Synthesis of Polylactic Acid Oligomers for Broad-Spectrum Antimicrobials
Abstract
:1. Introduction
2. Materials and Methods
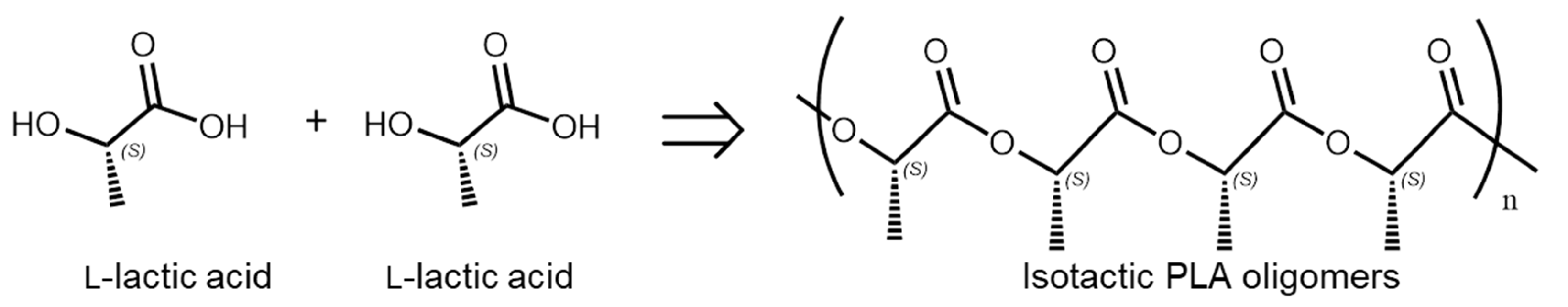
2.1. Synthesis of PLA Oligomers
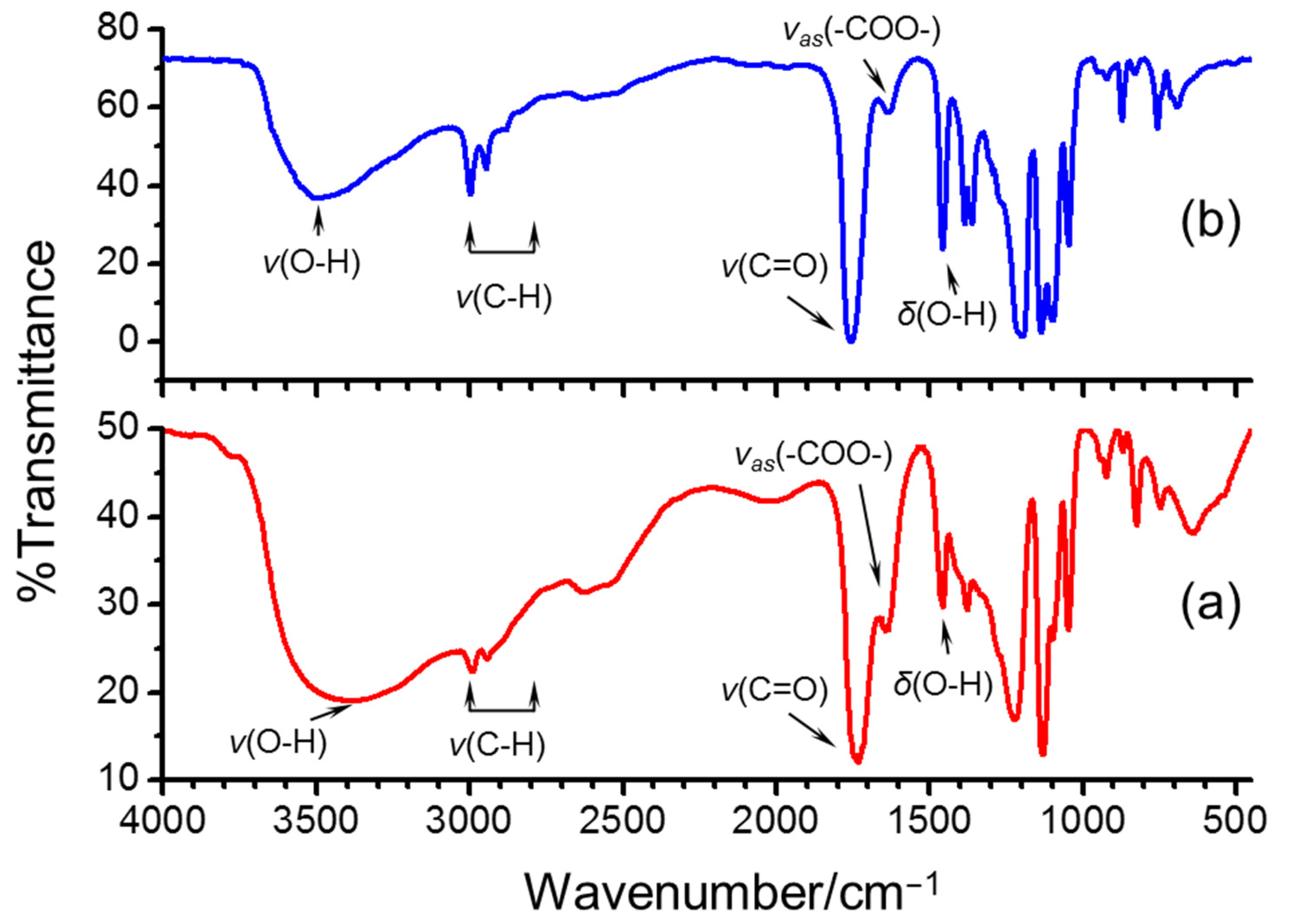
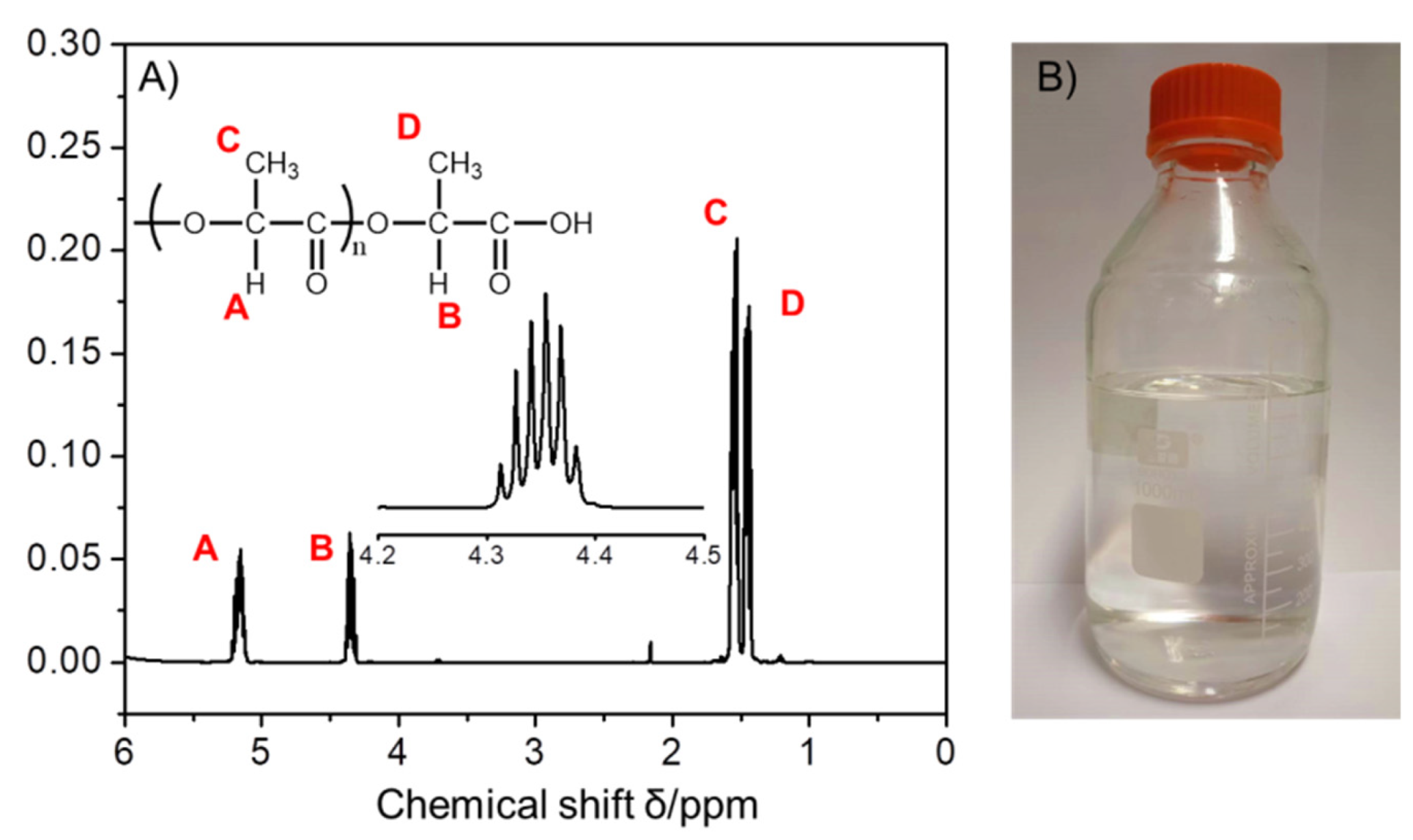
2.2. Characterization of PLA Oligomers
2.3. Assessment of Antibacterial/Antifungal Activity
2.4. Assessment of Virucidal Activity
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Enserink, M. Avian Flu Outbreak Sets Off Alarm Bells. Science 2003, 300, 718. [Google Scholar] [CrossRef] [PubMed]
- Kupferschmidt, K. Amid panic, a chance to learn about MERS. Science 2015, 348, 1183–1184. [Google Scholar] [CrossRef] [PubMed]
- Panáček, A.; Kvítek, L.; Smékalová, M.; Večeřová, R.; Kolář, M.; Röderová, M.; Dyčka, F.; Šebela, M.; Prucek, R.; Tomanec, O.; et al. Bacterial resistance to silver nanoparticles and how to overcome it. Nat. Nanotechnol. 2018, 13, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Levin-Reisman, I.; Ronin, I.; Gefen, O.; Braniss, I.; Shoresh, N.; Balaban, N.Q. Antibiotic tolerance facilitates the evolution of resistance. Science 2017, 355, 826–830. [Google Scholar] [CrossRef]
- Neu, H.C. The Crisis in Antibiotic Resistance. Science 1992, 257, 1064–1073. [Google Scholar] [CrossRef] [Green Version]
