Design of Potent and Salt-Insensitive Antimicrobial Branched Peptides
Abstract
:1. Introduction
2. Materials and Methods
3. Results
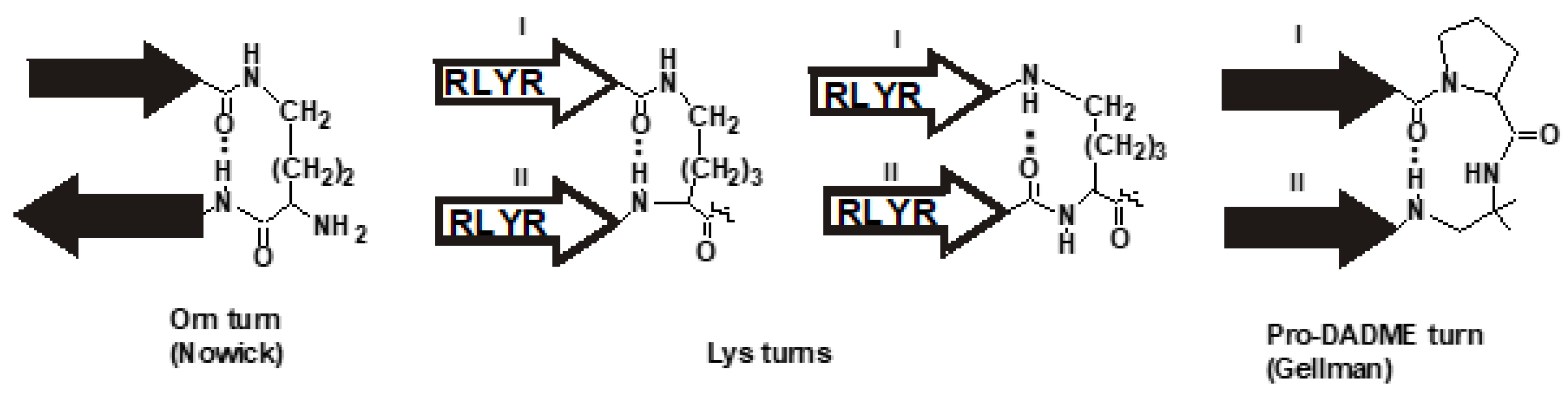
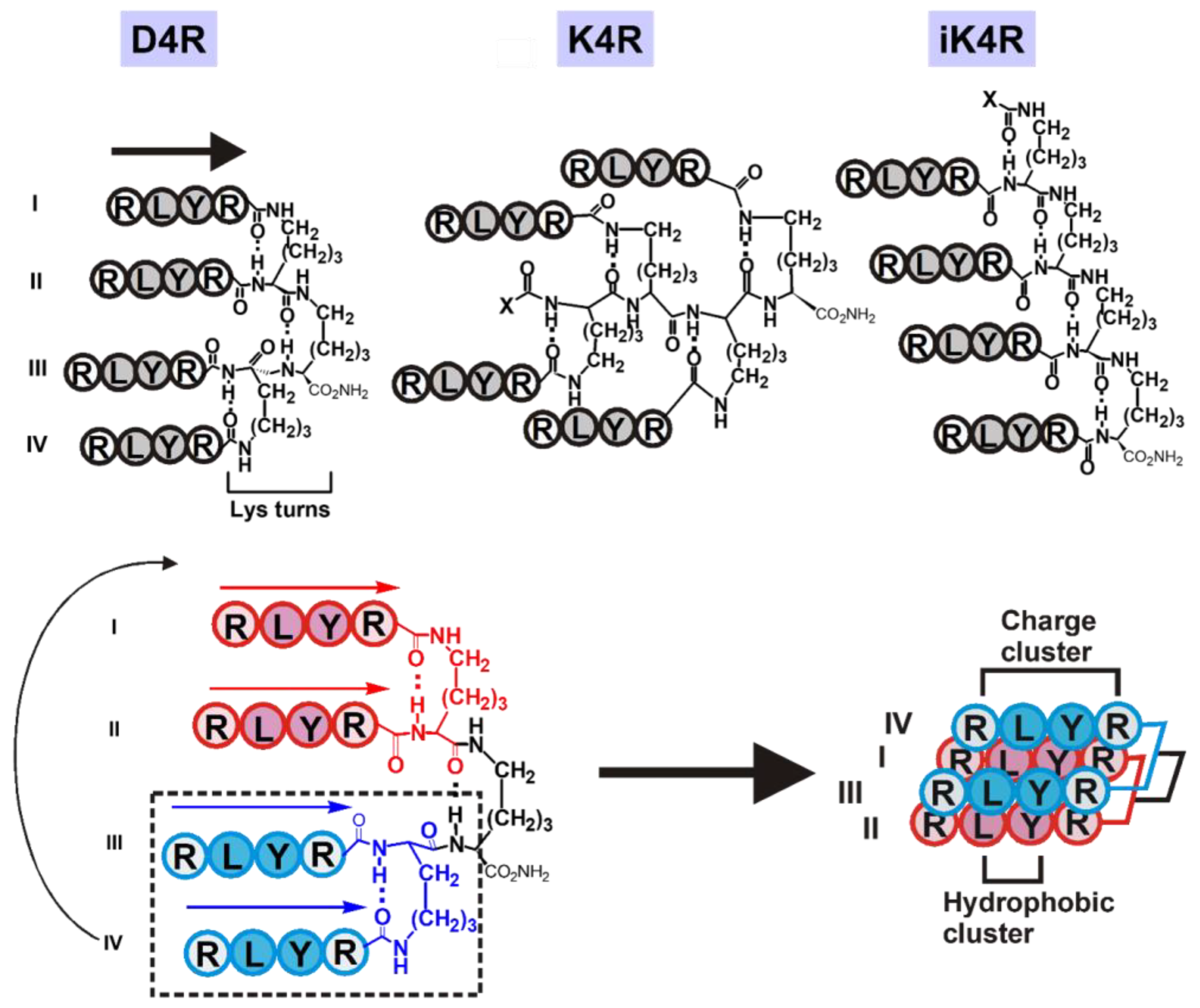
3.1. Hypothesis and Design
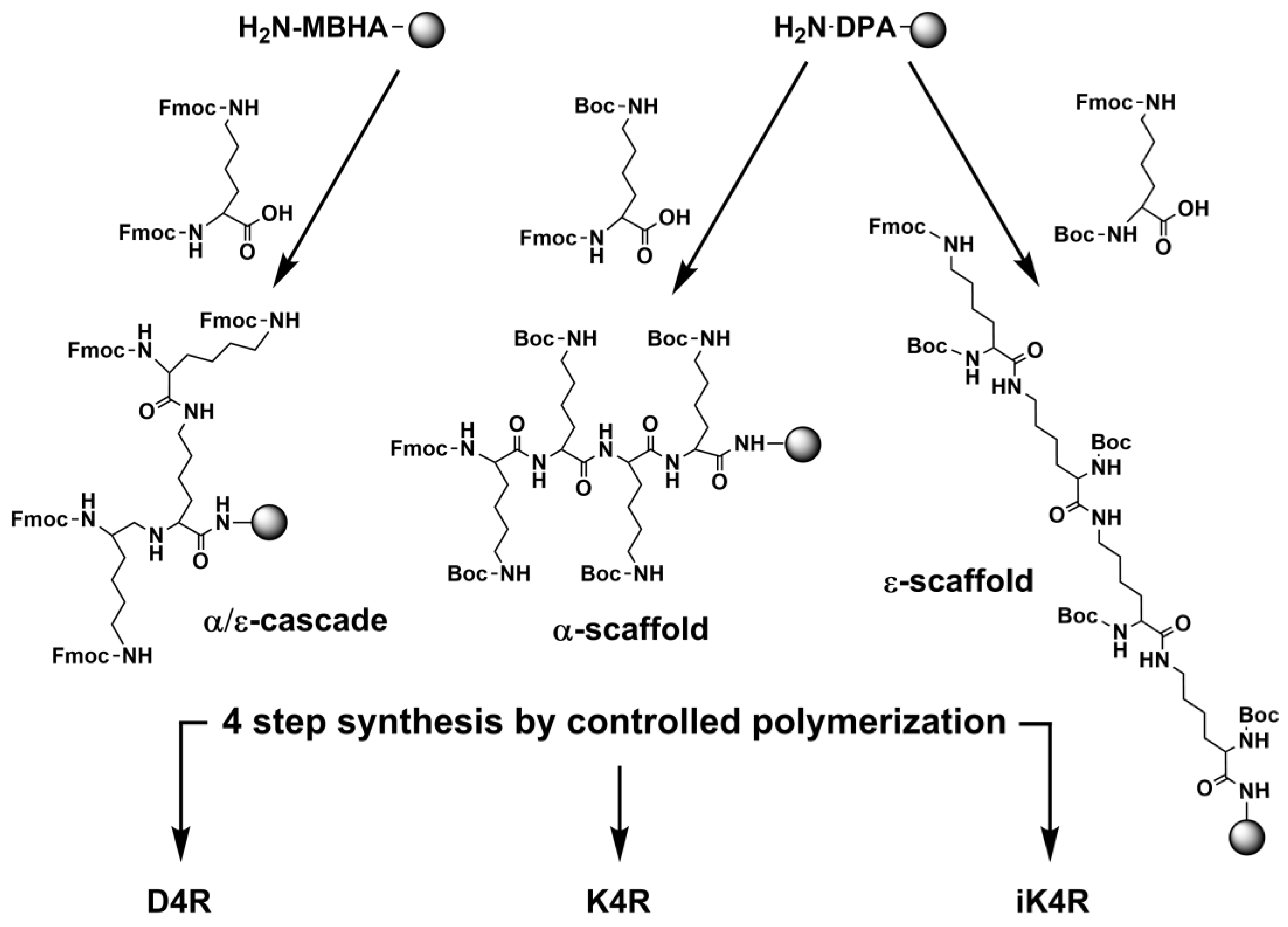
3.2. Synthesis by Controlled Polymerization
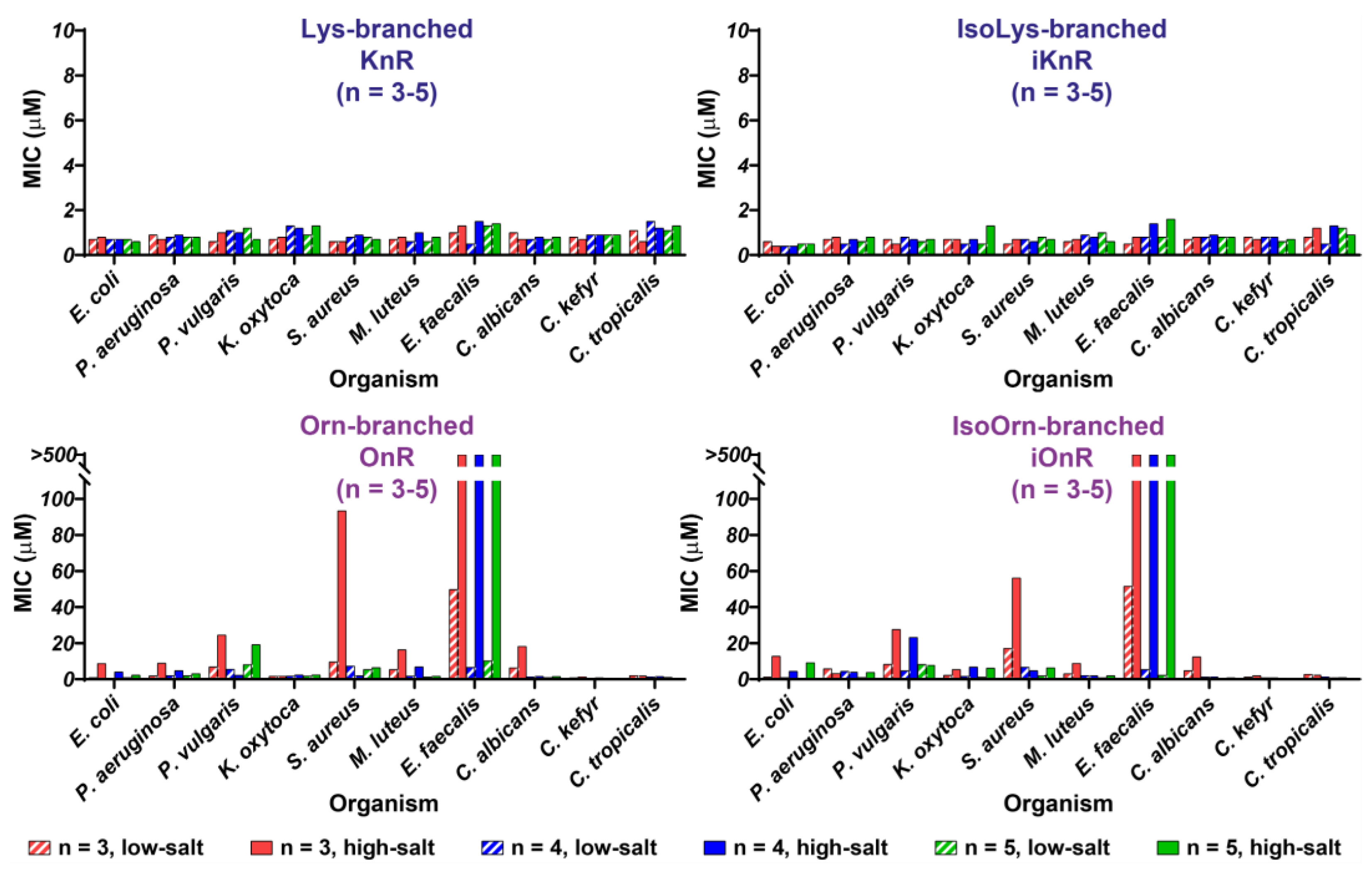
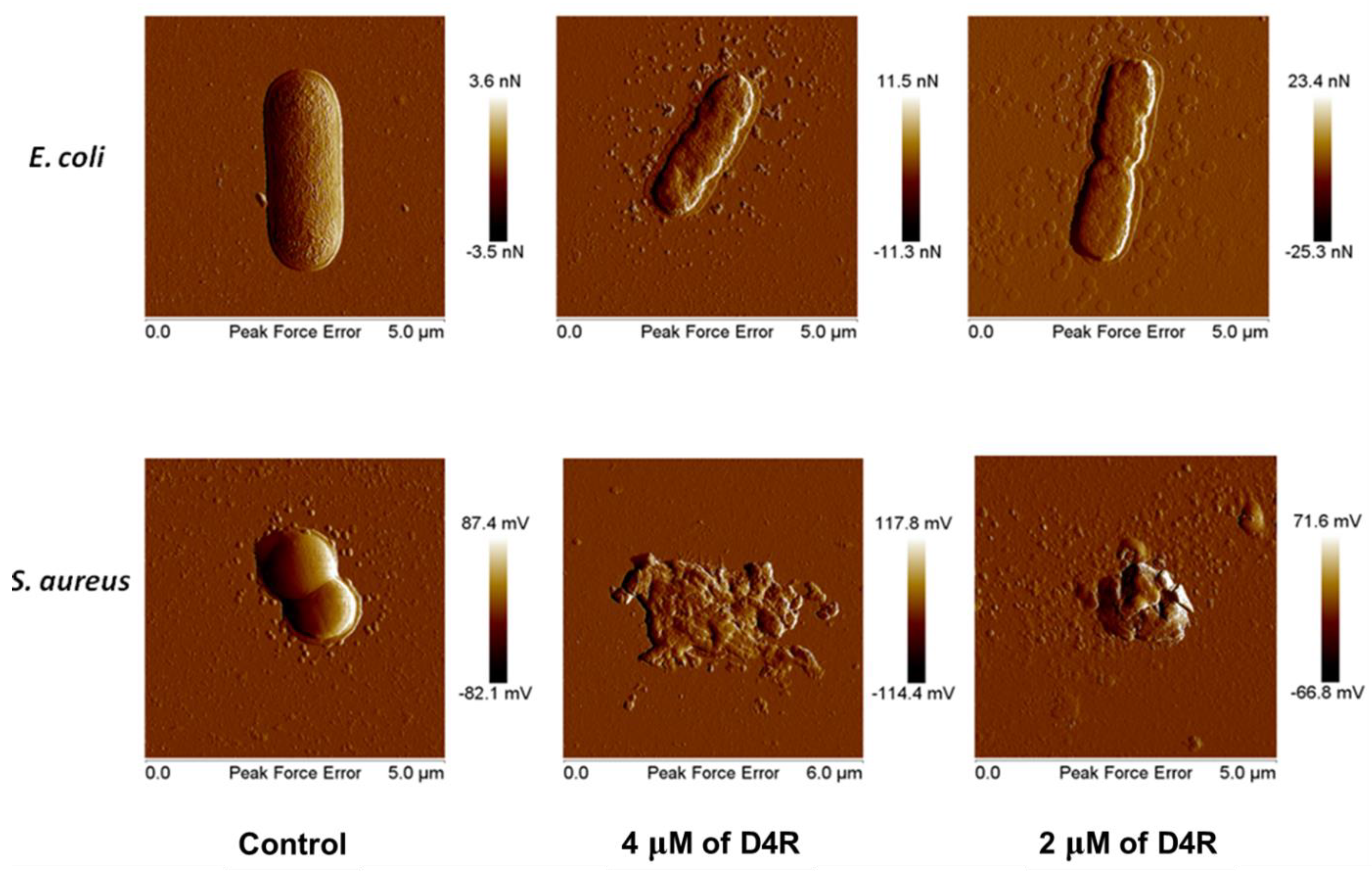
3.3. Antimicrobial Profiles of Orn- and Lys-Scaffold Branched Peptides
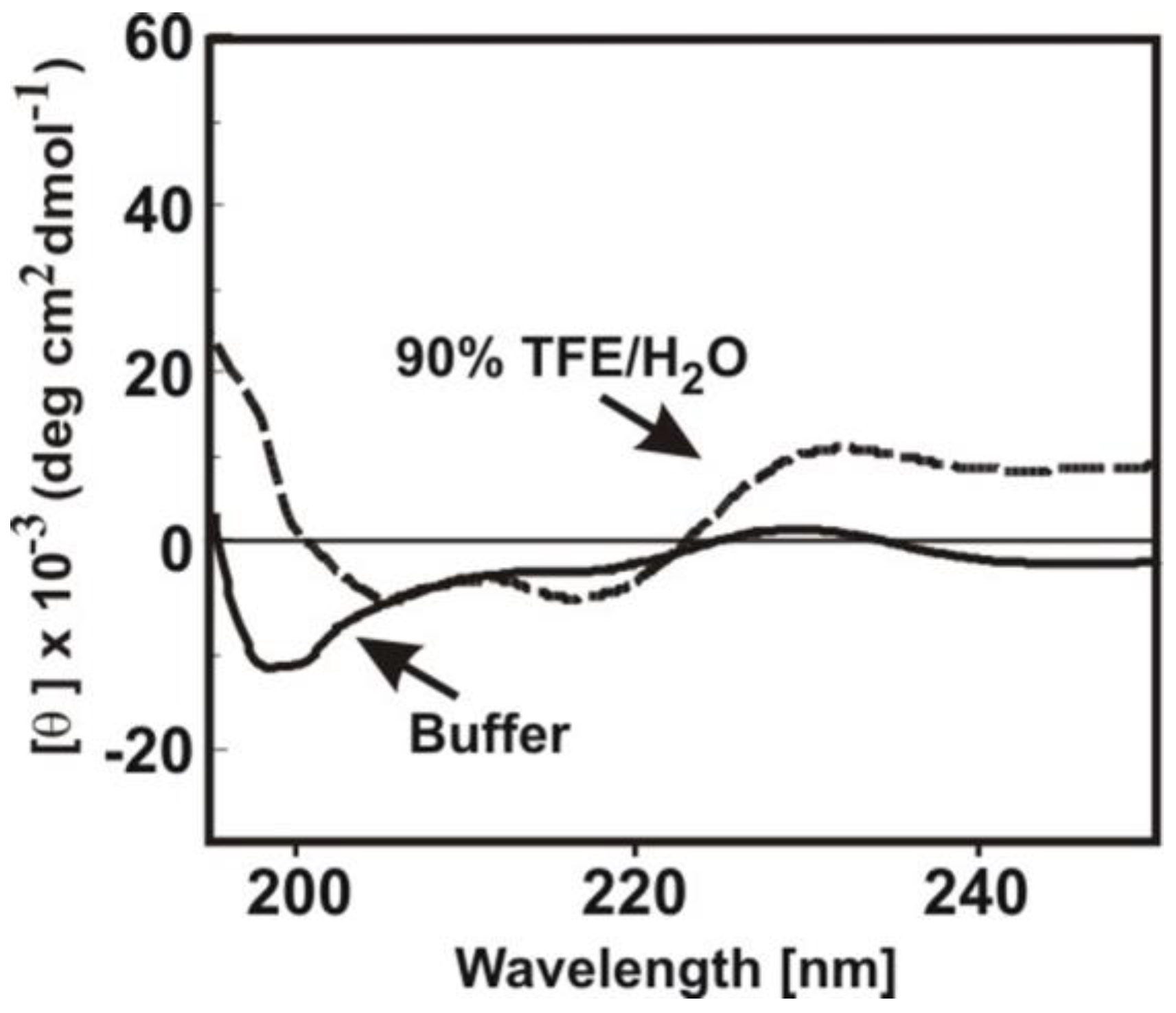
3.4. Structure-Activity Relationships
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Ganz, T.; Lehrer, R.I. Antimicrobial peptides of vertebrates. Curr. Opin. Immunol. 1998, 10, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Z.; Yang, P.; Lei, J.; Zhao, J. Biological Function of Antimicrobial Peptides on Suppressing Pathogens and Improving Host Immunity. Antibiotics 2023, 12, 1037. [Google Scholar] [CrossRef] [PubMed]
- Moretta, A.; Scieuzo, C.; Petrone, A.M.; Salvia, R.; Manniello, M.D.; Franco, A.; Lucchetti, D.; Vassallo, A.; Vogel, H.; Sgambato, A.; et al. Antimicrobial Peptides: A New Hope in Biomedical and Pharmaceutical Fields. Front. Cell. Infect. Microbiol. 2021, 11, 668632. [Google Scholar] [CrossRef]
- Montalbán-López, M.; Scott, T.A.; Ramesh, S.; Rahman, I.R.; Van Heel, A.J.; Viel, J.H.; Bandarian, V.; Dittmann, E.; Genilloud, O.; Goto, Y.; et al. New developments in RiPP discovery, enzymology and engineering. Nat. Prod. Rep. 2021, 38, 130–239. [Google Scholar] [CrossRef]
