Preparation and Optimization of Ibrutinib-Loaded Nanoliposomes Using Response Surface Methodology
Abstract
1. Introduction
2. Experimental
2.1. Materials
2.2. Methods
2.2.1. Preparation of Ibrutinib Nanoliposomes
2.2.2. HPLC Analysis
2.2.3. Encapsulation Efficiency
2.2.4. Determination of Particle Size and Zeta Potential
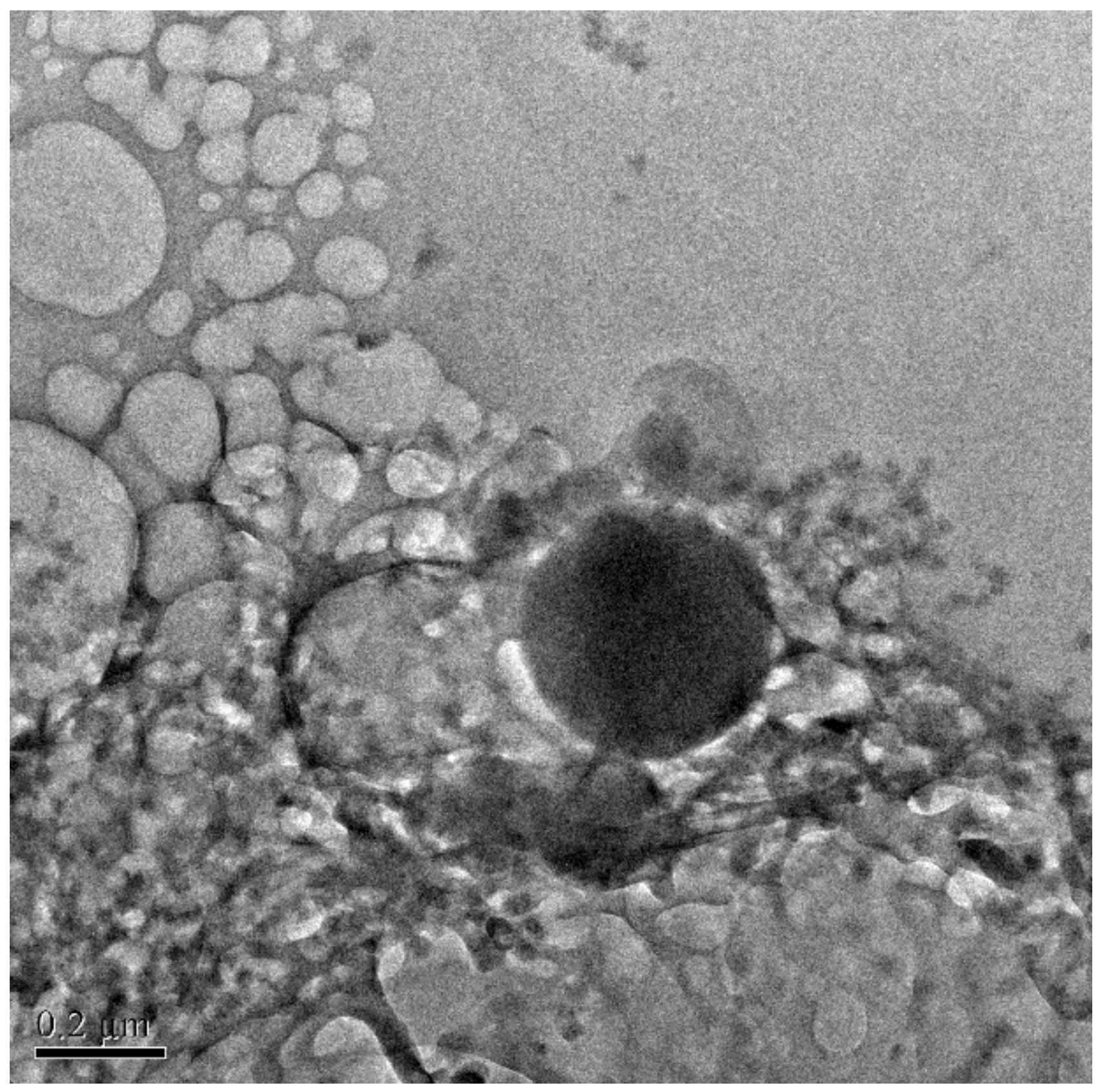
2.2.5. Surface Morphology Observation by TEM
2.2.6. Design of Experiments
2.2.7. Data Analysis
- Y—response parameter
- A0—intercept
- A1–A14—regression coefficients
- X1, X2, X3 and X4—main influencing factors
- X1×2—interactive effect
- —quadratic effect
2.2.8. Optimization
2.2.9. Stability of Ibrutinib Nanoliposomes
pH Stability
- EEt—encapsulation efficiency of the test sample solution at a specific pH and
- EEs—encapsulation efficiency of the standard sample at pH 7.
