Copolymer-Green-Synthesized Copper Oxide Nanoparticles Enhance Folate-Targeting in Cervical Cancer Cells In Vitro
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Sample Collection and Preparation
2.2.1. Sample Collection
2.2.2. Preparation of M. azedarach Leaf Extract
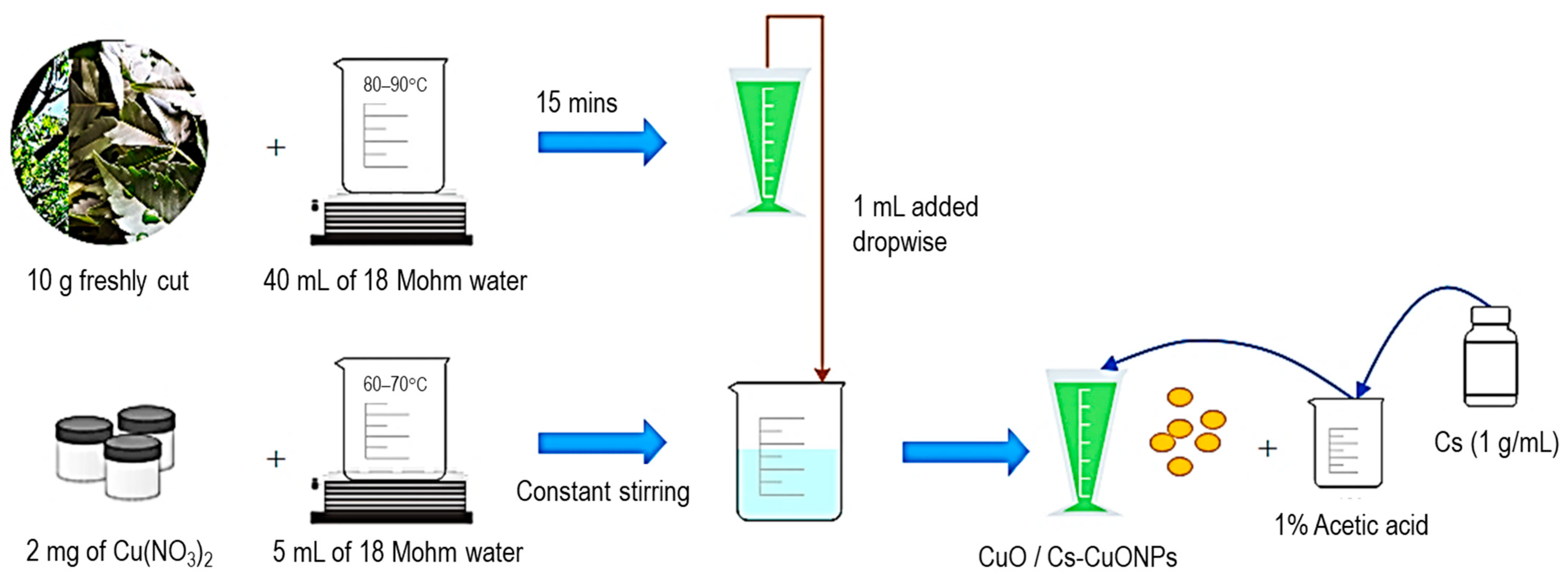
2.3. Synthesis and Chitosan (Cs) Functionalization of Copper Oxide Nanoparticles (CuONPs)
2.4. Formation of Folate–Chitosan–Copper Oxide (F-Cs-CuONPs) and PEG-F–Chitosan–Copper Oxide Nanoparticles (PEG-F-Cs-CuONPs)
2.5. Characterization of Nanoparticles and Nanocomplexes
2.5.1. UV-Vis Spectroscopy and X-ray Diffraction (XRD)
2.5.2. Fourier-Transform Infrared Spectroscopy (FTIR)
2.5.3. Transmission Electron Microscopy (TEM)
2.5.4. Nanoparticle Tracking Analysis (NTA)
2.6. Nucleic Acid Binding Studies
2.6.1. Electrophoretic Mobility Shift Assay
2.6.2. Nuclease Protection Assay
2.6.3. Ethidium Bromide Intercalation Assay
2.7. MTT Cell Viability Assay
2.8. Gene Expression Studies
2.9. Statistical Analysis
3. Results and Discussion
3.1. Characterization of Nanoparticles and Nanocomplexes
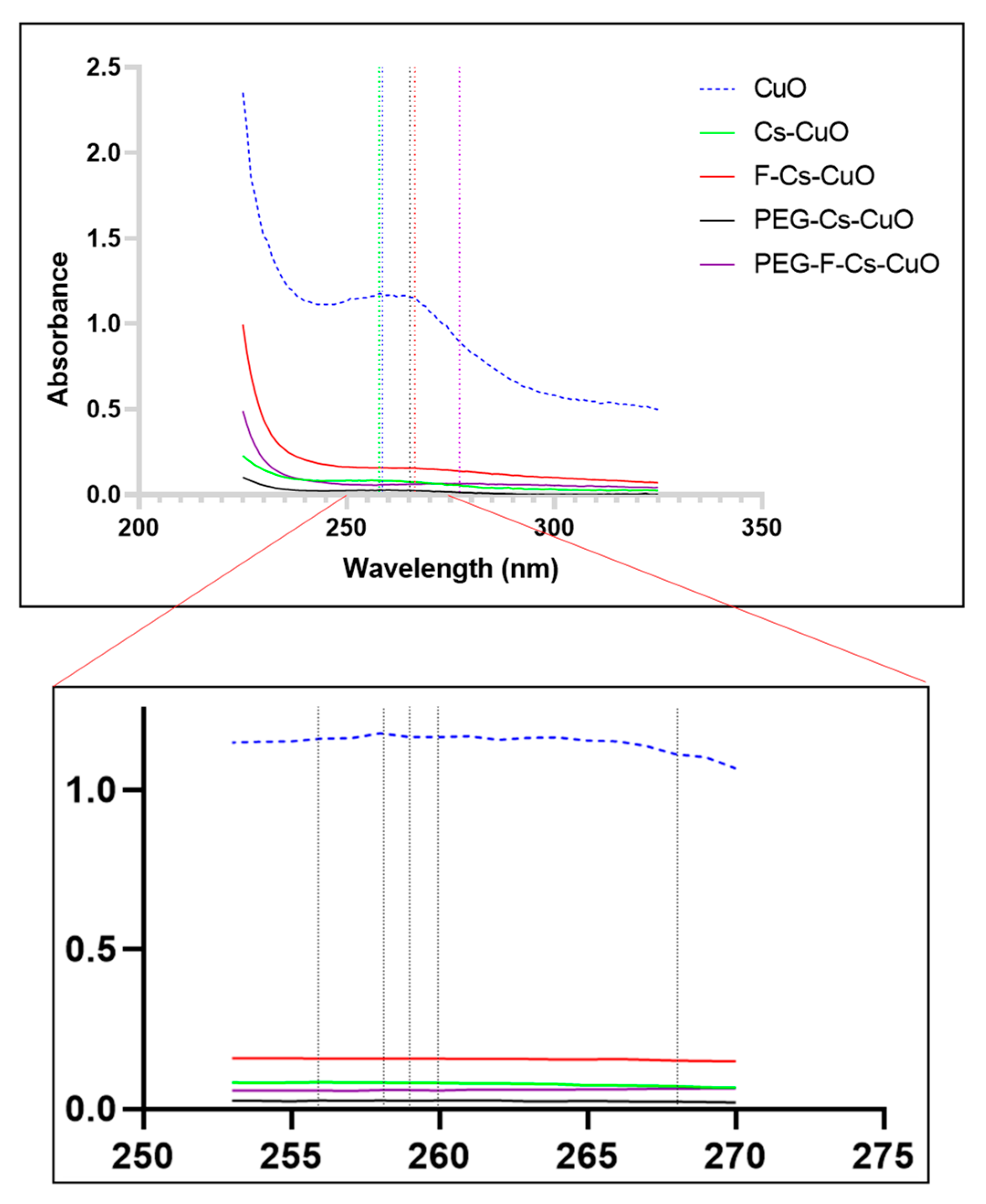
3.1.1. UV-Visible Spectroscopy and X-ray Diffraction (XRD)
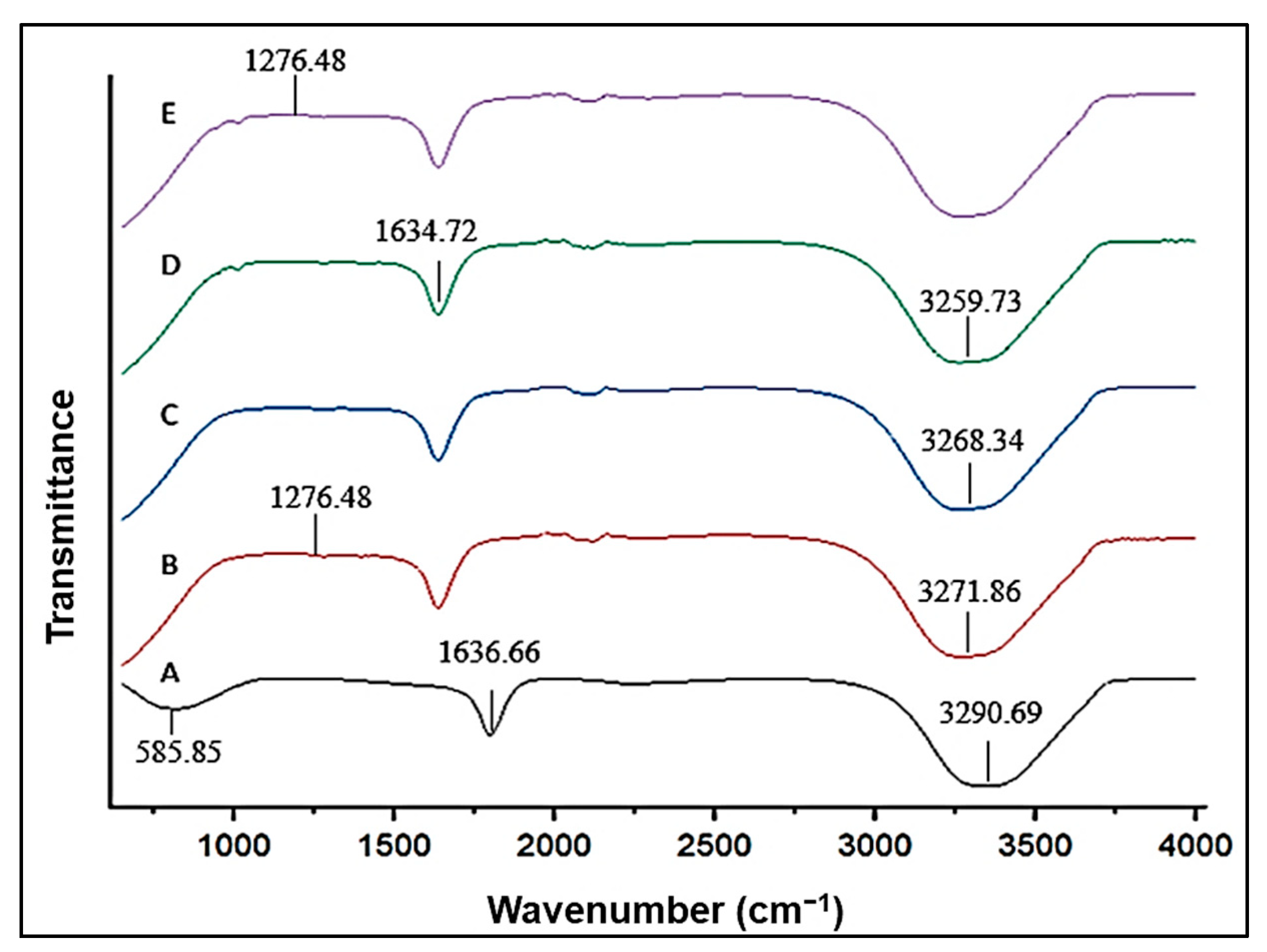
3.1.2. Fourier-Transform Infrared Spectroscopy (FTIR)
3.1.3. Transmission Electron Microscopy (TEM)
3.1.4. Nanoparticle Tracking Analysis (NTA)
3.2. DNA Binding Studies
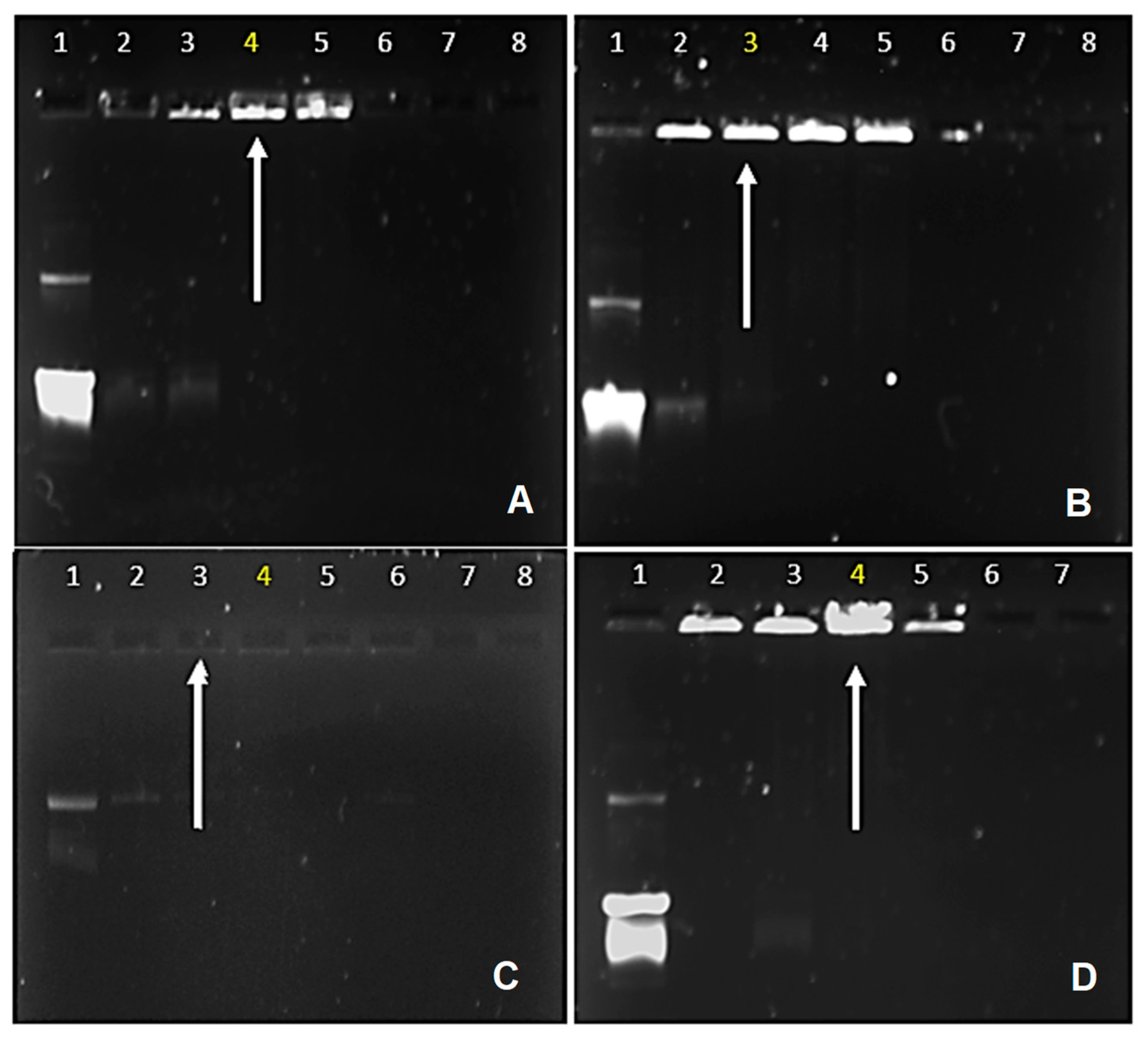
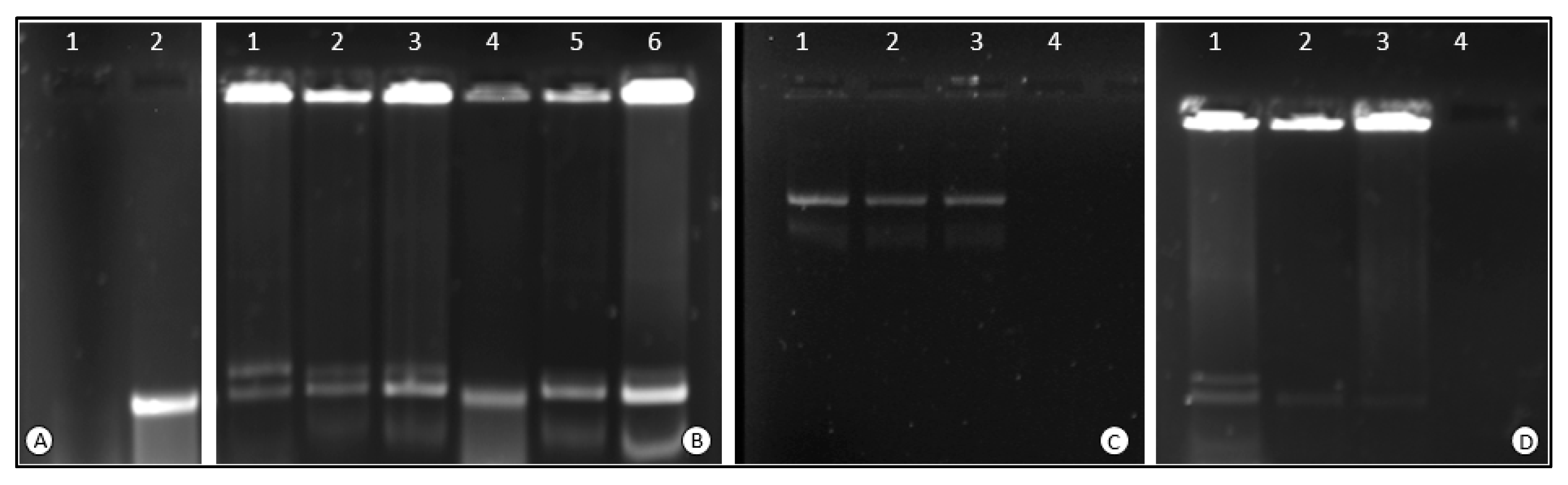
3.2.1. Electrophoretic Mobility Shift Assay
3.2.2. Nuclease Protection Assay
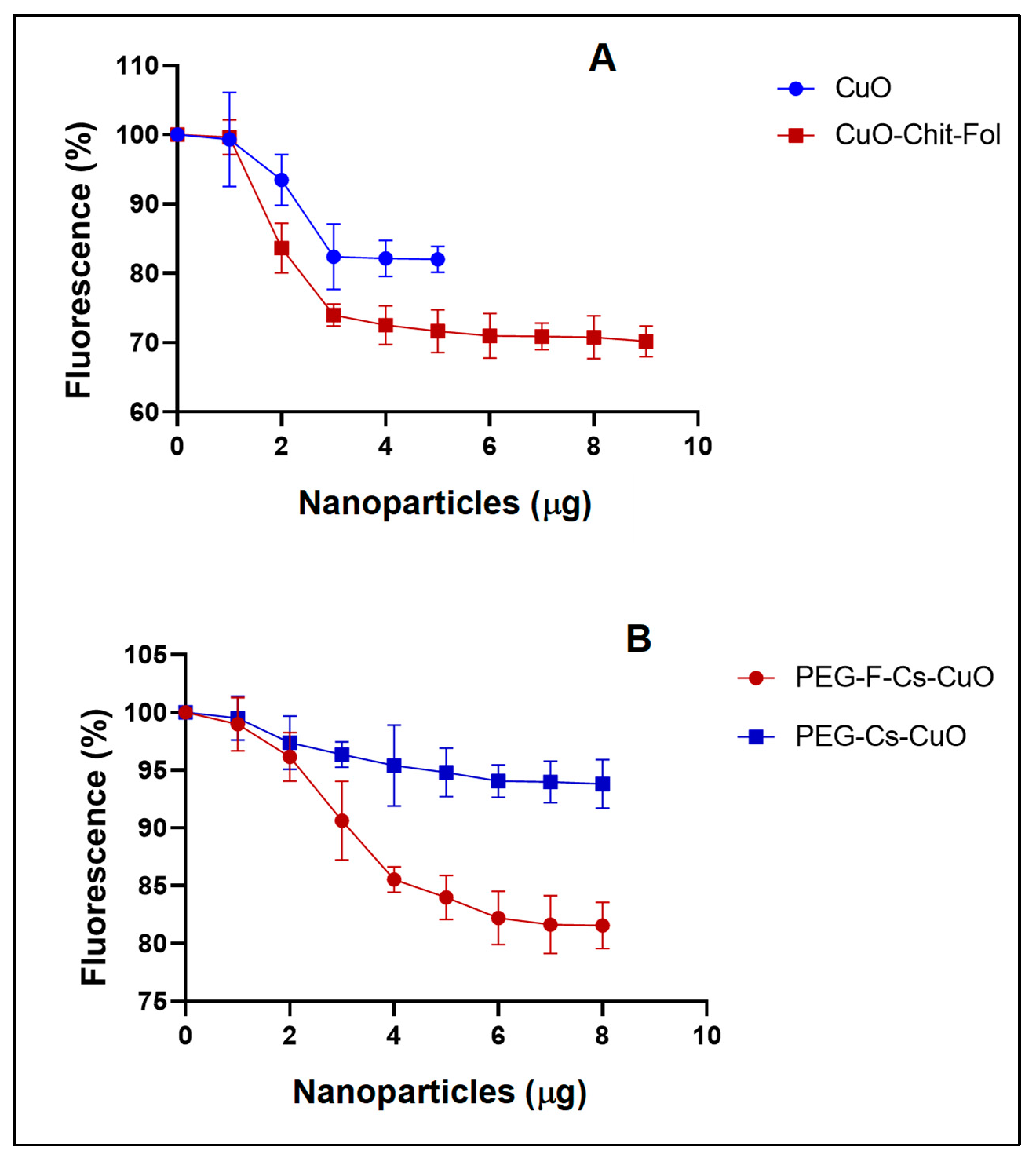
3.2.3. Ethidium Bromide Intercalation Assay
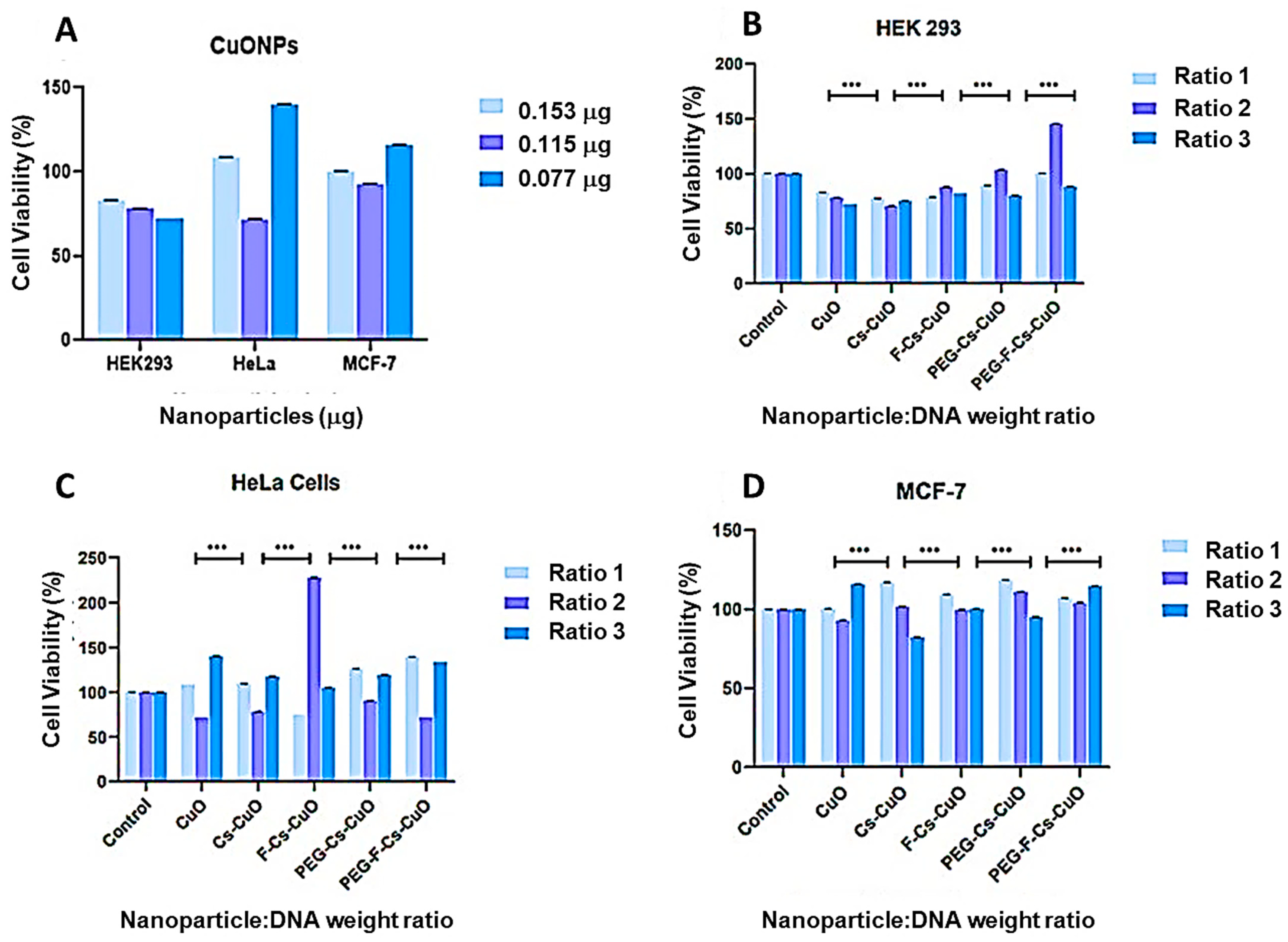
3.3. MTT Cell Viability Assay
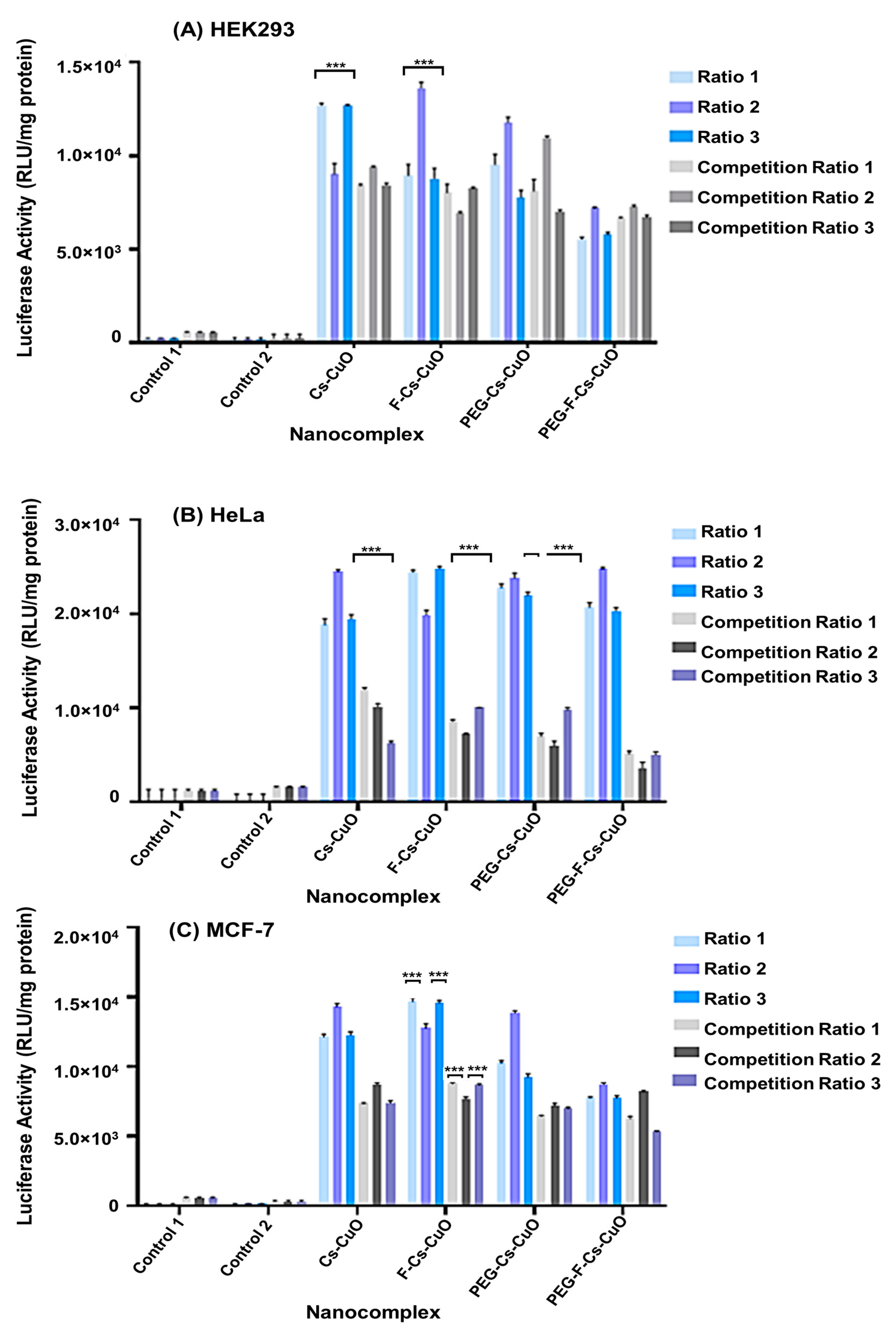
3.4. Gene Expression Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Cervical Cancer. Available online: https://www.who.int/health-topics/cervical-cancer#tab=tab_1 (accessed on 20 February 2023).
- Fang, J.H.; Yu, X.M.; Zhang, S.H.; Yang, Y. Effect of smoking on high-grade cervical cancer in women on the basis of human papillomavirus infection studies. J. Cancer Res. Ther. 2018, 14, 184–189. [Google Scholar] [CrossRef]
- Venkatas, J.; Singh, M. Cervical Cancer: A meta-analysis, Therapy and future of Nanomedicine. Ecancermedicalscience 2020, 14, 1111. [Google Scholar] [CrossRef]