- Chen, N.; Jiang, C. Antimicrobial peptides: Structure, mechanism, and modification. Eur. J. Med. Chem. 2023, 255, 115377. [Google Scholar] [CrossRef]
- Meister, M.; Lemaitre, B.; Hoffmann, J.A. Antimicrobial peptide defense in Drosophila. BioEssays 1997, 19, 1019–1026. [Google Scholar] [CrossRef]
- Ouellette, A.J.; Selsted, M.E. Paneth cell defensins: Endogenous peptide components of intestinal host defense. FASEB J. 1996, 10, 1280–1289. [Google Scholar] [CrossRef]
- Nicolas, P.; Mor, A. Peptides as weapons against microorganisms in the chemical defense system of vertebrates. Annu. Rev. Microbiol. 1995, 49, 277–304. [Google Scholar] [CrossRef]
- Danders, W.; Marahiel, M.A.; Krause, M.; Kosui, N.; Kato, T.; Izumiya, N.; Kleinkauf, H. Antibacterial action of gramicidin S and tyrocidines in relation to active transport, in vitro transcription, and spore outgrowth. Antimicrob. Agents Chemother. 1982, 22, 785–790. [Google Scholar] [CrossRef]
- Bernhard, W.; Avrameas, S. Ultrastructural visualization of cellular carbohydrate components by means of concanavalin A. Exp. Cell Res. 1971, 64, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Vogler, K.; Studer, R.O. The chemistry of the polymyxin antibiotics. Experientia 1966, 22, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.Q.; Yuan, J.; Ösapay, G.; Ösapay, K.; Tran, D.; Miller, C.J.; Ouellette, A.J.; Selsted, M.E. A cyclic antimicrobial peptide produced in primate leukocytes by the ligation of two truncated α-defensins. Science 1999, 286, 498–502. [Google Scholar] [CrossRef]
