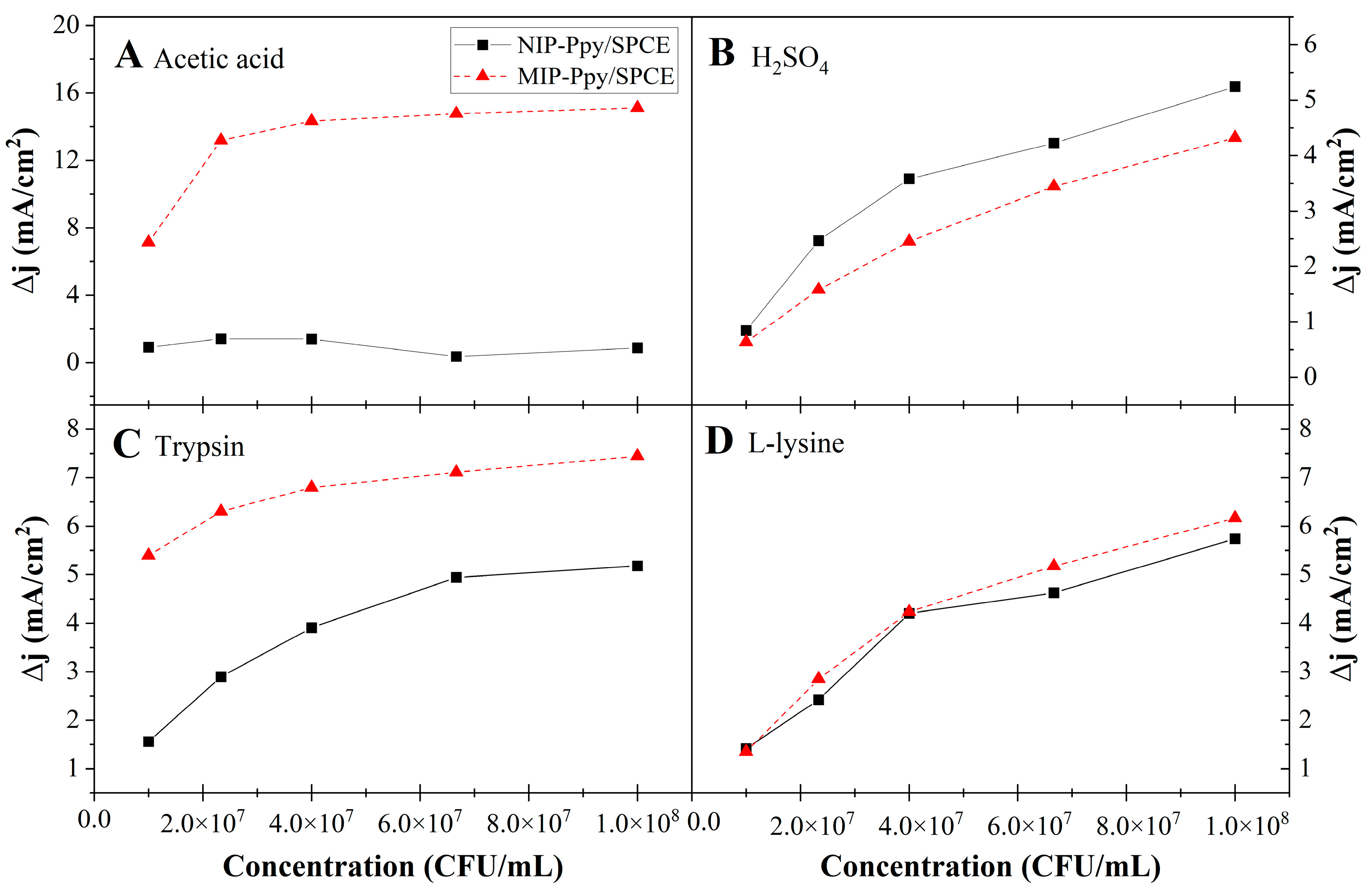
3.3.1. Assessment of first Electrochemical System
3.3.2. Assessment of MIP-Ppy/Pt- and NIP-Ppy/Pt-Based Electrodes
3.4. Determination of Limit of Detection and Limit of Quantification
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vizzini, P.; Braidot, M.; Vidic, J.; Manzano, M. Electrochemical and Optical Biosensors for the Detection of Campylobacter and Listeria: An Update Look. Micromachines 2019, 10, 500. [Google Scholar] [CrossRef] [PubMed]
- Batt, C.A. LISTERIA|Listeria monocytogenes. In Encyclopedia of Food Microbiology, 2nd ed.; Batt, C.A., Tortorello, M.L., Eds.; Academic Press: Oxford, UK, 2014; pp. 490–493. [Google Scholar]
- Matle, I.; Mbatha Khanyisile, R.; Madoroba, E. A review of Listeria monocytogenes from meat and meat products: Epidemiology, virulence factors, antimicrobial resistance and diagnosis. Onderstepoort J. Vet. Res. 2020, 87, a1869. [Google Scholar] [CrossRef] [PubMed]
- Lepe, J.A. Current aspects of listeriosis. Med. Clínica 2020, 154, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.H.; Park, Y.J.; Kim, M.; Seo, Y.H.; Kim, Y.A.; Choi, J.Y.; Yong, D.; Jeong, S.H.; Lee, K. Increasing Incidence of Listeriosis and Infection-associated Clinical Outcomes. Ann. Lab. Med. 2018, 38, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Valenti, M.; Ranganathan, N.; Moore, L.S.; Hughes, S. Listeria monocytogenes infections: Presentation, diagnosis and treatment. Br. J. Hosp. Med. 2021, 82, 1–6. [Google Scholar] [CrossRef]
- Freitag, I.G.R.; Pereira, R.d.C.L.; Machado, E.S.; Hofer, E.; Vallim, D.C.; Hofer, C.B. Seroprevalence of Listeria monocytogenes in HIV infected pregnant women from Brazil. Braz. J. Infect. Dis. 2021, 25, 101635. [Google Scholar] [CrossRef]
- Craig, A.M.; Dotters-Katz, S.; Kuller, J.A.; Thompson, J.L. Listeriosis in Pregnancy: A Review. Obstet. Gynecol. Surv. 2019, 74, 362–368. [Google Scholar] [CrossRef]
- Soni, D.K.; Ahmad, R.; Dubey, S.K. Biosensor for the detection of Listeria monocytogenes: Emerging trends. Crit. Rev. Microbiol. 2018, 44, 590–608. [Google Scholar] [CrossRef]
- EFSA Panel on Biological Hazards (BIOHAZ); Ricci, A.; Allende, A.; Bolton, D.; Chemaly, M.; Davies, R.; Fernández Escámez, P.S.; Girones, R.; Herman, L.; Koutsoumanis, K.; et al. Listeria monocytogenes contamination of ready-to-eat foods and the risk for human health in the EU. EFSA J. 2018, 16, e05134. [Google Scholar] [CrossRef]
- Jadhav, S.; Bhave, M.; Palombo, E.A. Methods used for the detection and subtyping of Listeria monocytogenes. J. Microbiol. Methods 2012, 88, 327–341. [Google Scholar] [CrossRef]