- Stabryla, L.M.; Johnston, K.A.; Diemler, N.A.; Cooper, V.S.; Millstone, J.E.; Haig, S.-J.; Gilbertson, L.M. Role of bacterial motility in differential resistance mechanisms of silver nanoparticles and silver ions. Nat. Nanotechnol. 2021, 16, 996–1003. [Google Scholar] [CrossRef]
- Wang, H.; Wang, M.; Xu, X.; Gao, P.; Xu, Z.; Zhang, Q.; Li, H.; Yan, A.; Kao, R.Y.-T.; Sun, H. Multi-target mode of action of silver against Staphylococcus aureus endows it with capability to combat antibiotic resistance. Nat. Commun. 2021, 12, 3331. [Google Scholar] [CrossRef]
- Bao, Q.; Zhang, D.; Qi, P. Synthesis and characterization of silver nanoparticle and graphene oxide nanosheet composites as a bactericidal agent for water disinfection. J. Colloid Interface Sci. 2011, 360, 463–470. [Google Scholar] [CrossRef]
- AshaRani, P.V.; Low Kah Mun, G.; Hande, M.P.; Valiyaveettil, S. Cytotoxicity and Genotoxicity of Silver Nanoparticles in Human Cells. ACS Nano 2009, 3, 279–290. [Google Scholar] [CrossRef]
- Sharifi, S.; Behzadi, S.; Laurent, S.; Laird Forrest, M.; Stroeve, P.; Mahmoudi, M. Toxicity of nanomaterials. Chem. Soc. Rev. 2012, 41, 2323–2343. [Google Scholar] [CrossRef] [Green Version]
- Ricke, S.C. Perspectives on the use of organic acids and short chain fatty acids as antimicrobials. Poult. Sci. 2003, 82, 632–639. [Google Scholar] [CrossRef]
- Worley, S.; Sun, G. Biocidal polymers. Trends Polym. Sci. 1996, 11, 364–370. [Google Scholar]