- Seebach, D.; Matthews, J.L. β-peptides: A surprise at every turn. Chem. Commun. 1997, 2015–2022. [Google Scholar] [CrossRef]
- Gellman, S.H. Foldamers: A Manifesto. Acc. Chem. Res. 1998, 31, 173–180. [Google Scholar] [CrossRef]
- Liu, D.; DeGrado, W.F. De novo design, synthesis, and characterization of antimicrobial β-peptides. J. Am. Chem. Soc. 2001, 123, 7553–7559. [Google Scholar] [CrossRef]
- Hintermann, T.; Gademann, K.; Jaun, B.; Seebach, D. γ-Peptides Forming More Stable Secondary Structures than α-Peptides: Synthesis and Helical NMR-Solution Structure of the γ-Hexapeptide Analog of H-(Val-Ala-Leu)2-OH. Helv. Chim. Acta 1998, 81, 983–1002. [Google Scholar] [CrossRef]
- Karig, G.; Fuchs, A.; Büsing, A.; Brandstetter, T.; Scherer, S.; Bats, J.W.; Eschenmoser, A.; Quinkert, G. δ-Peptide analogues of pyranosyl-RNA. Part 1. Nucleo-δ-peptides derived from conformationally constrained nucleo-δ-amino acids: Preparation of monomers. Helv. Chim. Acta 2000, 83, 1049–1078. [Google Scholar] [CrossRef]
- Shima, S.; Matsuoka, H.; Iwamoto, T.; Sakai, H. Antimicrobial action of epsilon-poly-L-lysine. J. Antibiot. 1984, 37, 1449–1455. [Google Scholar] [CrossRef]
- Szókén, G.; Almas, M.; Krizsán, K.; Khlafulla, A.R.; Tyihák, E.; Szende, B. Structure determination and synthesis of lysine isopeptides influencing on cell proliferation. Biopolym. Nucleic Acid Sci. Sect. 1997, 42, 305–318. [Google Scholar] [CrossRef]
- Lee, D.U.; Park, Y.J.; Yu, H.H.; Jung, S.C.; Park, J.H.; Lee, D.H.; Lee, N.K.; Paik, H.D. Antimicrobial and Antibiofilm Effect of ε-Polylysine against Salmonella Enteritidis, Listeria monocytogenes, and Escherichia coli in Tryptic Soy Broth and Chicken Juice. Foods 2021, 10, 2211. [Google Scholar] [CrossRef] [PubMed]