- Small, W.; Bacon, M.; Bajaj, A.; Chuang, L.; Fisher, B.; Harkenrider, M.; Jhingran, A.; Kitchener, H.; Mileshkin, L.; Viswanathan, A.; et al. Cervical cancer: A global health crisis. Cancer 2017, 123, 2404–2412. [Google Scholar] [CrossRef]
- Baetke, S.; Lammers, T.; Kiessling, F. Applications of nanoparticles for diagnosis and therapy of cancer. Br. J. Radiol. 2015, 88, 20150207. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Kudgus, R.A.; Bhattacharya, R.; Mukherjee, P. Inorganic Nanoparticles in Cancer Therapy. Pharm. Res. 2011, 28, 237–259. [Google Scholar] [CrossRef]
- Ge, E.J.; Bush, A.I.; Casini, A.; Cobine, P.A.; Cross, J.R.; DeNicola, G.M.; Dou, Q.P.; Franz, K.J.; Gohil, V.M.; Gupta, S.; et al. Connecting copper and cancer: From transition metal signalling to metalloplasia. Nat. Rev. Cancer 2022, 22, 102–113. [Google Scholar] [CrossRef]
- Ingle, A.P.; Duran, N.; Rai, M. Bioactivity, mechanism of action, and cytotoxicity of copper-based nanoparticles: A review. Appl. Microbiol. Biotechnol. 2014, 98, 1001–1009. [Google Scholar] [CrossRef] [PubMed]
- Wang, L. Synthetic methods of CuS nanoparticles and their applications for imaging and cancer therapy. RSC Adv. 2016, 6, 82596–82615. [Google Scholar] [CrossRef]
- Chudobova, D.; Cihalova, K.; Kopel, P.; Melichar, L.; Ruttkay-Nedecky, B.; Vaculovicova, M.; Adam, V.; Kizek, R. Complexes of Metal-Based Nanoparticles with Chitosan Suppressing the Risk of Staphylococcus aureus and Escherichia coli Infections. In Nanotechnology in Diagnosis, Treatment and Prophylaxis of Infectious Diseases; Rai, M., Kon, K., Eds.; Academic Press: London, UK, 2015; pp. 217–232. [Google Scholar]
- Tadjarodi, A.; Roshani, R. A green synthesis of copper oxide nanoparticles by mechanochemical method. Curr. Chem. Lett. 2014, 3, 215–220. [Google Scholar] [CrossRef]
- Sankar, R.; Maheswari, R.; Karthik, S.; Shivashangari, K.; Ravikumar, V. Anticancer activity of Ficus religiosa engineered copper oxide nanoparticles. Mater. Sci. Eng. C 2014, 44, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Hang, X.; Peng, H.; Song, H.; Qi, Z.; Miao, X.; Xu, W. Antiviral activity of cuprous oxide nanoparticles against Hepatitis C Virus in vitro. J. Virol. Meth. 2015, 222, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Muhammad Imran Din, M.I.; Rehan, R. Synthesis, Characterization, and Applications of Copper Nanoparticles. Anal. Lett. 2017, 50, 50–62. [Google Scholar]
- Jagaran, K.; Singh, M. Nanomedicines for COVID-19: Potential of Copper nanoparticles. Biointerface Res. Appl.Chem. 2021, 11, 10716–10728. [Google Scholar]