- Silva, N.F.D.; Neves, M.M.P.S.; Magalhães, J.M.C.S.; Freire, C.; Delerue-Matos, C. Emerging electrochemical biosensing approaches for detection of Listeria monocytogenes in food samples: An overview. Trends Food Sci. Technol. 2020, 99, 621–633. [Google Scholar] [CrossRef]
- Tasbasi, B.B.; Guner, B.C.; Sudagidan, M.; Ucak, S.; Kavruk, M.; Ozalp, V.C. Label-free lateral flow assay for Listeria monocytogenes by aptamer-gated release of signal molecules. Anal. Biochem. 2019, 587, 113449. [Google Scholar] [CrossRef] [PubMed]
- Wachiralurpan, S.; Chansiri, K.; Lieberzeit, P.A. Direct detection of Listeria monocytogenes DNA amplification products with quartz crystal microbalances at elevated temperatures. Sens. Actuat. B-Chem. 2020, 308, 127678. [Google Scholar] [CrossRef]
- Jiang, X.; Ding, W.; Lv, Z.; Rao, C. Highly Sensitive Electrochemical Immunosensing for Listeria Monocytogenes Based on 3,4,9,10-Perylene Tetracarboxylic Acid/Graphene Ribbons as a Sensing Platform and Ferrocene/Gold Nanoparticles as an Amplifier. Anal. Sci. 2021, 37, 1701–1706. [Google Scholar] [CrossRef] [PubMed]
- Crowley, E.L.; O’Sullivan, C.K.; Guilbault, G.G. Increasing the sensitivity of Listeria monocytogenes assays: Evaluation using ELISA and amperometric detection. Analyst 1999, 124, 295–299. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Luz, L.; Mendonça, M.; Bernardes Fogaça, M.; Kipnis, A.; Bhunia, A.K.; Bührer-Sékula, S. Listeria monocytogenes: Review of pathogenesis and virulence determinants-targeted immunological assays. Crit. Rev. Microbiol. 2021, 47, 647–666. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Peng, Y.; Bai, J.; Zhang, X.; Liu, Y.; Fan, X.; Ning, B.; Gao, Z. Rapid detection of Listeria monocytogenes in milk by self-assembled electrochemical immunosensor. Sens. Actuat. B-Chem. 2014, 190, 900–906. [Google Scholar] [CrossRef]
- Zolti, O.; Suganthan, B.; Maynard, R.; Asadi, H.; Locklin, J.; Ramasamy, R.P. Electrochemical Biosensor for Rapid Detection of Listeria monocytogenes. J. Electrochem. Soc. 2022, 169, 067510. [Google Scholar] [CrossRef]
- Lee, B.E.; Kang, T.; Jenkins, D.; Li, Y.; Wall, M.M.; Jun, S. A single-walled carbon nanotubes-based electrochemical impedance immunosensor for on-site detection of Listeria monocytogenes. J. Food Sci. 2022, 87, 280–288. [Google Scholar] [CrossRef]
- Rivas-Macho, A.; Eletxigerra, U.; Diez-Ahedo, R.; Merino, S.; Sanjuan, A.; Bou-Ali, M.M.; Ruiz-Rubio, L.; del Campo, J.; Vilas-Vilela, J.L.; Goñi-de-Cerio, F.; et al. Design and 3D printing of an electrochemical sensor for Listeria monocytogenes detection based on loop mediated isothermal amplification. Heliyon 2023, 9, e12637. [Google Scholar] [CrossRef]
- Mosbach, K. Molecular imprinting. Trends Biochem. Sci. 1994, 19, 9–14. [Google Scholar] [CrossRef]