- Matthyssen, T.; Li, W.; Holden, J.A.; Lenzo, J.C.; Hadjigol, S.; O’Brien-Simpson, N.M. The Potential of Modified and Multimeric Antimicrobial Peptide Materials as Superbug Killers. Front. Chem. 2021, 9, 795433. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.-Y.; Yan, Z.-B.; Meng, Y.-M.; Hong, X.-Y.; Shao, G.; Ma, J.-J.; Cheng, X.-R.; Liu, J.; Kang, J.; Fu, C.-Y. Antimicrobial peptides: Mechanism of action, activity and clinical potential. Mil. Med. Res. 2021, 8, 48. [Google Scholar] [CrossRef]
- Muhle, S.A.; Tam, J.P. Design of Gram-negative selective antimicrobial peptides. Biochemistry 2001, 40, 5777–5785. [Google Scholar] [CrossRef] [PubMed]
- Tam, J.P.; Lu, Y.A.; Yang, J.L. Design of salt-insensitive glycine-rich antimicrobial peptides with cyclic tricystine structures. Biochemistry 2000, 39, 7159–7169. [Google Scholar] [CrossRef]
- Tam, J.P.; Lu, Y.A.; Yang, J.L. Marked increase in membranolytic selectivity of novel cyclic tachyplesins constrained with an antiparallel two-β strand cystine knot framework. Biochem. Biophys. Res. Commun. 2000, 267, 783–790. [Google Scholar] [CrossRef]
- Tam, J.P.; Lu, Y.A.; Yang, J.L.; Chiu, K.W. An unusual structural motif of antimicrobial peptides containing end-to-end macrocycle and cystine-knot disulfides. Proc. Natl. Acad. Sci. USA 1999, 96, 8913–8918. [Google Scholar] [CrossRef]
- Hemu, X.; Taichi, M.; Qiu, Y.; Liu, D.X.; Tam, J.P. Biomimetic synthesis of cyclic peptides using novel thioester surrogates. Biopolymers 2013, 100, 492–501. [Google Scholar] [CrossRef]
