Immobilization of BoPAL3 Phenylalanine Ammonia-Lyase on Electrospun Nanofibrous Membranes of Polyvinyl Alcohol/Nylon 6/Chitosan Crosslinked with Dextran Polyaldehyde
Abstract
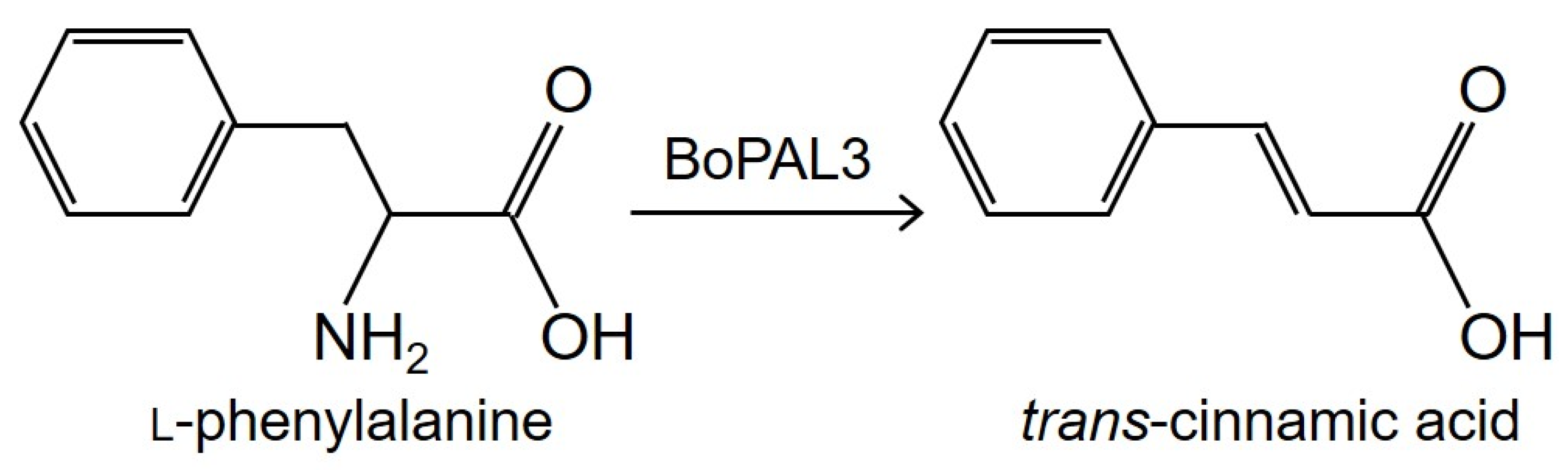
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Expressions of Recombinant BoPAL3 Protein in Escherichia coli
2.3. Preparation of Recombinant BoPAL3 Protein
2.4. Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis
2.5. Preparation of Nanofibrous Membrane by Electrospinning Method
2.6. Preparation of Dextran Polyaldehyde Crosslinker
2.7. Immobilized BoPAL3 Protein on Nanofiber Membrane by Crosslinking
2.8. Biochemical Properties Analysis of Free and Immobilized BoPAL3 Proteins
2.9. Determination of Reusability and Denaturant Tolerance of Immobilized BoPAL3 Protein
2.10. Data Statistical Analysis
3. Results and Discussion
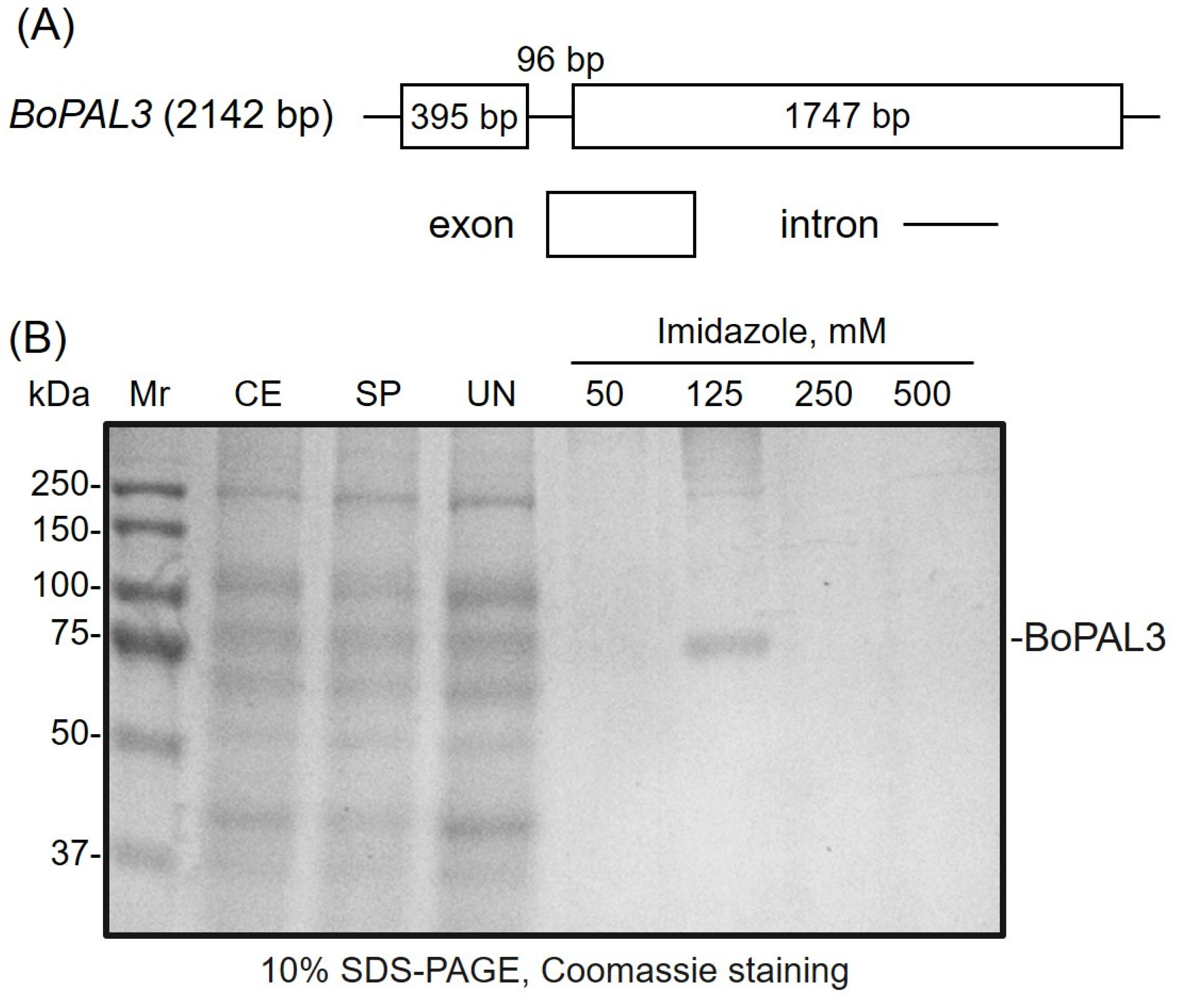
3.1. Preparation of Recombinant BoPAL3 Protein by Affinity Chromatography
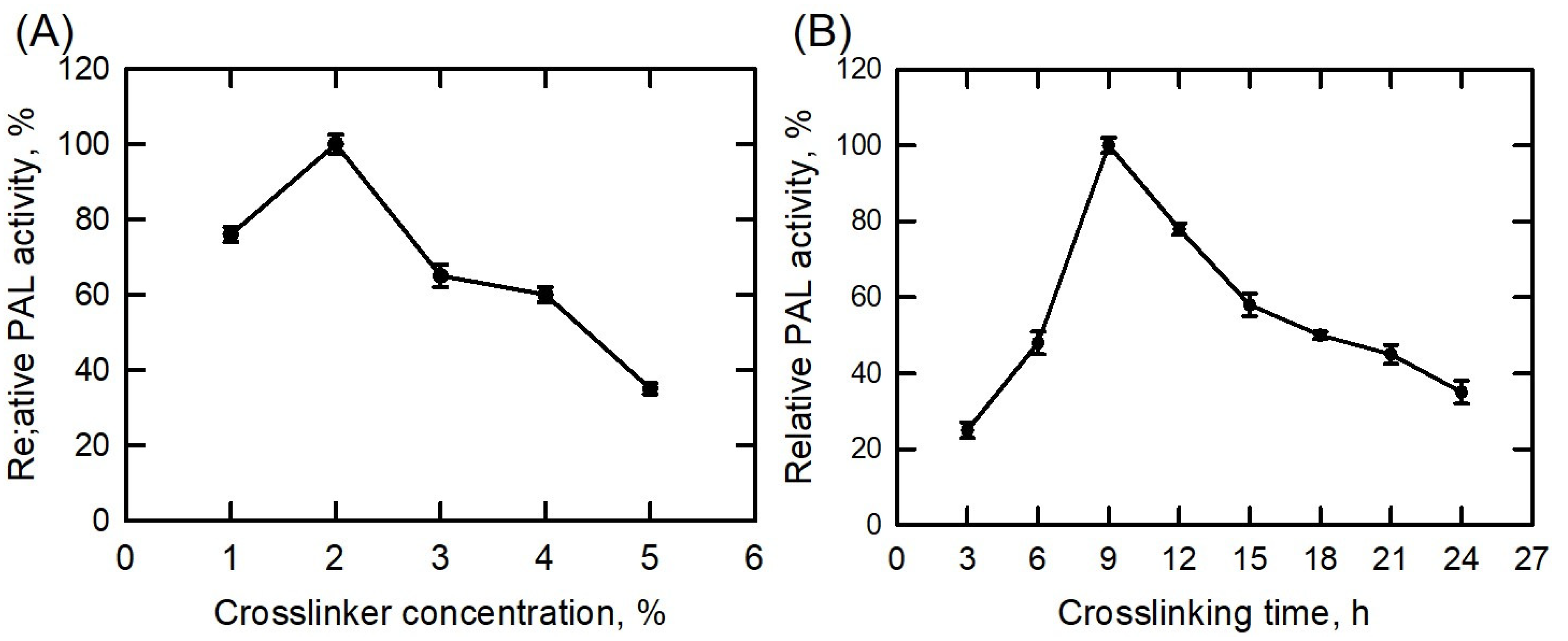
3.2. Crosslinking Condition Optimization
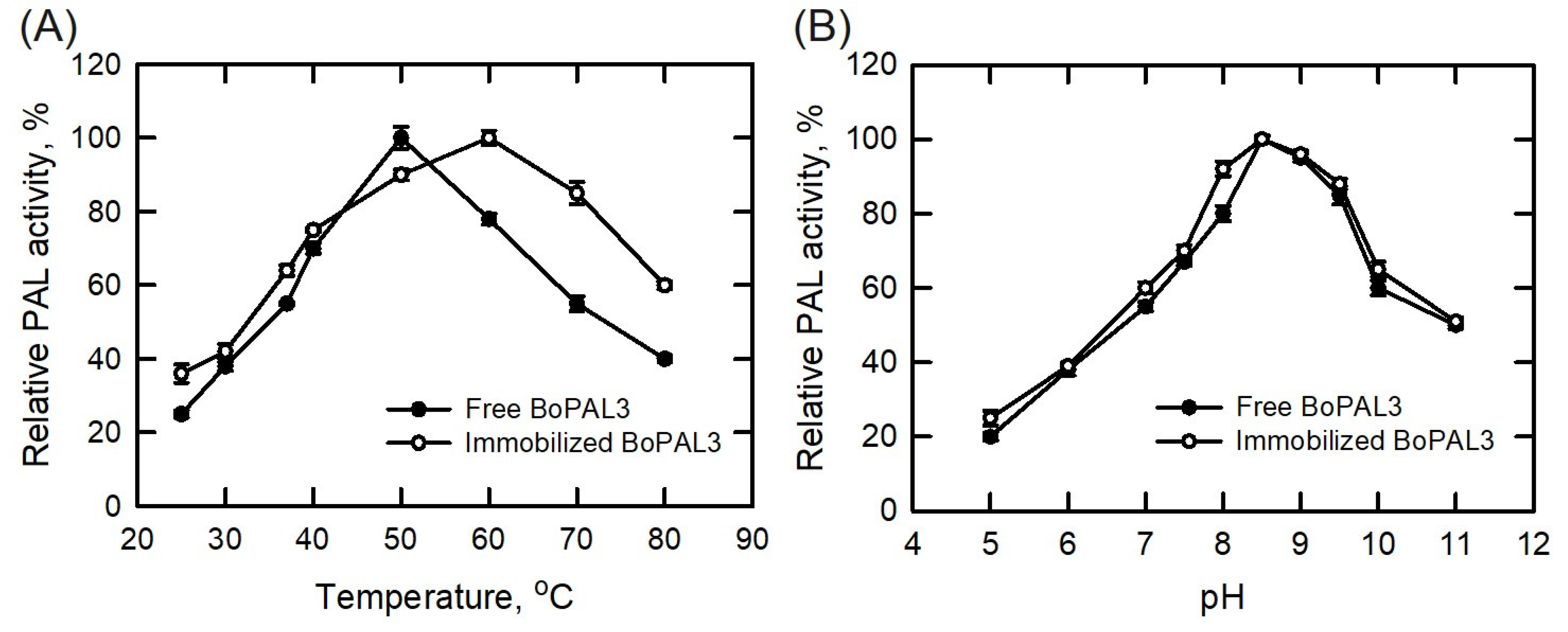
3.3. Temperature and pH Stability of Free and Immobilized BoPAL3 Proteins
3.4. Kinetic Parameters of Free and Immobilized BoPAL3 Proteins
3.5. Reusability and Storage Stability of Free and Immobilized BoPAL3 Proteins
3.6. Denaturant Tolerance Assay between Free and Immobilized BoPAL3 Proteins
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Koukol, J.; Conn, E.E. Metabolism of aromatic compounds in higher plants. IV. Purification and properties of phenylalanine deaminase of Hordeum vulgare. J. Biol. Chem. 1961, 236, 2692–2698. [Google Scholar] [CrossRef] [PubMed]
- Bate, N.J.; Orr, J.; Ni, W.; Meromi, A.; Nadler-Hassar, T.; Doerner, P.W.; Elkind, Y. Quantitative relationship between phenylalanine ammonia-lyase levels and phenylpropanoid accumulation in transgenic tobacco identifies a rate-determining step in natural product synthesis. Proc. Natl Acad. Sci. USA 1994, 91, 7608–7612. [Google Scholar] [CrossRef]