- Tretjakov, A.; Syritski, V.; Reut, J.; Boroznjak, R.; Öpik, A. Molecularly imprinted polymer film interfaced with Surface Acoustic Wave technology as a sensing platform for label-free protein detection. Anal. Chim. Acta 2016, 902, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Ramanavicius, S.; Jagminas, A.; Ramanavicius, A. Advances in Molecularly Imprinted Polymers Based Affinity Sensors (Review). Polymers 2021, 13, 974. [Google Scholar] [CrossRef] [PubMed]
- Ratautaite, V.; Boguzaite, R.; Brazys, E.; Ramanaviciene, A.; Ciplys, E.; Juozapaitis, M.; Slibinskas, R.; Bechelany, M.; Ramanavicius, A. Molecularly Imprinted Polypyrrole based Sensor for the Detection of SARS-CoV-2 Spike Glycoprotein. Electrochim. Acta 2022, 403, 139581. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.; Liu, F.; Kan, X. Voltammetric dopamine sensor based on three-dimensional electrosynthesized molecularly imprinted polymers and polypyrrole nanowires. Microchim. Acta 2017, 184, 2515–2522. [Google Scholar] [CrossRef]
- Ratautaite, V.; Nesladek, M.; Ramanaviciene, A.; Baleviciute, I.; Ramanavicius, A. Evaluation of Histamine Imprinted Polypyrrole Deposited on Boron Doped Nanocrystalline Diamond. Electroanalysis 2014, 26, 2458–2464. [Google Scholar] [CrossRef]
- Nguy, T.P.; Van Phi, T.; Tram, D.T.N.; Eersels, K.; Wagner, P.; Lien, T.T.N. Development of an impedimetric sensor for the label-free detection of the amino acid sarcosine with molecularly imprinted polymer receptors. Sens. Actuat. B-Chem. 2017, 246, 461–470. [Google Scholar] [CrossRef]
- Ratautaite, V.; Boguzaite, R.; Brazys, E.; Plausinaitis, D.; Ramanavicius, S.; Samukaite-Bubniene, U.; Bechelany, M.; Ramanavicius, A. Evaluation of the interaction between SARS-CoV-2 spike glycoproteins and the molecularly imprinted polypyrrole. Talanta 2023, 253, 123981. [Google Scholar] [CrossRef]
- Piletsky, S.; Canfarotta, F.; Poma, A.; Bossi, A.M.; Piletsky, S. Molecularly Imprinted Polymers for Cell Recognition. Trends Biotechnol. 2020, 38, 368–387. [Google Scholar] [CrossRef]
- Dar, K.K.; Shao, S.; Tan, T.; Lv, Y. Molecularly imprinted polymers for the selective recognition of microorganisms. Biotechnol. Adv. 2020, 45, 107640. [Google Scholar] [CrossRef]
- Zhao, X.; Cui, Y.; Wang, J.; Wang, J. Preparation of Fluorescent Molecularly Imprinted Polymers via Pickering Emulsion Interfaces and the Application for Visual Sensing Analysis of Listeria Monocytogenes. Polymers 2019, 11, 984. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Guo, Z.; Qiu, X.; Lu, W.; Yang, W.; Wang, Q.; Wu, Q. Simple electrochemical detection of Listeria monocytogenes based on a surface-imprinted polymer-modified electrode. Anal. Methods 2021, 13, 4864–4870. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Lv, Z.; Ding, W.; Zhang, Y.; Lin, F. Pathogen-Imprinted Polymer Film Integrated probe/Ti3C2Tx MXenes Electrochemical Sensor for Highly Sensitive Determination of Listeria Monocytogenes. J. Electrochem. Sci. Technol. 2022, 13, 431–437. [Google Scholar] [CrossRef]