- Kokryakov, V.N.; Harwig, S.S.L.; Panyutich, E.A.; Shevchenko, A.A.; Aleshina, G.M.; Shamova, O.V.; Korneva, H.A.; Lehrer, R.I. Protegrins: Leukocyte antimicrobial peptides that combine features of corticostatic defensins and tachyplesins. FEBS Lett. 1993, 327, 231–236. [Google Scholar] [CrossRef]
- Nakamura, T.; Furunaka, H.; Miyata, T.; Tokunaga, F.; Muta, T.; Iwanaga, S.; Niwa, M.; Takao, T.; Shimonishi, Y. Tachyplesin, a class of antimicrobial peptide from the hemocytes of the horseshoe crab (Tachypleus tridentatus). Isolation and chemical structure. J. Biol. Chem. 1988, 263, 16709–16713. [Google Scholar] [CrossRef]
- Tam, J.P. Synthetic peptide vaccine design: Synthesis and properties of a high-density multiple antigenic peptide system. Proc. Natl. Acad. Sci. USA 1988, 85, 5409–5413. [Google Scholar] [CrossRef] [PubMed]
- Posnett, D.N.; McGrath, H.; Tam, J.P. A novel method for producing anti-peptide antibodies. Production of site-specific antibodies to the T cell antigen receptor β-chain. J. Biol. Chem. 1988, 263, 1719–1725. [Google Scholar] [CrossRef] [PubMed]
- Tam, J.P. Recent advances in multiple antigen peptides. J. Immunol. Methods 1996, 196, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Hemu, X.; Tam, J.P. Macrocyclic Antimicrobial Peptides Engineered from ω-Conotoxin. Curr. Pharm. Des. 2017, 23, 2131–2138. [Google Scholar] [CrossRef]
- Merrifield, R.B. Solid Phase Peptide Synthesis. I. The Synthesis of a Tetrapeptide. J. Am. Chem. Soc. 1963, 85, 2149–2154. [Google Scholar] [CrossRef]