- Huang, Y.H.; You, W.C.; Hong, P.Y.; Chang, Y.T.; Ciou, J.Y.; Hsieh, L.S. Molecular characterization of the Bambusa oldhamii BoPAL3-encoded phenylalanine ammonia-lyase. Phytochem. Lett. 2022, 48, 15–18. [Google Scholar] [CrossRef]
- Wanner, L.A.; Li, G.; Ware, D.; Somssich, I.E.; Davis, K.R. The phenylalanine ammonia-lyase gene family in Arabidopsis thaliana. Plant Mol. Biol. 1995, 27, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Cochrane, F.C.; Davin, L.B.; Lewis, N.G. The Arabidopsis phenylalanine ammonia-lyase gene family: Kinetic characterization of the four PAL isoforms. Phytochemistry 2004, 65, 1557–1564. [Google Scholar] [CrossRef]
- Hsieh, L.S.; Yeh, C.S.; Pan, H.C.; Cheng, C.Y.; Yang, C.C.; Lee, P.D. Cloning and expression of a phenylalanine ammonia-lyase gene (BoPAL2) from Bambusa oldhamii in Escherichia coli and Pichia pastoris. Protein Expr. Purif. 2010, 71, 224–230. [Google Scholar] [CrossRef]
- Hsieh, L.S.; Ma, G.J.; Yang, C.C.; Lee, P.D. Cloning, expression, site-directed mutagenesis and immunolocalization of phenylalanine ammonia-lyase in Bambusa oldhamii. Phytochemistry 2010, 71, 1999–2009. [Google Scholar] [CrossRef]
- Hsieh, L.S.; Hsieh, Y.L.; Yeh, C.S.; Cheng, C.Y.; Yang, C.C.; Lee, P.D. Molecular characterization of a phenylalanine ammonia-lyase gene (BoPAL1) from Bambusa oldhamii. Mol. Biol. Rep. 2011, 38, 283–290. [Google Scholar] [CrossRef]
- Hsieh, C.Y.; Huang, Y.H.; Lin, Z.Y.; Hsieh, L.S. Insights into the substrate selectivity of Bambusa oldhamii phenylalanine ammonia-lyase 1 and 2 through mutational analysis. Phytochem. Lett. 2020, 38, 140–143. [Google Scholar] [CrossRef]
- Hsieh, C.Y.; Huang, Y.H.; Yeh, H.H.; Hong, P.Y.; Hsiao, C.J.; Hsieh, L.S. Phenylalanine, tyrosine, and DOPA are bona fide substrates for Bambusa oldhamii BoPAL4. Catalysts 2021, 11, 1263. [Google Scholar] [CrossRef]
- Moffitt, M.C.; Louie, G.V.; Bowman, M.E.; Pence, J.; Noel, J.P.; Moore, B.S. Discovery of the cyanobacteria phenylalanine ammonia-lyases: Kinetic and structural characterization. Biochemistry 2007, 46, 4286–4289. [Google Scholar] [CrossRef]