- Cui, F.; Zhou, Z.; Zhou, H.S. Molecularly Imprinted Polymers and Surface Imprinted Polymers Based Electrochemical Biosensor for Infectious Diseases. Sensors 2020, 20, 996. [Google Scholar] [CrossRef]
- Ramanavicius, A.; Oztekin, Y.; Ramanaviciene, A. Electrochemical formation of polypyrrole-based layer for immunosensor design. Sens. Actuat. B-Chem. 2014, 197, 237–243. [Google Scholar] [CrossRef]
- Trček, J.; Mira, N.P.; Jarboe, L.R. Adaptation and tolerance of bacteria against acetic acid. Appl. Microbiol. Biotechnol. 2015, 99, 6215–6229. [Google Scholar] [CrossRef]
- Miura, C.; Ohta, T.; Ozaki, Y.; Tanaka, H.; Miura, T. Trypsin is a multifunctional factor in spermatogenesis. Proc. Natl. Acad. Sci. USA 2009, 106, 20972–20977. [Google Scholar] [CrossRef]
- Grenier, D. Effect of proteolytic enzymes on the lysis and growth of oral bacteria. Oral Microbiol. Immunol. 1994, 9, 224–228. [Google Scholar] [CrossRef]
- Zhou, J.; Meng, X.; Han, Q.; Huang, Y.; Huo, L.; Lei, Y. An in vitro study on the degradation of multispecies biofilm of periodontitis-related microorganisms by bovine trypsin. Front. Microbiol. 2022, 13, 951291. [Google Scholar] [CrossRef]
- Yarman, A.; Scheller, F.W. How Reliable Is the Electrochemical Readout of MIP Sensors? Sensors 2020, 20, 2677. [Google Scholar] [CrossRef]
- Conte, M.P.; Petrone, G.; Biase, A.M.D.; Longhi, C.; Penta, M.; Tinari, A.; Superti, F.; Fabozzi, G.; Visca, P.; Seganti, L. Effect of Acid Adaptation on the Fate of Listeria monocytogenes in THP-1 Human Macrophages Activated by Gamma Interferon. Infect. Immun. 2002, 70, 4369–4378. [Google Scholar] [CrossRef] [PubMed]
- Kamal Ahmed, R.; Saad, E.M.; Fahmy, H.M.; El Nashar, R.M. Design and application of molecularly imprinted Polypyrrole/Platinum nanoparticles modified platinum sensor for the electrochemical detection of Vardenafil. Microchem. J. 2021, 171, 106771. [Google Scholar] [CrossRef]
- Wu, J.; Wang, R.; Lu, Y.; Jia, M.; Yan, J.; Bian, X. Facile Preparation of a Bacteria Imprinted Artificial Receptor for Highly Selective Bacterial Recognition and Label-Free Impedimetric Detection. Anal. Chem. 2019, 91, 1027–1033. [Google Scholar] [CrossRef] [PubMed]
- Ratautaite, V.; Brazys, E.; Ramanaviciene, A.; Ramanavicius, A. Electrochemical Sensors based on L-Tryptophan Molecularly Imprinted Polypyrrole and Polyaniline. J. Electroanal. Chem. 2022, 917, 116389. [Google Scholar] [CrossRef]
- Mustafa, Y.L.; Keirouz, A.; Leese, H.S. Molecularly imprinted polymers in diagnostics: Accessing analytes in biofluids. J. Mater. Chem. B 2022, 10, 7418–7449. [Google Scholar] [CrossRef]