- Castro, B.; Dormoy, J.; Evin, G.; Selve, C. Reactifs de couplage peptidique I (1)-l’hexafluorophosphate de benzotriazolyl N-oxytrisdimethylamino phosphonium (BOP). Tetrahedron Lett. 1975, 16, 1219–1222. [Google Scholar] [CrossRef]
- Lehrer, R.I.; Rosenman, M.; Harwig, S.S.; Jackson, R.; Eisenhauer, P. Ultrasensitive assays for endogenous antimicrobial polypeptides. J. Immunol. Methods 1991, 137, 167–173. [Google Scholar] [CrossRef]
- Matsuzaki, K.; Yoneyama, S.; Fujii, N.; Miyajima, K.; Yamada, K.-i.; Kirino, Y.; Anzai, K. Membrane permeabilization mechanisms of a cyclic antimicrobial peptide, tachyplesin I, and its linear analog. Biochemistry 1997, 36, 9799–9806. [Google Scholar] [CrossRef]
- Fisk, J.D.; Gellman, S.H. A parallel β-sheet model system that folds in water. J. Am. Chem. Soc. 2001, 123, 343–344. [Google Scholar] [CrossRef]
- Woll, M.G.; Lai, J.R.; Guzei, I.A.; Taylor, S.J.C.; Smith, M.E.B.; Gellman, S.H. Parallel Sheet Secondary Structure in γ-Peptides. J. Am. Chem. Soc. 2001, 123, 11077–11078. [Google Scholar] [CrossRef]
- Nowick, J.S.; Brower, J.O. A New Turn Structure for the Formation of β-Hairpins in Peptides. J. Am. Chem. Soc. 2003, 125, 876–877. [Google Scholar] [CrossRef] [PubMed]
- Gross, E.; Morell, J.L. The Structure of Nisin. J. Am. Chem. Soc. 1971, 93, 4634–4635. [Google Scholar] [CrossRef] [PubMed]
- Woody, R.W. Circular dichroism. Methods Enzym. 1995, 246, 34–71. [Google Scholar] [CrossRef]