- De Santis, E.; Alkassem, H.; Lamarre, B.; Faruqui, N.; Bella, A.; Noble, J.E.; Micale, N.; Ray, S.; Burns, J.R.; Yon, A.R.; et al. Antimicrobial peptide capsids of de novo design. Nat. Commun. 2017, 8, 2263. [Google Scholar] [CrossRef] [Green Version]
- Bao, Q.; Zhang, D.; Wan, Y. 2-Mercaptobenzothiazole doped chitosan/11-alkanethiolate acid composite coating: Dual function for copper protection. Appl. Surf. Sci. 2011, 257, 10529–10534. [Google Scholar] [CrossRef]
- Dong, A.; Wang, Y.-J.; Gao, Y.; Gao, T.; Gao, G. Chemical Insights into Antibacterial N-Halamines. Chem. Rev. 2017, 117, 4806–4862. [Google Scholar] [CrossRef]
- Liu, L.; Courtney, K.C.; Huth, S.W.; Rank, L.A.; Weisblum, B.; Chapman, E.R.; Gellman, S.H. Beyond Amphiphilic Balance: Changing Subunit Stereochemistry Alters the Pore-Forming Activity of Nylon-3 Polymers. J. Am. Chem. Soc. 2021, 143, 3219–3230. [Google Scholar] [CrossRef]
- Ma, L.; Zhang, Z.; Li, J.; Yang, X.; Fei, B.; Leung, P.H.M.; Tao, X. A New Antimicrobial Agent: Poly (3-hydroxybutyric acid) Oligomer. Macromol. Biosci. 2019, 19, 1800432. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, J.; Ma, L.; Yang, X.; Fei, B.; Leung, P.H.M.; Tao, X. Mechanistic Study of Synergistic Antimicrobial Effects between Poly (3-hydroxybutyrate) Oligomer and Polyethylene Glycol. Polymers 2020, 12, 2735. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A Simple Method of Estimating Fifty Per Cent Endpoints12. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Lei, C.; Yang, J.; Hu, J.; Sun, X. On the Calculation of TCID(50) for Quantitation of Virus Infectivity. Virol. Sin. 2021, 36, 141–144. [Google Scholar] [CrossRef]
- Yadav, N.; Nain, L.; Khare, S.K. Studies on the degradation and characterization of a novel metal-free polylactic acid synthesized via lipase-catalyzed polymerization: A step towards curing the environmental plastic issue. Environ. Technol. Innov. 2021, 24, 101845. [Google Scholar] [CrossRef]
- Korsman, S.N.J.; Preiser, W.; Nutt, L.; Andersson, M.I. Virology: An Illustrated Colour Text; Churchill Livingstone Elsevier: London, UK, 2012. [Google Scholar]
- Dadonaite, B.; Gilbertson, B.; Knight, M.L.; Trifkovic, S.; Rockman, S.; Laederach, A.; Brown, L.E.; Fodor, E.; Bauer, D.L.V. The structure of the influenza A virus genome. Nat. Microbiol. 2019, 4, 1781–1789. [Google Scholar] [CrossRef] [PubMed]
- Weisberg, E.; Sattler, M.; Yang, P.L.; Parent, A.; Gray, N.; Griffin, J.D. Current therapies under investigation for COVID-19: Potential COVID-19 treatments. Can. J. Physiol. Pharmacol. 2020, 98, 483–489. [Google Scholar] [CrossRef] [PubMed]
- WHO Coronavirus (COVID-19) Dashboard—Situation by Region, Country, Territory & Area. Available online: https://covid19.who.int/table (accessed on 21 September 2020).