- Ayerdurai, V.; Cieplak, M.; Kutner, W. Molecularly imprinted polymer-based electrochemical sensors for food contaminants determination. TRAC-Trends Anal. Chem. 2023, 158, 116830. [Google Scholar] [CrossRef]
- Li, F.; Ye, Q.; Chen, M.; Zhou, B.; Zhang, J.; Pang, R.; Xue, L.; Wang, J.; Zeng, H.; Wu, S.; et al. An ultrasensitive CRISPR/Cas12a based electrochemical biosensor for Listeria monocytogenes detection. Biosens. Bioelectron. 2021, 179, 113073. [Google Scholar] [CrossRef]
- Lu, Y.; Liu, Y.; Zhao, Y.; Li, W.; Qiu, L.; Li, L. A Novel and Disposable Enzyme-Labeled Amperometric Immunosensor Based on MWCNT Fibers for Listeria monocytogenes Detection. J. Nanomater. 2016, 2016, 3895920. [Google Scholar] [CrossRef]
- Chai, C.; Lee, J.; Oh, S.-W.; Takhistov, P. Impedimetric Characterization of Adsorption of Listeria monocytogenes on the Surface of an Aluminum-Based Immunosensor. J. Food Sci. 2014, 79, E2266–E2271. [Google Scholar] [CrossRef]
- Pintavirooj, C.; Vongmanee, N.; Sukjee, W.; Sangma, C.; Visitsattapongse, S. Biosensors for Klebsiella pneumoniae with Molecularly Imprinted Polymer (MIP) Technique. Sensors 2022, 22, 4638. [Google Scholar] [CrossRef]
- Sharma, R.; Lakshmi, G.B.V.S.; Kumar, A.; Solanki, P. Polypyrrole Based Molecularly Imprinted Polymer Platform for Klebsiella pneumonia Detection. ECS Sensors Plus 2022, 1, 010603. [Google Scholar] [CrossRef]
- Wang, O.; Jia, X.; Liu, J.; Sun, M.; Wu, J. Rapid and simple preparation of an MXene/polypyrrole-based bacteria imprinted sensor for ultrasensitive Salmonella detection. J. Electroanal. Chem. 2022, 918, 116513. [Google Scholar] [CrossRef]
- Tokonami, S.; Nakadoi, Y.; Takahashi, M.; Ikemizu, M.; Kadoma, T.; Saimatsu, K.; Dung, L.Q.; Shiigi, H.; Nagaoka, T. Label-Free and Selective Bacteria Detection Using a Film with Transferred Bacterial Configuration. Anal. Chem. 2013, 85, 4925–4929. [Google Scholar] [CrossRef] [PubMed]
- Tokonami, S.; Nakadoi, Y.; Nakata, H.; Takami, S.; Kadoma, T.; Shiigi, H.; Nagaoka, T. Recognition of gram-negative and gram-positive bacteria with a functionalised conducting polymer film. Res. Chem. Intermed. 2014, 40, 2327–2335. [Google Scholar] [CrossRef]
- Tokonami, S.; Shimizu, E.; Tamura, M.; Iida, T. Mechanism in External Field-mediated Trapping of Bacteria Sensitive to Nanoscale Surface Chemical Structure. Sci. Rep. 2017, 7, 16651. [Google Scholar] [CrossRef] [PubMed]
- Idil, N.; Hedström, M.; Denizli, A.; Mattiasson, B. Whole cell based microcontact imprinted capacitive biosensor for the detection of Escherichia coli. Biosens. Bioelectron. 2017, 87, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Yasmeen, N.; Etienne, M.; Sharma, P.S.; El-Kirat-Chatel, S.; Helú, M.B.; Kutner, W. Molecularly imprinted polymer as a synthetic receptor mimic for capacitive impedimetric selective recognition of Escherichia coli K-12. Anal. Chim. Acta 2021, 1188, 339177. [Google Scholar] [CrossRef]