- Hyun, M.W.; Yun, Y.H.; Kim, J.Y.; Kim, S.H. Fungal and plant phenylalanine ammonia-lyase. Mycobiology 2011, 39, 257–265. [Google Scholar] [CrossRef]
- Zhang, F.; Ren, J.; Zhan, J. Identification and characterization of an efficient phenylalanine ammonia-lyase from Photorhabdus luminescens. Appl. Biochem. Biotechnol. 2021, 193, 1099–1115. [Google Scholar] [CrossRef] [PubMed]
- Hahlbrock, K.; Scheel, D. Physiology and molecular biology of phenylpropanoid metabolism. Ann. Rev. Plant Physiol. Plant Mol. Biol. 1989, 40, 347–369. [Google Scholar] [CrossRef]
- Deng, Y.; Shanfa, L. Biosynthesis and regulation of phenylpropanoids in plants. CRC Crit. Rev. Plant Sci. 2017, 36, 257–290. [Google Scholar] [CrossRef]
- Chen, O.; Deng, L.; Ruan, C.; Yi, L.; Zeng, K. Pichia galeiformis induced resistance in postharvest citrus by activating the phenylpropanoid biosynthesis pathway. J. Agric. Food Chem. 2021, 69, 2619–2631. [Google Scholar] [CrossRef]
- Anderson, R.A.; Broadhurst, C.L.; Polansky, M.M.; Schmidt, W.F.; Khan, A.; Flanagan, V.P.; Graves, D.J. Isolation and characterization of polyphenol type-A polymers from cinnamon with insulin-like biological activity. J. Agric. Food Chem. 2004, 52, 65–70. [Google Scholar] [CrossRef]
- Soliman, M.M.; Attia, H.F.; El-Shazly, S.A.; Saleh, O.M. Biomedical effects of cinnamon extract on obesity and diabetes relevance in Wistar rats. Am. J. Biochem. Mol. Biol. 2012, 2, 133–145. [Google Scholar] [CrossRef]
- Yılmaz, S.; Ergün, S. Trans-cinnamic acid application for rainbow trout (Oncorhynchus mykiss): I. Effects on haematological, serum biochemical, non-specific immune and head kidney gene expression responses. Fish Shellfish Immunol. 2018, 78, 140–157. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, Y.; Wang, J.; Zhou, M.; Wang, M.; Feng, J. Antifungal activity and action mechanism of the natural product cinnamic acid against Sclerotinina sclerotiorum. Plant Dis. 2019, 103, 944–950. [Google Scholar] [CrossRef]
