PLA 3D Printing as a Straightforward and Versatile Fabrication Method for PDMS Molds
Abstract
1. Introduction
2. Materials and Methods
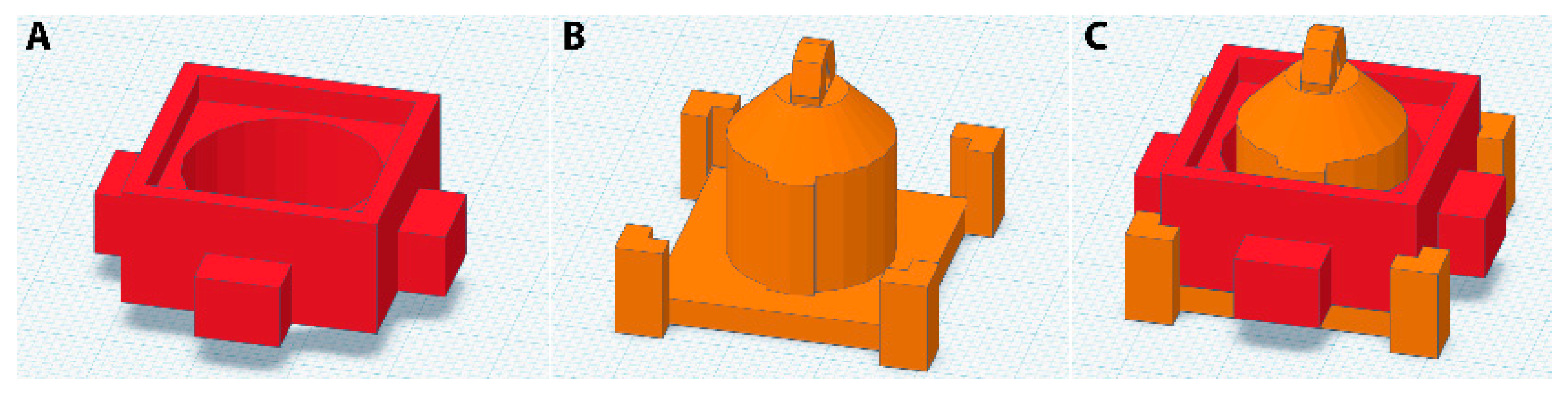
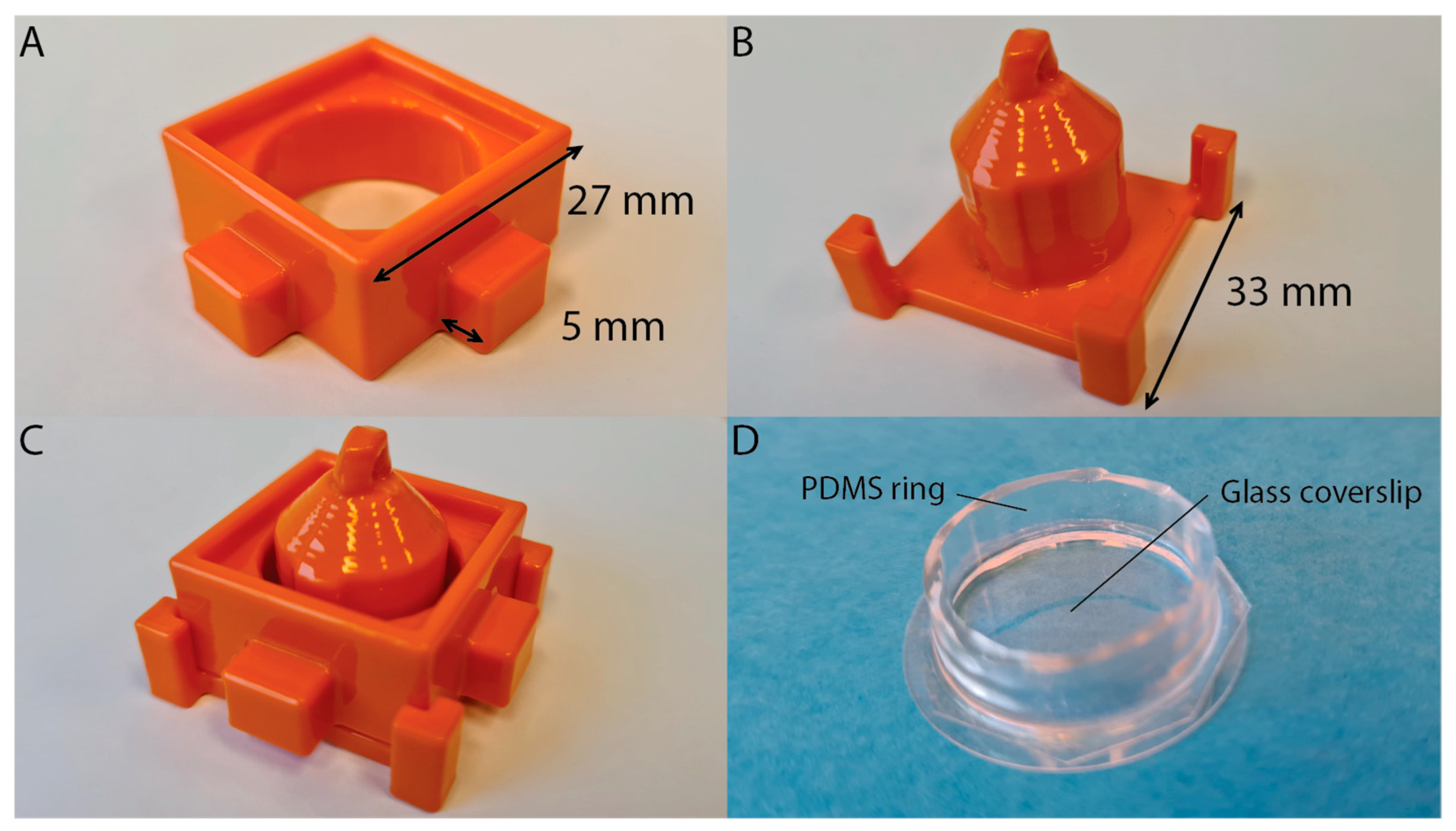
2.1. Mold Design
2.2. Printer and Settings
2.3. Mold Smoothing
2.4. Mold and PDMS Preparation
2.5. Liquid Well Preparation
2.6. Cell Culture
2.7. Confocal Microscopy
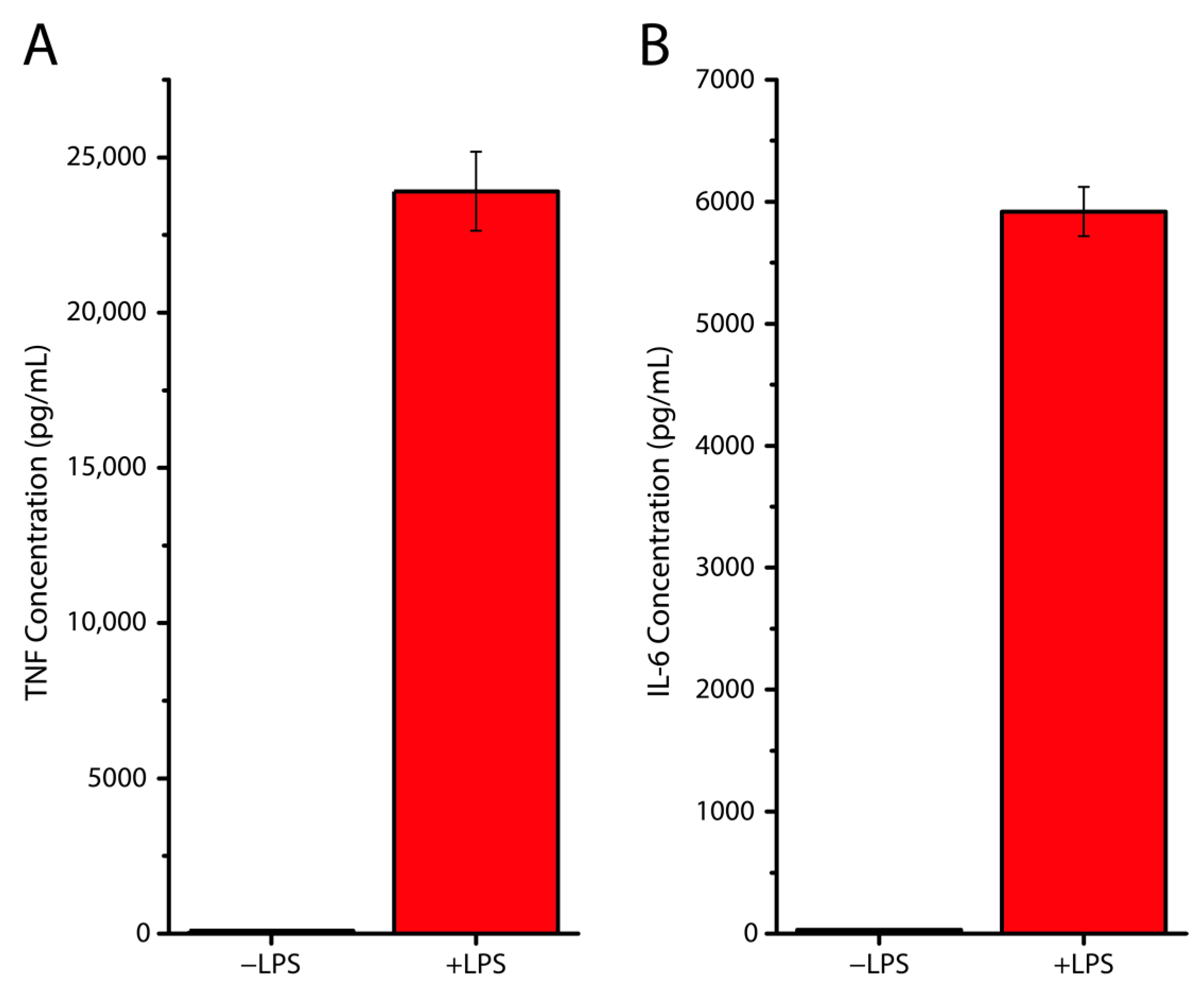
2.8. ELISA
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Mata, A.; Fleischman, A.J.; Roy, S. Characterization of polydimethylsiloxane (PDMS) properties for biomedical micro/nanosystems. Biomed. Microdevices 2005, 7, 281–293. [Google Scholar] [CrossRef] [PubMed]
- McDonald, J.C.; Whitesides, G.M. Poly(dimethylsiloxane) as a material for fabricating microfluidic devices. Acc. Chem. Res. 2002, 35, 491–499. [Google Scholar] [CrossRef]
- Xia, Y.; Whitesides, G.M. Soft Lithography. Angew. Chem. Int. Ed. Engl. 1998, 37, 550–575. [Google Scholar] [CrossRef]
- Folch, A.; Toner, M. Cellular micropatterns on biocompatible materials. Biotechnol. Prog. 1998, 14, 388–392. [Google Scholar] [CrossRef]
- Roos, W.; Ulmer, J.; Gräter, S.; Surrey, T.; Spatz, J.P. Microtubule gliding and cross-linked microtubule networks on micropillar interfaces. Nano Lett. 2005, 5, 2630–2634. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, M.L.; Tsai, S.; Bellan, L.M.; Sappington, R.; Xu, Y.; Li, D. The relationship between the Young’s modulus and dry etching rate of polydimethylsiloxane (PDMS). Biomed. Microdevices 2019, 21, 26. [Google Scholar] [CrossRef]