- Olawale, F.; Oladimeji, O.; Ariatti, M.; Singh, M. Emerging Roles of green synthesized Chalcogen and Chalcogenide nanoparticles in Cancer theranostics. J. Nanotechnol. 2022, 2022, 6176610. [Google Scholar] [CrossRef]
- Iravani, S. Green synthesis of metal nanoparticles using plants. Green Chem. 2011, 13, 2638. [Google Scholar] [CrossRef]
- Černík, M.; Thekkae, P.V. Green synthesis of copper oxide nanoparticles using gum karaya as a biotemplate and their antibacterial application. Int. J. Nanomed. 2013, 8, 889–898. [Google Scholar] [CrossRef]
- Vishnukanta, A.R. Melia Azedarach: A Phytopharmacological Review. Phcog. Rev. 2008, 2, 173–179. [Google Scholar]
- Nerome, K.; Ito-Kureha, T.; Paganini, T.; Fukuda, T.; Igarashi, Y.; Ashitomi, H.; Ikematsu, S.; Yamamoto, T. Potent and broad anticancer activities of leaf extracts from Melia azedarach L. of the subtropical Okinawa islands. Am. J. Cancer Res. 2020, 10, 581–594. [Google Scholar]
- Al-Marzoqi, A.H.; Imad Hadi Hameed, I.H.; Idan, S.A. Analysis of bioactive chemical components of two medicinal plants (Coriandrum sativum and Melia azedarach) leaves using gas chromatography-mass spectrometry (GC-MS). Afr. J. Biotechnol. 2015, 14, 2812–2830. [Google Scholar]
- Cao, S.; Bi, Z.; Li, Q.; Zhang, S.; Singh, M.; Chen, J.D. Shape memory and antibacterial chitosan-based cryogel with hemostasis and skin wound repair. Carbohyd. Polym. 2023, 305, 120545. [Google Scholar] [CrossRef]
- Wang, W.; Meng, Q.; Li, Q.; Liu, J.; Zhou, M.; Jin, Z.; Zhao, K. Chitosan derivatives and their application in biomedicine. Int. J. Mol. Sci. 2020, 21, 487. [Google Scholar] [CrossRef] [PubMed]
- Cheung, A.; Bax, H.J.; Josephs, D.H.; Ilieva, K.M.; Pellizzari, G.; Opzoomer, J.; Bloomfield, J.; Fittall, M.; Grigoriadis, A.; Figini, M.; et al. Targeting folate receptor alpha for cancer treatment. Oncotarget 2016, 7, 52553–52574. [Google Scholar] [CrossRef]
- Scaranti, M.; Cojocaru, E.; Banerjee, S.; Banerji, U. Exploiting the folate receptor α in oncology. Nat. Rev. Clin. Oncol. 2020, 17, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Suk, J.; Xu, Q.; Kim, N.; Hanes, J.; Ensign, L. PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug Deliv. Rev. 2016, 99, 28–51. [Google Scholar] [CrossRef]
- Sercombe, L.; Veerati, T.; Moheimani, F.; Wu, S.Y.; Sood, A.K.; Hua, S. Advances and Challenges of Liposome Assisted Drug Delivery. Front. Pharmacol. 2015, 6, 286. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Kanasty, R.; Eltoukhy, A.; Vegas, A.; Dorkin, J.; Anderson, D. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef]