- Silva, A.T.; Bento, C.M.; Pena, A.C.; Figueiredo, L.M.; Prudêncio, C.; Aguiar, L.; Teixeira, C. Cinnamic acid conjugates in the rescuing and repurposing of classical antimalarial drugs. Molecules 2019, 25, 66. [Google Scholar] [CrossRef]
- Kang, N.H.; Mukherjee, S.; Yun, J.W. Trans-cinnamic acid stimulates white fat browning and activates brown adipocytes. Nutrients 2019, 11, 577. [Google Scholar] [CrossRef]
- Wang, Z.; Ge, S.; Li, S.; Lin, H.; Lin, S. Anti-obesity effect of trans-cinnamic acid on HepG2 cells and HFD-fed mice. Food Chem. Toxicol. 2020, 137, 111148. [Google Scholar] [CrossRef]
- Gunia-Krzyżak, A.; Słoczyńska, K.; Popiół, J.; Koczurkiewicz, P.; Marona, H.; Pękala, E. Cinnamic acid derivatives in cosmetics: Current use and future prospects. Int. J. Cosmet. Sci. 2018, 40, 356–366. [Google Scholar] [CrossRef] [PubMed]
- De, P.; Baltas, M.; Bedos-Belval, F. Cinnamic acid derivatives as anticancer agents-a review. Curr. Med. Chem. 2011, 18, 1672–1703. [Google Scholar] [CrossRef] [PubMed]
- Sova, M. Antioxidant and antimicrobial activities of cinnamic acid derivatives. Mini-Rev. Med. Chem. 2012, 12, 749–767. [Google Scholar] [CrossRef] [PubMed]
- Ruwizhi, N.; Aderibigbe, B.A. Cinnamic acid derivatives and their biological efficacy. Int. J. Mol. Sci. 2020, 21, 5712. [Google Scholar] [CrossRef]
- Li, Y.H.; Wu, J.X.; He, Q.; Gu, J.; Zhang, L.; Niu, H.Z.; Zhang, X.W.; Zhao, H.T.; Xu, J.Y.; Qin, L.Q. Amelioration of radiation-induced liver damage by p-coumaric acid in mice. Food Sci. Biotechnol. 2022, 31, 1315–1323. [Google Scholar] [CrossRef]
- Ge, J.; Lu, D.; Liu, Z.; Liu, Z. Recent advances in nanostructured biocatalysts. Biochem. Eng. J. 2009, 44, 53–59. [Google Scholar] [CrossRef]
- Datta, S.; Christena, L.; Rajaram, Y. Enzyme immobilization: An overview on techniques and support materials. 3 Biotech 2013, 3, 1–9. [Google Scholar] [CrossRef]
- Huang, Z.M.; Zhang, Y.Z.; Kotaki, M.; Ramakrishna, S. A review on polymer nanofibers by electrospinning and their applications in nanocomposites. Compos. Sci. Technol. 2003, 63, 2223–2253. [Google Scholar] [CrossRef]