- da Silva, E.N.T.; Ferreira, V.S.; Lucca, B.G. Rapid and inexpensive method for the simple fabrication of PDMS-based electrochemical sensors for detection in microfluidic devices. Electrophoresis 2019, 40, 1322–1330. [Google Scholar] [CrossRef]
- Hattori, K.; Sugiura, S.; Kanamori, T. Microfluidic perfusion culture. Methods Mol. Biol. 2014, 1104, 251–263. [Google Scholar] [CrossRef]
- Siddique, A.; Meckel, T.; Stark, R.W.; Narayan, S. Improved cell adhesion under shear stress in PDMS microfluidic devices. Colloids Surf. B Biointerfaces 2017, 150, 456–464. [Google Scholar] [CrossRef]
- Roos, W.H.; Campas, O.; Montel, F.; Woehlke, G.; Spatz, J.P.; Bassereau, P.; Cappello, G. Dynamic kinesin-1 clustering on microtubules due to mutually attractive interactions. Phys. Biol. 2008, 5, 046004. [Google Scholar] [CrossRef]
- Kim, J.; An, H.; Seo, Y.; Jung, Y.; Lee, J.S.; Choi, N.; Bong, K.W. Flow lithography in ultraviolet-curable polydimethylsiloxane microfluidic chips. Biomicrofluidics 2017, 11, 024120. [Google Scholar] [CrossRef] [PubMed]
- Amin, R.; Knowlton, S.; Hart, A.; Yenilmez, B.; Ghaderinezhad, F.; Katebifar, S.; Messina, M.; Khademhosseini, A.; Tasoglu, S. 3D-printed microfluidic devices. Biofabrication 2016, 8, 022001. [Google Scholar] [CrossRef] [PubMed]
- Bonyár, A.; Sántha, H.; Varga, M.; Ring, B.; Vitéz, A.; Harsányi, G. Characterization of rapid PDMS casting technique utilizing molding forms fabricated by 3D rapid prototyping technology (RPT). Int. J. Mater. Form. 2012, 7, 189–196. [Google Scholar] [CrossRef]
- Hwang, Y.; OPaydar, H.; Candler, R.N. 3D printed molds for non-planar PDMS microfluidic channels. Sens. Actuators A Phys. 2015, 226, 137–142. [Google Scholar] [CrossRef]