- Sreerama, N.; Venyaminov, S.Y.; Woody, R.W. Estimation of the number of alpha-helical and beta-strand segments in proteins using circular dichroism spectroscopy. Protein Sci. 1999, 8, 370–380. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, D.; Mathur, K.; Balasubramanian, D. Poly (ε-L-lysine): Synthesis and conformation. Biopolym. Orig. Res. Biomol. 1980, 19, 219–229. [Google Scholar]
- Tam, J.P.; Lu, Y.A.; Yang, J.L. Antimicrobial dendrimeric peptides. Eur. J. Biochem. 2002, 269, 923–932. [Google Scholar] [CrossRef]
- Stanger, H.E.; Syud, F.A.; Espinosa, J.F.; Giriat, I.; Muir, T.; Gellman, S.H. Length-dependent stability and strand length limits in antiparallel β-sheet secondary structure. Proc. Natl. Acad. Sci. USA 2001, 98, 12015–12020. [Google Scholar] [CrossRef]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef]
- Dill, K.A. Dominant forces in protein folding. Biochemistry 1990, 29, 7133–7155. [Google Scholar] [CrossRef]
- Nelson, R.; Sawaya, M.R.; Balbirnie, M.; Madsen, A.Ø.; Riekel, C.; Grothe, R.; Eisenberg, D. Structure of the cross-β spine of amyloid-like fibrils. Nature 2005, 435, 773–778. [Google Scholar] [CrossRef]

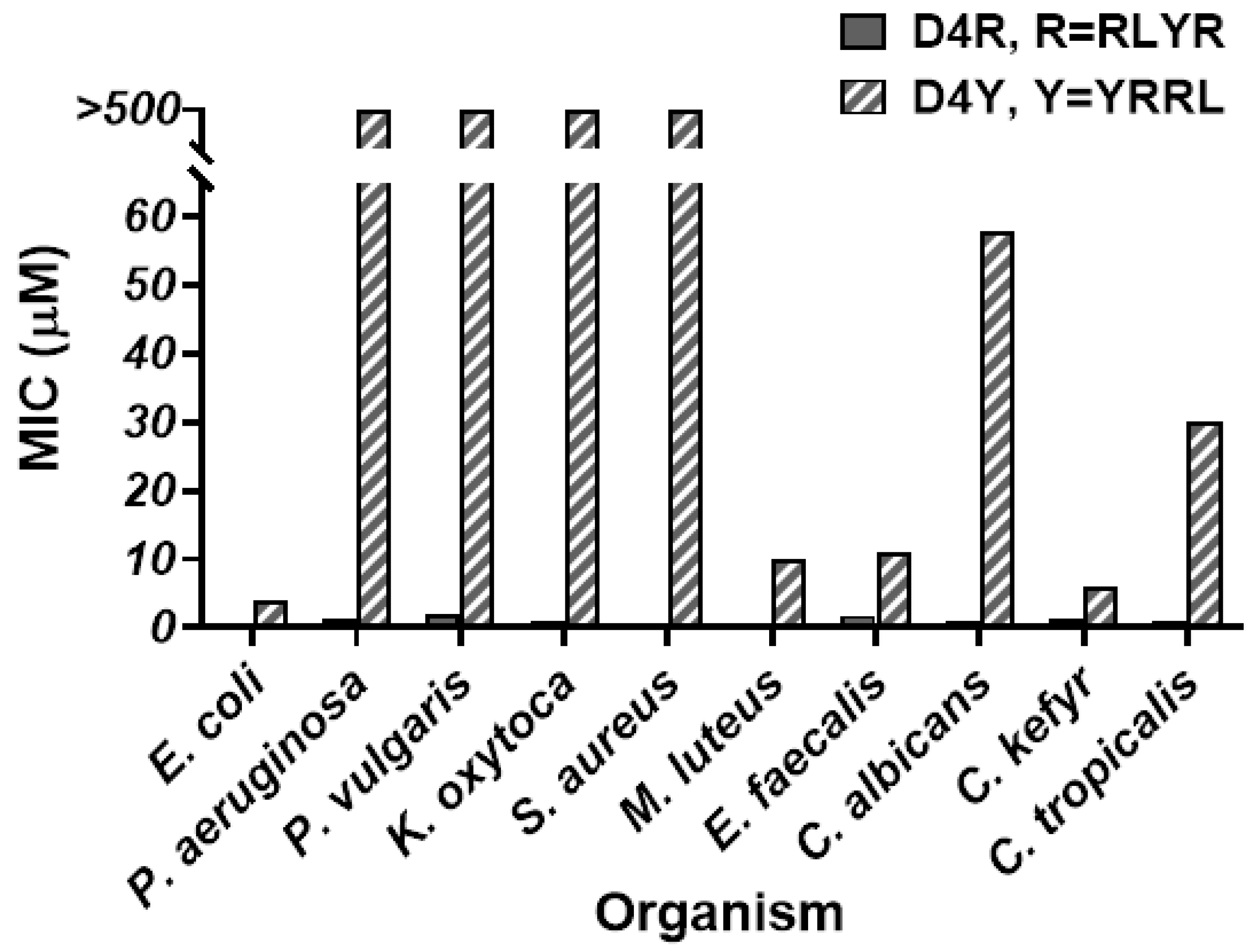
| MIC (µM) * | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Controls | Lys-Scaffolded † | Orn-Scaffolded † | ||||||||||
| Tachyplesin | D4R | K4R | iK4R | O4R | iO4R | |||||||
| Organism | L-Salt | H-Salt | L-Salt | H-Salt | L-Salt | H-Salt | L-Salt | H-Salt | L-Salt | H-Salt | L-Salt | H-Salt |
| Gram-negative | ||||||||||||
| E. coli | 0.3 | 0.4 | 0.6 | 0.7 | 0.7 | 0.7 | 0.4 | 0.4 | 0.7 | 4.1 | 0.9 | 4.4 |