- Garcia-Rubio, R.; de Oliveira, H.C.; Rivera, J.; Trevijano-Contador, N. The Fungal Cell Wall: Candida, Cryptococcus, and Aspergillus Species. Front. Microbiol. 2020, 10, 2993. [Google Scholar] [CrossRef]
- Wang, C.; Chang, T.; Yang, H.; Cui, M. Antibacterial mechanism of lactic acid on physiological and morphological properties of Salmonella Enteritidis, Escherichia coli and Listeria monocytogenes. Food Control 2015, 47, 231–236. [Google Scholar] [CrossRef]
- Benn, G.; Mikheyeva, I.V.; Inns, P.G.; Forster, J.C.; Ojkic, N.; Bortolini, C.; Ryadnov, M.G.; Kleanthous, C.; Silhavy, T.J.; Hoogenboom, B.W. Phase separation in the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. USA 2021, 118, e2112237118. [Google Scholar] [CrossRef]
- Benthin, S.; Villadsen, J. Different inhibition of Lactobacillus delbrueckii subsp. bulgaricus by D- and L-lactic acid: Effects on lag phase, growth rate and cell yield. J. Appl. Bacteriol. 1995, 78, 647–654. [Google Scholar] [CrossRef]
- Cherrington, C.A.; Hinton, M.; Mead, G.C.; Chopra, I. Organic Acids: Chemistry, Antibacterial Activity and Practical Applications. In Advances in Microbial Physiology; Rose, A.H., Tempest, D.W., Eds.; Academic Press: Cambridge, MA, USA, 1991; Volume 32, pp. 87–108. [Google Scholar]
- Cherrington, C.A.; Hinton, M.; Chopra, I. Effect of short-chain organic acids on macromolecular synthesis in Escherichia coli. J. Appl. Bacteriol. 1990, 68, 69–74. [Google Scholar] [CrossRef]