- Chinnasamy, G.; Chandrasekharan, S.; Bhatnagar, S. Biosynthesis of Silver Nanoparticles from Melia azedarach: Enhancement of Antibacterial, Wound Healing, Antidiabetic and Antioxidant Activities. Int. J. Nanomed. 2019, 14, 9823–9836. [Google Scholar] [CrossRef] [PubMed]
- Arunkumar, B.; Jeyakumar, S.J.; Jothibas, M. A sol-gel approach to the synthesis of CuO nanoparticles using Lantana camara leaf extract and their photo catalytic activity. Optik 2019, 183, 698–705. [Google Scholar] [CrossRef]
- Akinyelu, J.; Singh, M. Folate-tagged chitosan-functionalized gold nanoparticles for enhanced delivery of 5-fluorouracil to cancer cells. Appl. Nanosci. 2019, 9, 7–17. [Google Scholar] [CrossRef]
- Rana, S.; Shetake, N.G.; Barick, K.C.; Pandey, B.N.; Salunke, H.G.; Hassan, P.A. Folic acid conjugated Fe3O4 magnetic nanoparticles for targeted delivery of doxorubicin. Dalton Transact. 2016, 45, 17401–17408. [Google Scholar] [CrossRef]
- Daniels, A.N.; Singh, M. Sterically stabilized siRNA:gold nanocomplexes enhance c-MYC silencing in a breast cancer cell model. Nanomedicine 2019, 14, 1387–1401. [Google Scholar] [CrossRef]
- Malloy, A. Count, Size and Visualize Nanoparticles. Mater. Today 2011, 14, 170–173. [Google Scholar] [CrossRef]
- Singh, M. Assessing nucleic acid: Cationic nanoparticle interaction for gene delivery. In Bio-Carrier Vectors; Narayanan, K., Ed.; Springer: New York, NY, USA, 2021; Volume 2211, pp. 43–55. [Google Scholar]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Maiyo, F.C.; Mbatha, L.S.; Singh, M. Selenium Nanoparticles in Folate-Targeted delivery of the pCMV-Luc DNA Reporter Gene. Curr. Nanosci. 2021, 17, 871–880. [Google Scholar] [CrossRef]
- Naidoo, S.; Daniels, A.; Habib, S.; Singh, M. Poly-L-Lysine–Lactobionic Acid-Capped Selenium Nanoparticles for Liver-Targeted Gene Delivery. Int. J. Mol. Sci. 2022, 23, 1492. [Google Scholar] [CrossRef] [PubMed]
- Kumar, B.; Smita, K.; Cumbal, L.; Debut, A.; Angulo, Y. Biofabrication of copper oxide nanoparticles using Andean blackberry (Rubus glaucus Benth.) fruit and leaf. J. Saudi Chem. Soc. 2017, 21, S475–S480. [Google Scholar] [CrossRef]
- Haiss, W.; Thanh, N.T.; Aveyard, J.; Fernig, D.G. determination of size and concentration of gold nanoparticles from UV–Vis. spectra. Anal. Chem. 2007, 79, 4215–4221. [Google Scholar] [CrossRef]