| P. aeruginosa | 0.9 | 0.5 | 0.5 | 1.2 | 0.8 | 0.9 | 0.5 | 0.7 | 2.1 | 4.8 | 4.4 | 4 |
| P. vulgaris | 0.7 | 1 | 1 | 1.9 | 1.1 | 1 | 0.8 | 0.7 | 5.6 | 2.2 | 4.7 | 23.2 |
| K. oxytoca | 0.2 | 0.5 | 0.4 | 0.9 | 1.3 | 1.2 | 0.5 | 0.7 | 1.8 | 2.4 | 1.6 | 6.8 |
| Gram-positive | ||||||||||||
| S. aureus | 0.4 | 0.5 | 0.8 | 0.6 | 0.8 | 0.9 | 0.7 | 0.6 | 7.3 | 2 | 6.7 | 4.8 |
| M. luteus | 1 | 1.1 | 0.5 | 0.7 | 0.6 | 1 | 0.9 | 0.8 | 1.8 | 6.9 | 2.1 | 2 |
| E. faecalis | 0.3 | 0.4 | 0.8 | 1.8 | 0.5 | 1.5 | 0.8 | 1.4 | 6.6 | >500 | 5.4 | >500 |
| Fungi | ||||||||||||
| C. albicans | 0.7 | 0.9 | 0.8 | 0.8 | 0.7 | 0.8 | 0.8 | 0.9 | 1.4 | 1.6 | 1.3 | 1.4 |
| C. kefyr | 0.9 | 1.3 | 0.9 | 1.3 | 0.9 | 0.9 | 0.8 | 0.8 | 0.7 | 0.8 | 0.8 | 0.8 |
| C. tropicalis | 0.5 | 1 | 0.7 | 0.8 | 1.5 | 1.2 | 0.5 | 1.3 | 1.4 | 1.5 | 1.4 | 0.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
To, J.; Zhang, X.; Tam, J.P. Design of Potent and Salt-Insensitive Antimicrobial Branched Peptides. Polymers 2023, 15, 3594. https://doi.org/10.3390/polym15173594
To J, Zhang X, Tam JP. Design of Potent and Salt-Insensitive Antimicrobial Branched Peptides. Polymers. 2023; 15(17):3594. https://doi.org/10.3390/polym15173594
Chicago/Turabian StyleTo, Janet, Xiaohong Zhang, and James P. Tam. 2023. "Design of Potent and Salt-Insensitive Antimicrobial Branched Peptides" Polymers 15, no. 17: 3594. https://doi.org/10.3390/polym15173594
APA StyleTo, J., Zhang, X., & Tam, J. P. (2023). Design of Potent and Salt-Insensitive Antimicrobial Branched Peptides. Polymers, 15(17), 3594. https://doi.org/10.3390/polym15173594