- Hayden, O.; Dickert, F.L. Selective Microorganism Detection with Cell Surface Imprinted Polymers. Adv. Mater. 2001, 13, 1480–1483. [Google Scholar] [CrossRef]
- Golabi, M.; Kuralay, F.; Jager, E.W.H.; Beni, V.; Turner, A.P.F. Electrochemical bacterial detection using poly(3-aminophenylboronic acid)-based imprinted polymer. Biosens. Bioelectron. 2017, 93, 87–93. [Google Scholar] [CrossRef]
- Namvar, A.; Warriner, K. Microbial imprinted polypyrrole/poly(3-methylthiophene) composite films for the detection of Bacillus endospores. Biosens. Bioelectron. 2007, 22, 2018–2024. [Google Scholar] [CrossRef]



| Electrode | Detection Technique | Method Used | LOD, LOQ and LR | Ref. |
|---|---|---|---|---|
| Gold disk | RAA-based E-CRISPR 1 | Square wave voltammetry | LOD 26 CFU/mL; LR 2.6 × 101 to 2.6 × 109 CFU/mL | [48] |
| Gold electrode | Sandwich assay | CV, EIS | LR 102 to 106 CFU/ml | [18] |
| Multiwalled carbon nanotube electrode | Immunoassay | CV | LOD 1.07 × 102 CFU/mL; LR 102 to 105 CFU/mL | [49] |
| Aluminium disc | Immunoassay | EIS | LOD 1.3 log CFU/mL LR 1.3 to 4.3 log CFU/mL | [50] |
| Bacteria | Electrode | Polymer | Bacteria Extraction from the Polymer | Method Used | LOD, LOQ and Linear Range (LR) | Ref. |
|---|---|---|---|---|---|---|
| Listeria monocytogenes | - | Acryloyl-functionalised chitosan with CdTe quantum dots | 10% acetic acid, 1% SDS, water, and methanol | Fluorescence microscope | LOD 103 CFU/mL | [32] |
| Listeria monocytogenes | GCE | Poly(TPA) | SDS/AA (w/v, 5%) solution | DPV | LOD 6 CFU/mL; LR 10 to 106 CFU/mL | [33] |
| Listeria monocytogenes | GCE with MXenes nanoribbon (Ti3C2TxR) | Poly(Th) with | 0.5 M HCl | DPV | LOD 2 CFU/mL; LR 10 to 108 CFU/mL | [34] |
| Klebsiella pneumoniae | SPE | Acrylamide-based polymer with carbon or gold or rGO | 10% acetic acid for 30 min | CV in PBS with a redox probe | LOD of 0.012 CFU/mL and LOQ of 1.61 CFU/mL | [51] |
| Klebsiella pneumonia | ITO coated glass electrode | Ppy obtained by the interfacial oxidative polymerisation process | DI and ethanol | DPV and CV in PBS with a redox probe | LOD of 1.352 CFU/mL | [52] |
| Salmonella | GCE | Ppy with MXene | SDS/acetic acid (5%, w/v) for 5 min and washed three times | EIS | LOD of 23 CFU/mL | [53] |
| Pseudomonas aeruginosa | QCM electrode | Overoxidised Ppy | With lysozyme (10 mg/mL) for 2 h at 4 °C and 10% Triton X for 80 min | QCM | LOD 103 CFU/mL | [54] |
| Escherichia coli and Pseudomonas aeruginosa | QCM electrode | Overoxidised Ppy | Lysozyme (10 mg/mL) containing 10% Triton X and EDTA (200 µg/mL) for 1 day at room temperature | QCM | - | [55] |