- Gamedze, N.; Mthiyane, D.M.N.; Babalola, O.; Singh, M.; Onwudiwe, D.C. Physico-chemical characteristics and cytotoxicity evaluation of CuO and TiO2 nanoparticles biosynthesized using extracts of Mucuna pruriensutilis seeds. Heliyon 2022, 8, e10187. [Google Scholar] [CrossRef]
- Velsankar, K.; Suganya, S.; Muthumari, P.; Mohandoss, S.; Sudhahar, S. Ecofriendly green synthesis, characterization and biomedical applications of CuO nanoparticles synthesized using leaf extract of Capsicum frutescens. J. Environ. Chem. Eng. 2021, 9, 106299. [Google Scholar]
- Shigemassa, Y.; Matsuura, H.; Sashiwa, H.; Saimoto, H. Heavy metal contamination. Int. J. Biol. Macromol. 2006, 18, 237. [Google Scholar]
- Qu, J.; Hu, Q.; Shen, K.; Zhang, K.; Li, Y.; Li, H.; Zhang, Q.; Wang, J.; Quan, W. The preparation and characterization of chitosan rods modified with Fe3þ by a chelation mechanism. Carbohydr. Res. 2011, 346, 822–827. [Google Scholar] [CrossRef] [PubMed]
- Jalilian, A.R.; Hosseini-Salekdeh, S.L.; Mahmoudi, M.; Yousefnia, H.; Majdabadi, A.; Pouladian, M. Preparation and biological evaluation of radiolabeled-folate embedded superparamagnetic nanoparticles in wild-type rats. J. Radioanal. Nucl. Chem. 2011, 287, 119–127. [Google Scholar] [CrossRef]
- Laha, D.; Pramanik, A.; Chattopadhyay, S.; Dash, D.K.; Roy, S.; Pramanik, P.; Karmakar, P. Folic acid modified copper oxide nanoparticles for targeted delivery in in vitro and in vivo systems. RSC Adv. 2015, 5, 68169. [Google Scholar] [CrossRef]
- Filipe, V.; Hawe, A.; Jiskoot, W. Critical Evaluation of Nanoparticle Tracking Analysis (NTA) by NanoSight for the Measurement of Nanoparticles and Protein Aggregates. Pharm. Res. 2010, 27, 796–810. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, J.; Chakraborty, N.; Chatterjee, A.; Bhattacharjee, A.; Dasgupta, D.; Acharya, K. Green Synthesized Copper Oxide Nanoparticles Ameliorate Defence and Antioxidant Enzymes in Lens culinaris. Nanomaterials 2020, 10, 312. [Google Scholar] [CrossRef]
- Sarkar, J.; Dey, P.; Saha, S.; Acharya, K. Mycosynthesis of selenium nanoparticles. Micro Nano Lett. 2011, 6, 599. [Google Scholar] [CrossRef]
- Shi, L.; Zhang, J.; Zhao, M.; Tang, M.; Cheng, X.; Zhang, W.; Li, W.; Liu, X.; Peng, H.; Wang, Q. Effects of polyethylene glycol on the surface of nanoparticles for targeted drug delivery. Nanoscale 2021, 13, 10748–10764. [Google Scholar] [CrossRef]