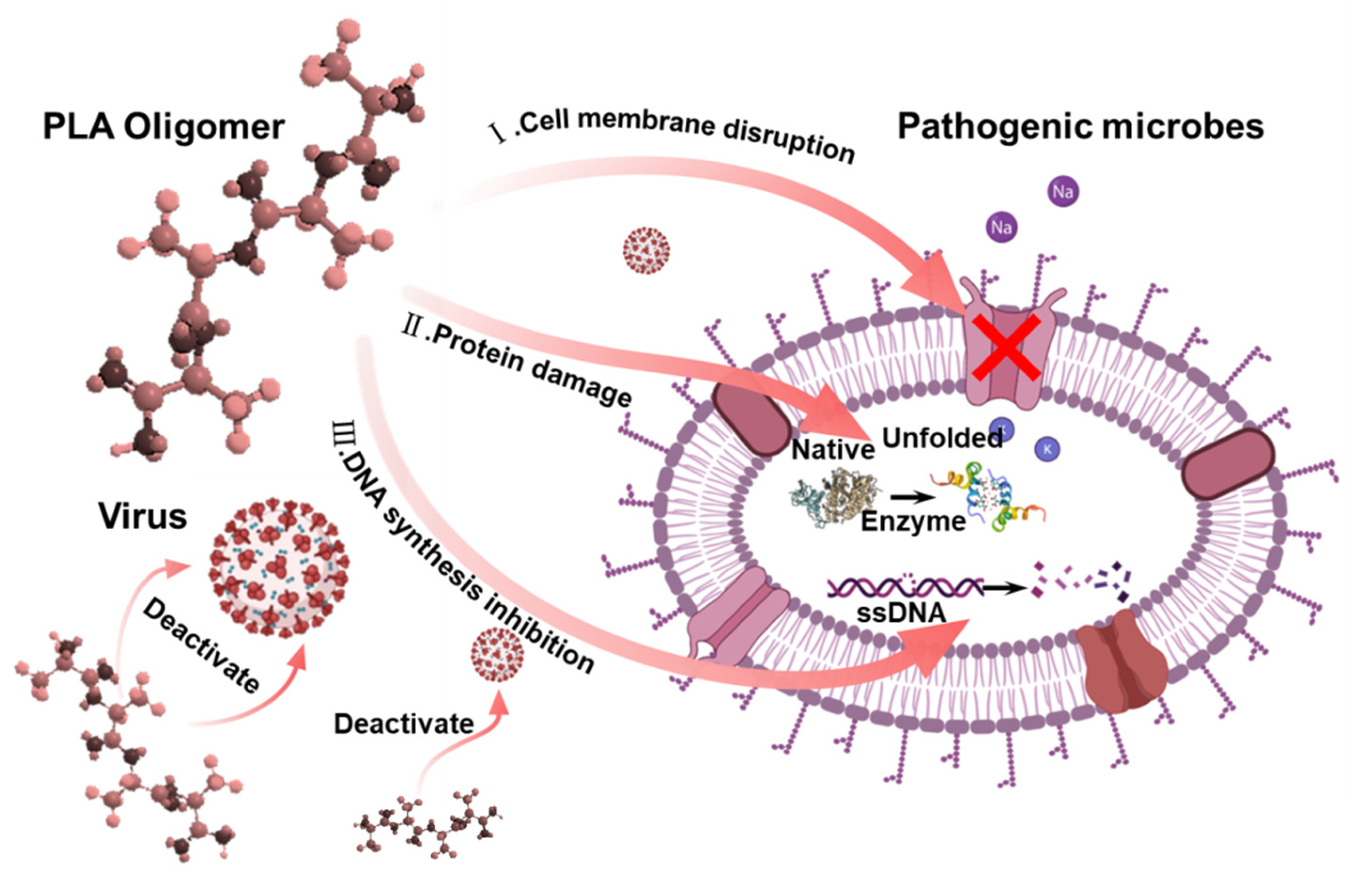
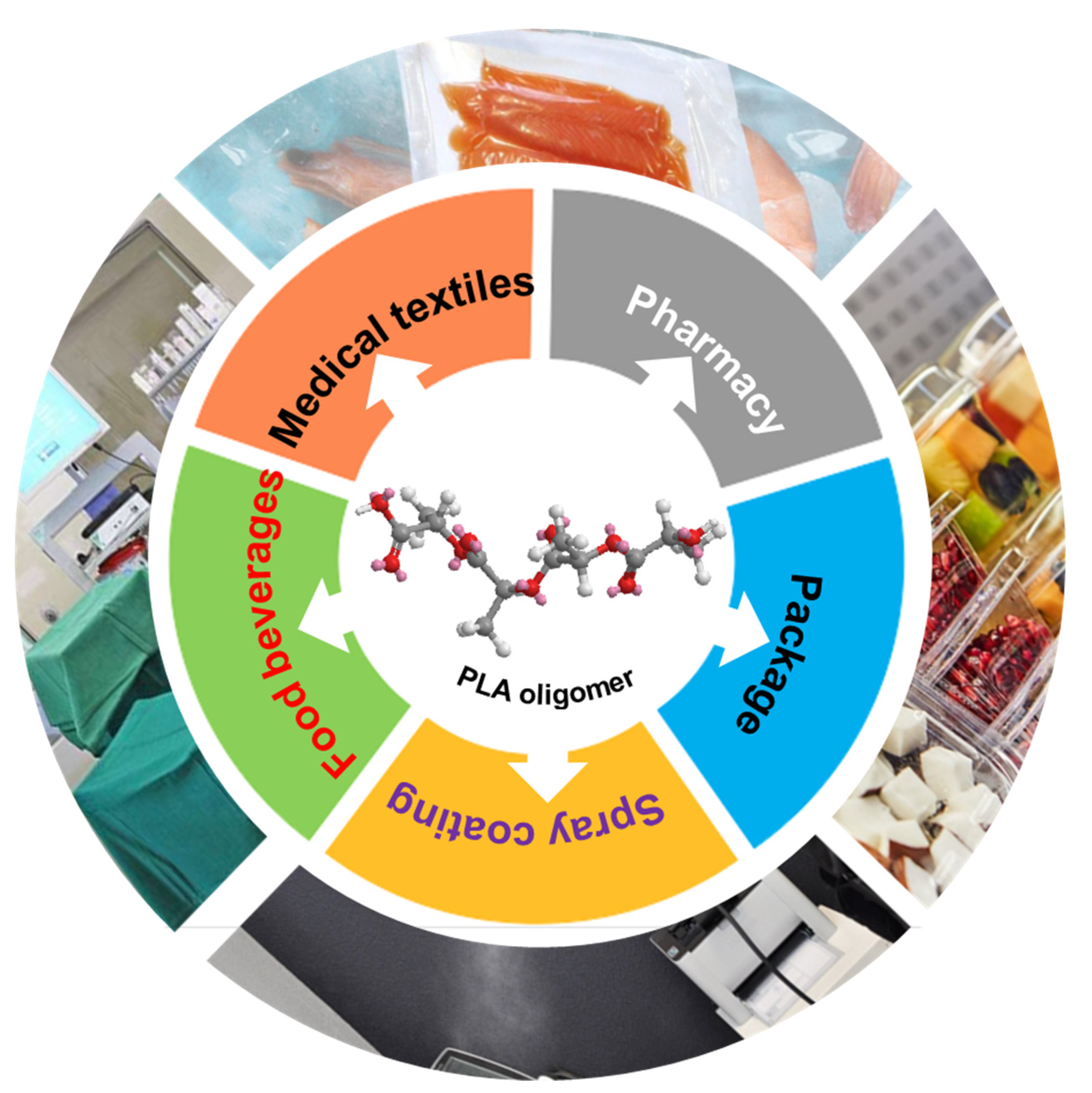
| Virus Species and Host Cell | Culture Concentration | Contact Time | Groups | LgTCID50/mL | Average TCID50/mL | Average LgTCID50/mL | Logarithm Reduction Value (KL) | Virus Inactivation Ratio (%) |
|---|---|---|---|---|---|---|---|---|
| Virus: Influenza A virus | 0 mg/mL | 1 h | Ctrl 1 | 5.80 | 5.89 × 105 | 5.77 | - | - |
| Ctrl 2 | 5.80 | |||||||
| Ctrl 3 | 5.71 | |||||||
| H1N1 (ATCC VR-1469) | 20 mg/mL | 1 h | PLA1 | <1.50 | <31.6 | <1.50 | >4.27 | >99.99 |
| PLA2 | <1.50 | |||||||
| PLA3 | <1.50 | |||||||
| Host cell: MDCK | 20 mg/mL | 1 h | PLA1D | 1.57 | 39.81 | 1.60 | 4.17 | >99.99 |
| PLA2D | 1.67 | |||||||
| PLA3D | 1.57 |
| Virus Species and Host Cell | Culture Concentration | Contact Time | Groups | LgTCID50/mL | Average TCID50/mL | Average LgTCID50/mL | Logarithm Reduction Value (KL) | Virus Inactivation Ratio (%) |
|---|---|---|---|---|---|---|---|---|
| Virus: Influenza A virus | 0 mg/mL | 10 min | Ctrl 1 | 5.90 | 6.85 × 105 | 5.83 | - | - |
| Ctrl 2 | 5.80 | |||||||
| Ctrl 3 | 5.80 | |||||||
| H3N2 (ATCC VR-1679) | 20 mg/mL | 10 min | PLA1 | <1.50 | <31.6 | <1.50 | >4.33 | >99.99 |
| PLA2 | <1.50 | |||||||
| PLA3 | <1.50 | |||||||
| Host cell: MDCK | 20 mg/mL | 10 min | PLA1D | 3.00 | 1.17 × 103 | 3.07 | 2.76 | 99.83 |
| PLA2D | 3.20 | |||||||
| PLA3D | 3.00 |