- Mi, S.; Du, Z.; Xu, Y.; Sun, W. The crossing and integration between microfluidic technology and 3D printing for organ-on-chips. J. Mater. Chem. B 2018, 6, 6191–6206. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.; Zhuang, X.; Tang, Z.; Chen, X. Polylactic acid (PLA): Research, development and industrialization. Biotechnol. J. 2010, 5, 1125–1136. [Google Scholar] [CrossRef]
- Singhvi, M.S.; Zinjarde, S.S.; Gokhale, D.V. Polylactic acid: Synthesis and biomedical applications. J. Appl. Microbiol. 2019, 127, 1612–1626. [Google Scholar] [CrossRef]
- Li, G.; Zhao, M.; Xu, F.; Yang, B.; Li, X.; Meng, X.; Teng, L.; Sun, F.; Li, Y. Synthesis and Biological Application of Polylactic Acid. Molecules 2020, 25, 5023. [Google Scholar] [CrossRef]
- Okolie, O.; Stachurek, I.; Kandasubramanian, B.; Njuguna, J. 3D Printing for Hip Implant Applications: A Review. Polymers 2020, 12, 2682. [Google Scholar] [CrossRef]
- Prasad, L.K.; Smyth, H. 3D Printing technologies for drug delivery: A review. Drug Dev. Ind. Pharm. 2016, 42, 1019–1031. [Google Scholar] [CrossRef]
- DeStefano, V.; Khan, S.; Tabada, A. Applications of PLA in modern medicine. Eng. Regen. 2020, 1, 76–87. [Google Scholar] [CrossRef]
- Alexander, A.E.; Wake, N.; Chepelev, L.; Brantner, P.; Ryan, J.; Wang, K.C. A guideline for 3D printing terminology in biomedical research utilizing ISO/ASTM standards. 3D Print Med. 2021, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Baranov, M.V.; Ter Beest, M.; Reinieren-Beeren, I.; Cambi, A.; Figdor, C.G.; van den Bogaart, G. Podosomes of dendritic cells facilitate antigen sampling. J. Cell Sci. 2014, 127 Pt 5, 1052–1064. [Google Scholar] [CrossRef] [PubMed]