- Garcia-Galan, C.; Berenguer-Murcia, Á.; Fernandez-Lafuente, R.; Rodrigues, R.C. Potential of different enzyme immobilization strategies to improve enzyme performance. Adv. Synth. Catal. 2011, 353, 2885–2904. [Google Scholar] [CrossRef]
- Homaei, A.A.; Sariri, R.; Vianello, F.; Stevanato, R. Enzyme immobilization: An update. J. Chem. Biol. 2013, 6, 185–205. [Google Scholar] [CrossRef]
- Basso, A.; Serban, S. Industrial applications of immobilized enzymes—A review. Mol. Catal. 2019, 479, 110607. [Google Scholar] [CrossRef]
- Chen, J.P.; Ho, K.H.; Chiang, Y.P.; Wu, K.W. Fabrication of electrospun poly(methyl methacrylate) nanofibrous membranes by statistical approach for application in enzyme immobilization. J. Membr. Sci. 2009, 340, 9–15. [Google Scholar] [CrossRef]
- Misson, M.; Dai, S.; Jin, B.; Chen, B.H.; Zhang, H. Manipulation of nanofiber-based β-galactosidase nanoenvironment for enhancement of galacto-oligosaccharide production. J. Biotechnol. 2016, 222, 56–64. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Berenguer-Murcia, Á.; Carballares, D.; Morellon-Sterling, R.; Fernandez-Lafuente, R. Stabilization of enzymes via immobilization: Multipoint covalent attachment and other stabilization strategies. Biotechnol. Adv. 2021, 52, 107821. [Google Scholar] [CrossRef] [PubMed]
- Hong, P.Y.; Huang, Y.H.; Lim, G.G.C.W.; Chen, Y.P.; Hsiao, C.J.; Chen, L.H.; Ciou, J.Y.; Hsieh, L.S. Production of trans-cinnamic acid by immobilization of the Bambusa oldhamii BoPAL1 and BoPAL2 phenylalanine ammonia-lyases on electrospun nanofibers. Int. J. Mol. Sci. 2021, 22, 11184. [Google Scholar] [CrossRef]
- Haider, A.; Haider, S.; Kang, I.-K. A comprehensive review summarizing the effect of electrospinning parameters and potential applications of nanofibers in biomedical and biotechnology. Arab. J. Chem. 2018, 11, 1165–1188. [Google Scholar] [CrossRef]