- Lu, T.; Low, P.S. Folate-mediated delivery of macromolecular anticancer therapeutic agents. Adv. Drug Deliv. Rev. 2002, 54, 675–693. [Google Scholar] [CrossRef]
- Honary, S.; Zahir, F. Effect of Zeta Potential on the Properties of Nano-Drug Delivery Systems—A Review (Part 2). Trop. J. Pharm. Res. 2013, 12, 265–273. [Google Scholar]
- Mansouri, S.; Cuie, Y.; Winnik, F.; Shi, Q.; Lavigne, P.; Benderdour, M.; Beaumont, E.; Fernandes, J. Characterization of folate-chitosan-DNA nanoparticles for gene therapy. Biomaterials 2006, 27, 2060–2065. [Google Scholar] [CrossRef]
- Obata, Y.; Saito, S.; Takeda, N.; Takeoka, S. Plasmid DNA-encapsulating liposomes: Effect of a spacer between the cationic head group and hydrophobic moieties of the lipids on gene expression efficiency. Biochim. Biophys. Acta Biomembr. 2009, 1788, 1148–1158. [Google Scholar] [CrossRef] [PubMed]
- Akinyelu, A.; Oladimeji, O.; Singh, M. Lactobionic Acid-Chitosan Functionalized Gold Coated Poly(lactide-co-glycolide) Nanoparticles for Hepatocyte Targeted Gene Delivery. Adv. Nat. Sci. Nanosci. Nanotechnol. 2020, 11, 045017. [Google Scholar] [CrossRef]
- Alishah, H.; Pourseyedi, S.; Ebrahimipour, Y.S.; Mahani, S.E.; Rafiei, N. Green synthesis of starch-mediated CuO nanoparticles: Preparation, characterization, antimicrobial activities and in vitro MTT assay against MCF-7 cell line. Rend. Fis. Acc. Lincei 2017, 28, 65–71. [Google Scholar] [CrossRef]
- Fard, M.Z.; Fatholahi, M.; Abyadeh, M.; Bakhtiarian, A.; Mousavi, S.E.; Falahati, M. The Investigation of the Cytotoxicity of Copper Oxide Nanoparticles on Peripheral Blood Mononuclear Cells. Nanomed. Res. J. 2020, 5, 364–368. [Google Scholar]
- Boca, S.C.; Potara, M.; Gabudean, A.; Juhem, A.; Baldeck, P.L.; Astilean, S. Chitosan-coated triangular silver nanoparticles as a novel class of biocompatible, highly effective photothermal transducers for in vitro cancer cell therapy. Cancer Lett. 2011, 311, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Rajam, M.; Sivasami, P.; Rose, C.; Mandal, A.B. Chitosan nanoparticles as a dual growth factor delivery system for tissue engineering applications. Int. J. Pharm. 2011, 410, 145–152. [Google Scholar] [CrossRef]
- Sivashankari, P.; Prabaharan, M. Prospects of chitosan-based scaffolds for growth factor release in tissueengineering. Int. J. Biol. Macromol. 2016, 93, 1382–1389. [Google Scholar] [CrossRef]