- Hambali, R.H.; Cheong, K.M.; Azizan, N. Analysis of the influence of chemical treatment to the strength and surface roughness of FDM. In IOP Conference Series: Materials Science and Engineering; IOP Publishing: Bristol, UK, 2017; Volume 210, p. 012063. [Google Scholar] [CrossRef]
- Valerga, A.P.; Batista, M.; Fernandez-Vidal, S.R.; Gamez, A.J. Impact of Chemical Post-Processing in Fused Deposition Modelling (FDM) on Polylactic Acid (PLA) Surface Quality and Structure. Polymers 2019, 11, 566. [Google Scholar] [CrossRef]
- Salazar, R.; Pizarro, F.; Vasquez, D.; Rajo-Iglesias, E. Assessment of 3D-printed waveguides using conductive filaments and a chloroform-based smoothing process. Addit. Manuf. 2022, 51, 102593. [Google Scholar] [CrossRef]
- Barrientos, L.; Bignon, A.; Gueguen, C.; de Chaisemartin, L.; Gorges, R.; Sandré, C.; Mascarell, L.; Balabanian, K.; Kerdine-Römer, S.; Pallardy, M.; et al. Neutrophil extracellular traps downregulate lipopolysaccharide-induced activation of monocyte-derived dendritic cells. J. Immunol. 2014, 193, 5689–5698. [Google Scholar] [CrossRef]
- Rajnavolgyi, E.; Laczik, R.; Kun, V.; Szente, L.; Fenyvesi, É. Effects of RAMEA-complexed polyunsaturated fatty acids on the response of human dendritic cells to inflammatory signals. Beilstein. J. Org. Chem. 2014, 10, 3152–3160. [Google Scholar] [CrossRef]
- Rana, D.; Duseja, A.; Dhiman, R.K.; Chawla, Y.; Arora, S.K. Maturation defective myeloid dendritic cells in nonalcoholic fatty liver disease patients release inflammatory cytokines in response to endotoxin. Hepatol. Int. 2013, 7, 562–569. [Google Scholar] [CrossRef]