- Kamoun, E.A.; Loutfy, S.A.; Hussein, Y.; Kenawy, E.-R.S. Recent advances in PVA-polysaccharide based hydrogels and electrospun nanofibers in biomedical applications: A review. Int. J. Biol. Macromol. 2021, 187, 755–768. [Google Scholar] [CrossRef]
- Misson, M.; Saallah, S.; Zhang, H. Nanofiber-immobilized β-galactosidase for dairy waste conversion into galacto-oligosaccharides. In Advances in Waste Processing Technology; Springer: Singapore, 2020; pp. 37–48. [Google Scholar] [CrossRef]
- Wu, L.; Yuan, X.; Sheng, J. Immobilization of cellulase in nanofibrous PVA membranes by electrospinning. J. Membr. Sci. 2005, 250, 167–173. [Google Scholar] [CrossRef]
- Dai, M.; Jin, S.; Nugen, S. Water-soluble electrospun nanofibers as a method for on-chip reagent storage. Biosensors 2012, 2, 388–395. [Google Scholar] [CrossRef]
- Bang, H.B.; Lee, K.; Lee, Y.J.; Jeong, K.J. High-level production of trans-cinnamic acid by fed-batch cultivation of Escherichia coli. Process Biochem. 2018, 68, 30–36. [Google Scholar] [CrossRef]
- Son, J.; Jang, J.H.; Choi, I.H.; Lim, C.G.; Jeon, E.J.; Bang, H.B.; Jeong, K.J. Production of trans-cinnamic acid by whole-cell bioconversion from L-phenylalanine in engineered Corynebacterium glutamicum. BMC Microb. Cell Factories 2021, 20, 145. [Google Scholar] [CrossRef]
- Koplányi, G.; Sánta-Bell, E.; Molnár, Z.; Tóth, G.D.; Józó, M.; Szilágyi, A.; Ender, F.; Pukánszky, B.; Vértessy, B.G.; Poppe, L.; et al. Entrapment of phenylalanine ammonia-lyase in nanofibrous polylactic acid matrices by emulsion electrospinning. Catalysts 2021, 11, 1149. [Google Scholar] [CrossRef]
- Hsiao, C.J.; Hsieh, C.Y.; Hsieh, L.S. Cloning and characterization of the Bambusa oldhamii BoMDH-encoded malate dehydrogenase. Protein Expr. Purif. 2020, 174, 105665. [Google Scholar] [CrossRef] [PubMed]
- Hsu, W.H.; Huang, Y.H.; Chen, P.R.; Hsieh, L.S. NLIP and HAD-like domains of Pah1 and Lipin 1 phosphatidate phosphatases are essential for their catalytic activities. Molecules 2021, 26, 5470. [Google Scholar] [CrossRef]