- Ebadi, M.; Rifqi Md Zain, A.; Tengku Abdul Aziz, T.H.; Mohammadi, H.; Tee, C.A.T.; Rahimi Yusop, M. Formulation and Characterization of Fe3O4@PEG Nanoparticles Loaded Sorafenib; Molecular Studies and Evaluation of Cytotoxicity in Liver Cancer Cell Lines. Polymers 2023, 15, 971. [Google Scholar] [CrossRef]
- Ramnandan, D.; Mokhosi, S.; Daniels, A.; Singh, M. Chitosan, Polyethylene glycol and Polyvinyl alcohol modified MgFe2O4 ferrite magnetic nanoparticles in Doxorubicin delivery: A comparative study in vitro. Molecules 2021, 26, 3893. [Google Scholar] [CrossRef]
- Patil, U.; Adireddy, S.; Jaiswal, A.; Mandava, S.; Lee, B.; Chrisey, D. In vitro/in vivo toxicity evaluation and quantification of iron oxide nanoparticles. Int. J. Mol. Sci. 2015, 16, 24417. [Google Scholar] [CrossRef]
- Zhu, M.; Nie, G.; Meng, H.; Xia, T.; Nel, A.; Zhao, Y. Physicochemical Properties Determine Nanomaterial Cellular Uptake, Transport, and Fate. Acc. Chem. Res. 2013, 46, 622–631. [Google Scholar] [CrossRef] [PubMed]
- Rejman, J.; Oberle, V.; Zuhorn, I.; Hoekstra, D. Size-dependent internalization of particles via the pathways of clathrin-and caveolae-mediated endocytosis. Biochem. J. 2004, 377, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Panariti, A.; Miserocchi, G.; Rivolta, I. The effect of nanoparticle uptake on cellular behavior: Disrupting or enabling functions? Nanotechnol. Sci. Appl. 2012, 5, 87–100. [Google Scholar]
- Foroozandeh, P.; Aziz, A.A. Insight into Cellular Uptake and Intracellular Trafficking of Nanoparticles. Nanoscale Res. Lett. 2018, 13, 339. [Google Scholar] [CrossRef] [PubMed]
- Chithrani, B.; Chan, W. Elucidating the Mechanism of Cellular Uptake and Removal of Protein-Coated Gold Nanoparticles of Different Sizes and Shapes. Nano Lett. 2007, 7, 1542–1550. [Google Scholar] [CrossRef] [PubMed]
- Lu, F.; Wu, S.; Hung, Y.; Mou, C. Size Effect on Cell Uptake in Well-Suspended, Uniform Mesoporous Silica Nanoparticles. Small 2009, 5, 1408–1413. [Google Scholar] [CrossRef]
- Xu, L.; Wang, Y.-Y.; Huang, J.; Chen, C.-Y.; Wang, Z.-X.; Xie, H. Silver nanoparticles: Synthesis, medical applications, and biosafety. Theranostics 2020, 10, 8996–9031. [Google Scholar] [CrossRef]
- Shang, L.; Nienhaus, K.; Nienhaus, G.U. Engineered nanoparticles interacting with cells: Size matters. J. Nanobiotechnol. 2014, 12, 5. [Google Scholar] [CrossRef]
- Shan, Y.; Ma, S.; Nie, L.; Shang, X.; Hao, X.; Tang, Z.; Wang, H. Size-dependent endocytosis of single gold nanoparticles. Chem. Comm. 2011, 47, 8091–8093. [Google Scholar] [CrossRef]