- Khan, Z.; Albalawi, H.; Valle-Perez, A.; Aldoukhi, A.; Hammad, N.; Herrera Ponce de Leon, E.; Abdelrahman, S.; Hauser, C. From 3D printed molds to bioprinted scaffolds: A hybrid material extrusion and vat polymerization bioprinting approach for soft matter constructs. Mater. Sci. Add. Manuf. 2022, 1, 7. [Google Scholar] [CrossRef]
- Goh, G.D.; Sing, S.L.; Lim, Y.F.; Thong, J.L.J.; Peh, Z.K.; Mogali, S.R.; Yeong, W.Y. Machine learning for 3D printed multi-materials tissue-mimicking anatomical models. Mater. Design 2021, 211, 110125. [Google Scholar] [CrossRef]
- Venzac, B.; Deng, S.; Mahmoud, Z.; Lenferink, A.; Costa, A.; Bray, F.; Otto, C.; Rolando, C.; Le Gac, S. PDMS Curing Inhibition on 3D-Printed Molds: Why? Also, How to Avoid It? Anal. Chem. 2021, 93, 7180–7187. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
van der Borg, G.; Warner, H.; Ioannidis, M.; van den Bogaart, G.; Roos, W.H. PLA 3D Printing as a Straightforward and Versatile Fabrication Method for PDMS Molds. Polymers 2023, 15, 1498. https://doi.org/10.3390/polym15061498
van der Borg G, Warner H, Ioannidis M, van den Bogaart G, Roos WH. PLA 3D Printing as a Straightforward and Versatile Fabrication Method for PDMS Molds. Polymers. 2023; 15(6):1498. https://doi.org/10.3390/polym15061498
Chicago/Turabian Stylevan der Borg, Guus, Harry Warner, Melina Ioannidis, Geert van den Bogaart, and Wouter H. Roos. 2023. "PLA 3D Printing as a Straightforward and Versatile Fabrication Method for PDMS Molds" Polymers 15, no. 6: 1498. https://doi.org/10.3390/polym15061498
APA Stylevan der Borg, G., Warner, H., Ioannidis, M., van den Bogaart, G., & Roos, W. H. (2023). PLA 3D Printing as a Straightforward and Versatile Fabrication Method for PDMS Molds. Polymers, 15(6), 1498. https://doi.org/10.3390/polym15061498