- Huang, Y.H.; You, W.C.; Chen, Y.J.; Ciou, J.Y.; Hsieh, L.S. Insight into the substrate specificity of Lactobacillus paracasei aspartate ammonia-lyase. Fermentation 2023, 9, 49. [Google Scholar] [CrossRef]
- Michaelis, L.; Menten, M.L. Die kinetik der invertinwirkung. Biochem. Z. 1913, 49, 333–369. [Google Scholar] [CrossRef]
- Lineweaver, H.; Burk, D. The determination of enzyme dissociation constants. J. Am. Chem. Soc. 1934, 56, 658–666. [Google Scholar] [CrossRef]
- Sood, A.; Gupta, A.; Agrawal, G. Recent advances in polysaccharides based biomaterials for drug delivery and tissue engineering applications. Carbohydr. Polym. Technol. Appl. 2021, 2, 100067. [Google Scholar] [CrossRef]
- Sojitra, U.V.; Nadar, S.S.; Rathod, V.K. Immobilization of pectinase onto chitosan magnetic nanoparticles by macromolecular cross-linker. Carbohydr. Polym. 2017, 157, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Hardian, R.; Alammar, A.; Holtzl, T.; Szekely, G. Fabrication of sustainable organic solvent nanofiltration membranes using cellulose-chitosan biopolymer blends. J. Membr. Sci. 2022, 658, 120743. [Google Scholar] [CrossRef]
- Bouyer, D.; Méricq, J.-P.; Wlodarczyk, D.; Soussan, L.; Faur, C. How mass and heat transfers could affect chitosan membrane formation via an enzymatic gelation. J. Membr. Sci. 2022, 669, 121298. [Google Scholar] [CrossRef]
- Wang, B.; Cheng, F.; Lu, Y.; Ge, W.; Zhang, M.; Yue, B. Immobilization of pectinase from Penicillium oxalicum F67 onto magnetic cornstarch microspheres: Characterization and application in juice production. J. Mol. Catal. B Enzym. 2013, 97, 137–143. [Google Scholar] [CrossRef]
- Fatarella, E.; Spinelli, D.; Ruzzante, M.; Pogni, R. Nylon 6 film and nanofiber carriers: Preparation and laccase immobilization performance. J. Mol. Catal. B Enzym. 2014, 102, 41–47. [Google Scholar] [CrossRef]
- Nadar, S.S.; Rathod, V.K. Magnetic macromolecular cross linked enzyme aggregates (CLEAs) of glucoamylase. Enzym. Microb. Technol. 2016, 83, 78–87. [Google Scholar] [CrossRef]