| Virus Species and Host Cell | Culture Concentration | Contact Time | Groups | TCID50/mL | Average TCID50/mL | Average LgTCID50/mL | Logarithm Reduction Value (KL) | Virus Inactivation Ratio (%) |
|---|---|---|---|---|---|---|---|---|
| Virus: SARS-CoV-2 Host cell: Vero E6 | 0 mg/mL | 1.5 h | Ctrl 1 | 1.78 × 107 | 9.68 × 106 | 6.99 | - | - |
| Ctrl 2 | 5.62 × 106 | |||||||
| Ctrl 3 | 5.62 × 106 | |||||||
| 10 mg/mL | 1.5 h | PLA1 | <3.16 × 102 | <3.16 × 102 | <2.50 | >4.49 | >99.99 | |
| PLA2 | <3.16 × 102 | |||||||
| PLA3 | <3.16 × 102 | |||||||
| 20 mg/mL | 1.5 h | PLA1 | <3.16 × 102 | <3.16 × 102 | <2.54 | >4.49 | >99.99 | |
| PLA2 | <3.16 × 102 | |||||||
| PLA3 | <3.16 × 102 |
| Virus Species and Host Cell | Culture Concentration | Contact Time | Groups | TCID50/mL | Average TCID50/mL | Average LgTCID50/mL | Logarithm Reduction Value (KL) | Virus Inactivation Ratio (%) |
|---|---|---|---|---|---|---|---|---|
| Virus: SARS-CoV-2 Host cell: Vero E6 | 0 mg/mL | 20 min | Ctrl 1 | 3.16 × 107 | 1.89 × 107 | 7.28 | - | - |
| Ctrl 2 | 2.04 × 107 | |||||||
| Ctrl 3 | 4.64 × 106 | |||||||
| 10 mg/mL | 20 min | PLA1 | <3.16 × 102 | <3.16 × 102 | <2.50 | >4.78 | >99.99 | |
| PLA2 | 3.98 × 102 | |||||||
| PLA3 | <3.16 × 102 | |||||||
| 20 mg/mL | 20 min | PLA1 | <3.16 × 102 | <3.16 × 102 | <2.54 | >4.78 | >99.99 | |
| PLA2 | <3.16 × 102 | |||||||
| PLA3 | <3.16 × 102 |
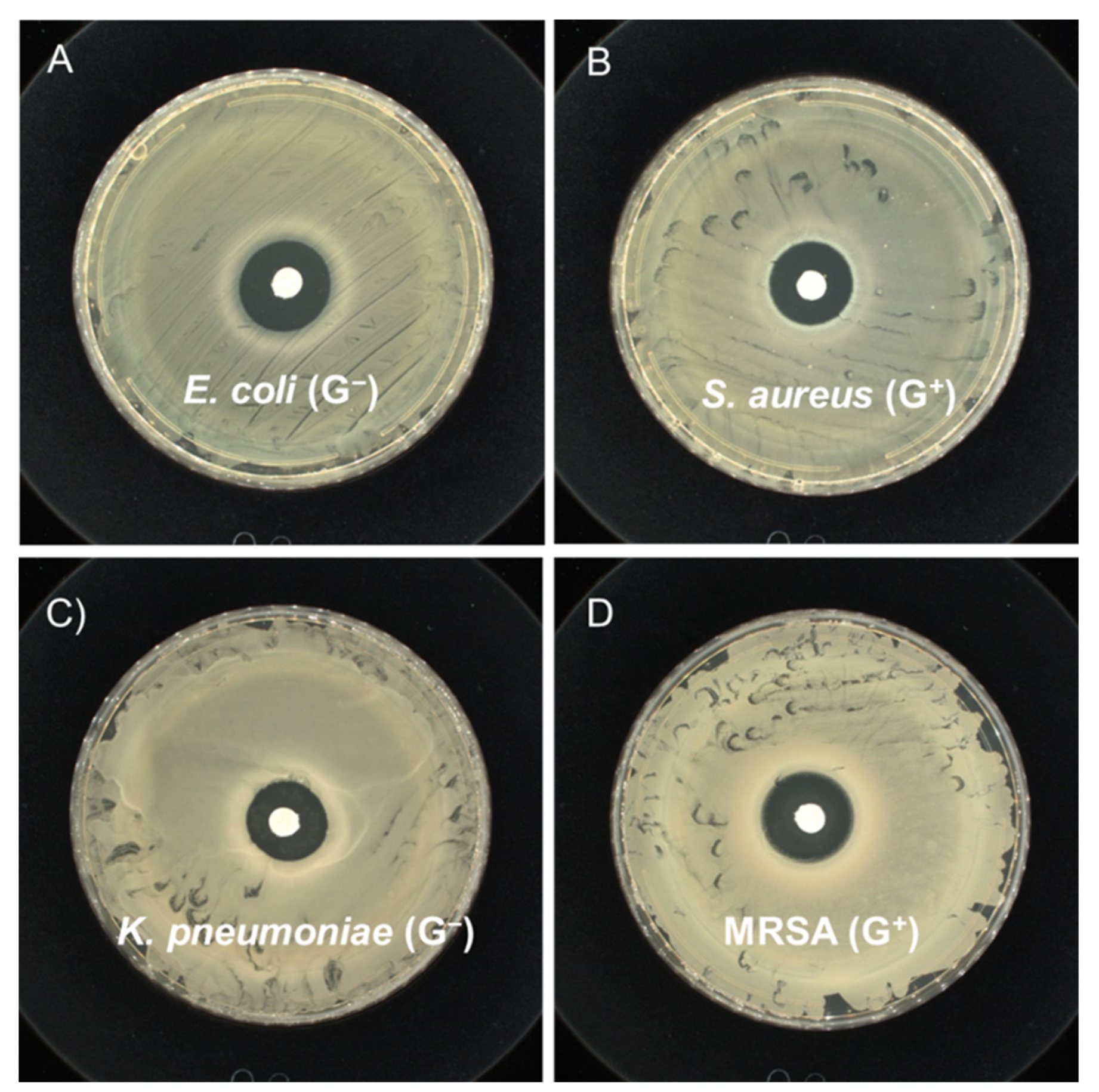