- Huang, K.; Ma, H.; Liu, J.; Huo, S.; Kumar, A.; Wei, T.; Zhang, X.; Jin, S.; Gan, Y.; Wang, P.C.; et al. Size-dependent localization and penetration of ultrasmall gold nanoparticles in cancer cells, multicellular spheroids, and tumors in vivo. ACS Nano 2012, 6, 4483–4493. [Google Scholar] [CrossRef]
- Marshalek, J.P.; Sheeran, P.S.; Ingram, P.; Dayton, P.A.; Witte, R.S.; Matsunaga, T.O. Intracellular delivery and ultrasonic activation of folate receptor-targeted phase-change contrast agents in breast cancer cells in vitro. J. Control. Release 2016, 243, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Feng, D.; Song, Y.; Shi, W.; Li, X.; Ma, H. Distinguishing Folate-Receptor-Positive Cells from Folate-ReceptorNegative Cells Using a Fluorescence off–on Nanoprobe. Anal. Chem. 2013, 85, 6530–6535. [Google Scholar] [CrossRef] [PubMed]


| Nanoparticles | Particle Size (nm) ± SE |
|---|---|
| CuO | 62.8 ± 12.8 |
| Cs-CuO | 85.7 ± 26.9 |
| F-Cs-CuO | 114.4 ± 11.9 |
| PEG-Cs-CuO | 91.8 ± 9.4 |
| PEG-F-Cs-CuO | 119.3 ± 17.9 |
| Sample | Particle Size (nm) ± SE | ζ Potential (mV) ± SE | ||
|---|---|---|---|---|
| Nanoparticle | Nanocomplex (NP + pDNA) | Nanoparticle | Nanocomplex (NP + pDNA) | |
| CuO | 78.2 ± 20.7 | - | −9 mV ± 0.1 | - |
| Cs-CuO | 103.9 ± 14.8 | 159.3 ± 6.7 | 45.3 mV ± 0.1 | 24.2 mV ± 0.1 |
| F-Cs-CuO | 128.0 ± 9.4 | 161.5 ± 9.5 | 55.1 mV ± 0.1 | 30.3 mV ± 0.2 |
| PEG-Cs-CuO | 148.8 ± 2.3 | 178.3 ± 3.7 | 42.3 mV ± 0.2 | 19.7 mV ± 0.1 |
| PEG-F-Cs-CuO | 197.1 ± 7.9 | 209.0 ± 9.8 | 55.5 mV ± 0.1 | 35.1 mV ± 0.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jagaran, K.; Singh, M. Copolymer-Green-Synthesized Copper Oxide Nanoparticles Enhance Folate-Targeting in Cervical Cancer Cells In Vitro. Polymers 2023, 15, 2393. https://doi.org/10.3390/polym15102393
Jagaran K, Singh M. Copolymer-Green-Synthesized Copper Oxide Nanoparticles Enhance Folate-Targeting in Cervical Cancer Cells In Vitro. Polymers. 2023; 15(10):2393. https://doi.org/10.3390/polym15102393
Chicago/Turabian StyleJagaran, Keelan, and Moganavelli Singh. 2023. "Copolymer-Green-Synthesized Copper Oxide Nanoparticles Enhance Folate-Targeting in Cervical Cancer Cells In Vitro" Polymers 15, no. 10: 2393. https://doi.org/10.3390/polym15102393
APA StyleJagaran, K., & Singh, M. (2023). Copolymer-Green-Synthesized Copper Oxide Nanoparticles Enhance Folate-Targeting in Cervical Cancer Cells In Vitro. Polymers, 15(10), 2393. https://doi.org/10.3390/polym15102393