- Cui, J.; Zhao, Y.; Feng, Y.; Lin, T.; Zhong, C.; Tan, Z.; Jia, S. Encapsulation of spherical cross-linked phenylalanine ammonia lyase aggregates in mesoporous biosilica. J. Agri. Food Chem. 2017, 65, 618–625. [Google Scholar] [CrossRef]
- Cui, J.; Zhao, Y.; Tan, Z.; Zhong, C.; Han, P.; Jia, S. Mesoporous phenylalanine ammonia lyase microspheres with improved stability through calcium carbonate templating. Int. J. Biol. Macromol. 2017, 98, 887–896. [Google Scholar] [CrossRef]


| Enzymes | kcat (s−1) | Km (µM) | kcat/Km (s−1 µM−1) | Reference | |||
|---|---|---|---|---|---|---|---|
| Free | Immobilized | Free | Immobilized | Free | Immobilized | ||
| BoPAL1 | 1.01 | 1.21 | 518 | 534 | 1.91 × 10−3 | 2.2 × 10−3 | [38] |
| BoPAL2 | 4.02 | 3.99 | 329 | 379 | 1.23 × 10−2 | 1.05 × 10−2 | [38] |
| BoPAL3 | 1.09 | 1.28 | 200 | 268 | 5.45 × 10−3 | 5.29 × 10−3 | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsieh, C.-Y.; Hong, P.-Y.; Hsieh, L.-S. Immobilization of BoPAL3 Phenylalanine Ammonia-Lyase on Electrospun Nanofibrous Membranes of Polyvinyl Alcohol/Nylon 6/Chitosan Crosslinked with Dextran Polyaldehyde. Polymers 2023, 15, 3699. https://doi.org/10.3390/polym15183699
Hsieh C-Y, Hong P-Y, Hsieh L-S. Immobilization of BoPAL3 Phenylalanine Ammonia-Lyase on Electrospun Nanofibrous Membranes of Polyvinyl Alcohol/Nylon 6/Chitosan Crosslinked with Dextran Polyaldehyde. Polymers. 2023; 15(18):3699. https://doi.org/10.3390/polym15183699
Chicago/Turabian StyleHsieh, Chun-Yen, Pei-Yu Hong, and Lu-Sheng Hsieh. 2023. "Immobilization of BoPAL3 Phenylalanine Ammonia-Lyase on Electrospun Nanofibrous Membranes of Polyvinyl Alcohol/Nylon 6/Chitosan Crosslinked with Dextran Polyaldehyde" Polymers 15, no. 18: 3699. https://doi.org/10.3390/polym15183699
APA StyleHsieh, C.-Y., Hong, P.-Y., & Hsieh, L.-S. (2023). Immobilization of BoPAL3 Phenylalanine Ammonia-Lyase on Electrospun Nanofibrous Membranes of Polyvinyl Alcohol/Nylon 6/Chitosan Crosslinked with Dextran Polyaldehyde. Polymers, 15(18), 3699. https://doi.org/10.3390/polym15183699