| PLA Oligomer Parameters | E. coli (G−) | S. aureus (G+) | K. pneumoniae (G−) | MRSA (G+) |
|---|---|---|---|---|
| Loading density (mg/mm2) | 0.64 | 0.60 | 0.60 | 0.64 |
| Inhibiting diameter (cm) | 1.90 ± 0.15 | 1.85 ± 0.15 | 1.70 ± 0.05 | 1.80 ± 0.12 |
| PLA Oligomer Concentration | E. coli (G−) | S. aureus (G+) | K. pneumoniae (G−) | MRSA (G+) |
|---|---|---|---|---|
| 10 mg/mL | >99.99% | >99.99% | >99.99% | >80.73% |
| 20 mg/mL | >99.99% | >99.99% | >99.99% | >84.44% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bao, Q.; Zhang, Z.; Yu, B.; Sun, H.; Leung, P.H.-m.; Tao, X. Synthesis of Polylactic Acid Oligomers for Broad-Spectrum Antimicrobials. Polymers 2022, 14, 4399. https://doi.org/10.3390/polym14204399
Bao Q, Zhang Z, Yu B, Sun H, Leung PH-m, Tao X. Synthesis of Polylactic Acid Oligomers for Broad-Spectrum Antimicrobials. Polymers. 2022; 14(20):4399. https://doi.org/10.3390/polym14204399
Chicago/Turabian StyleBao, Qi, Ziheng Zhang, Baocheng Yu, Huize Sun, Polly Hang-mei Leung, and Xiaoming Tao. 2022. "Synthesis of Polylactic Acid Oligomers for Broad-Spectrum Antimicrobials" Polymers 14, no. 20: 4399. https://doi.org/10.3390/polym14204399
APA StyleBao, Q., Zhang, Z., Yu, B., Sun, H., Leung, P. H.-m., & Tao, X. (2022). Synthesis of Polylactic Acid Oligomers for Broad-Spectrum Antimicrobials. Polymers, 14(20), 4399. https://doi.org/10.3390/polym14204399