| Escherichia coli (serotypes O157:H7 and O26:H11) | QCM sensor | Overoxidised Ppy | Lysozyme (30 mg/mL) and 5% SDS for 48 h at 30 °C | QCM | - | [56] |
| Escherichia coli | Gold electrode | Polymer of MAH, HEMA, and EGDMA | 10 mM sodium phosphate buffer (pH 7.4) and treated with 10 mg/mL lysozyme solution (in 10 mM Tris-HCl buffer, pH 8.0, with 1 mM EDTA) for 30 min | Capacitance measurements in a continuous flow system | LOD 70 CFU/mL, LR 1.0 × 102–1.0 × 107 CFU/mL | [57] |
| Escherichia coli K-12 | GCE | Polymer of 2-APBA and ANI | 2 h treated with 2 mg mL−1 lysozyme enzyme in PBS (pH ¼ 7.4), 10% Triton X, water, and then overoxidised | DPV, EIS | - | [58] |
| Saccharomyces cerevisiae (Bakers’ yeast) | QCM | Polyurethane | Hot water | QCM | LOD 1 × 104 cells/mL | [59] |
| Staphylococcus epidermidis | Gold electrode | Poly(3-APBA) | 30 min with fructose (20 mM), plenty of water, and phosphate solution (pH 2.2) for 20 min | EIS | LR 103–107 CFU/mL | [60] |
| Bacillus subtilis endospore | GCE | polypyrrole/poly(3-methylthiophene) | In DMSO for 10 min at room temperature | CV and EIS | - | [61] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liustrovaite, V.; Pogorielov, M.; Boguzaite, R.; Ratautaite, V.; Ramanaviciene, A.; Pilvenyte, G.; Holubnycha, V.; Korniienko, V.; Diedkova, K.; Viter, R.; et al. Towards Electrochemical Sensor Based on Molecularly Imprinted Polypyrrole for the Detection of Bacteria—Listeria monocytogenes. Polymers 2023, 15, 1597. https://doi.org/10.3390/polym15071597
Liustrovaite V, Pogorielov M, Boguzaite R, Ratautaite V, Ramanaviciene A, Pilvenyte G, Holubnycha V, Korniienko V, Diedkova K, Viter R, et al. Towards Electrochemical Sensor Based on Molecularly Imprinted Polypyrrole for the Detection of Bacteria—Listeria monocytogenes. Polymers. 2023; 15(7):1597. https://doi.org/10.3390/polym15071597
Chicago/Turabian StyleLiustrovaite, Viktorija, Maksym Pogorielov, Raimonda Boguzaite, Vilma Ratautaite, Almira Ramanaviciene, Greta Pilvenyte, Viktoriia Holubnycha, Viktoriia Korniienko, Kateryna Diedkova, Roman Viter, and et al. 2023. "Towards Electrochemical Sensor Based on Molecularly Imprinted Polypyrrole for the Detection of Bacteria—Listeria monocytogenes" Polymers 15, no. 7: 1597. https://doi.org/10.3390/polym15071597
APA StyleLiustrovaite, V., Pogorielov, M., Boguzaite, R., Ratautaite, V., Ramanaviciene, A., Pilvenyte, G., Holubnycha, V., Korniienko, V., Diedkova, K., Viter, R., & Ramanavicius, A. (2023). Towards Electrochemical Sensor Based on Molecularly Imprinted Polypyrrole for the Detection of Bacteria—Listeria monocytogenes. Polymers, 15(7), 1597. https://doi.org/10.3390/polym15071597