Thermal Stability
- EET—encapsulation efficiency of the test sample solution at a specific temperature and
- EEs—encapsulation efficiency at the standard temperature (4 °C).
Effect of Ultrasound Time on Stability of Nanoliposomes
2.2.10. Influence of Dissolution Medium on In Vitro Drug Dissolution
2.2.11. Fourier Transformed Infrared Spectroscopy
2.2.12. Thermal Analysis
2.2.13. X-ray Diffraction Study
2.2.14. Statistical Analysis
3. Results and Discussion
3.1. RSM Optimization
3.1.1. Statistical Treatment
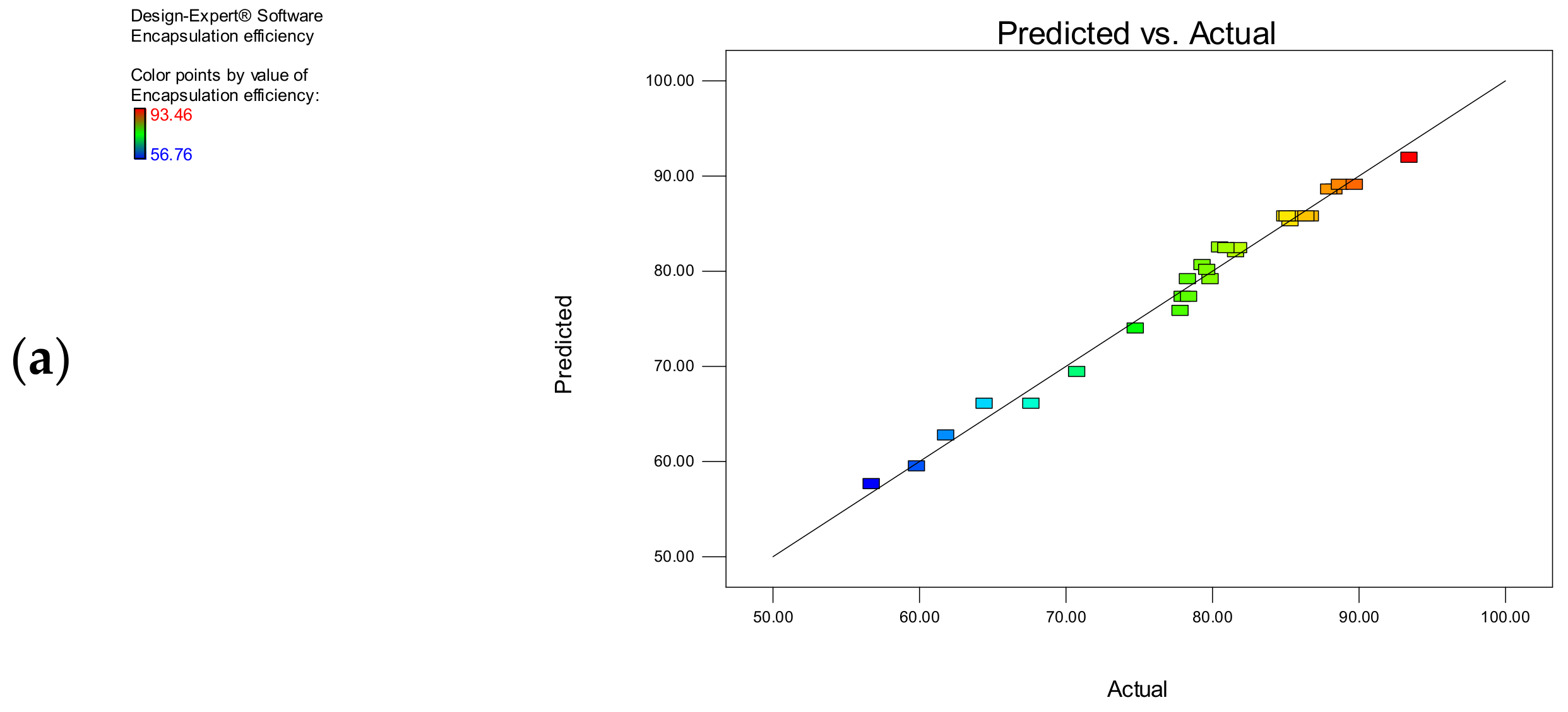
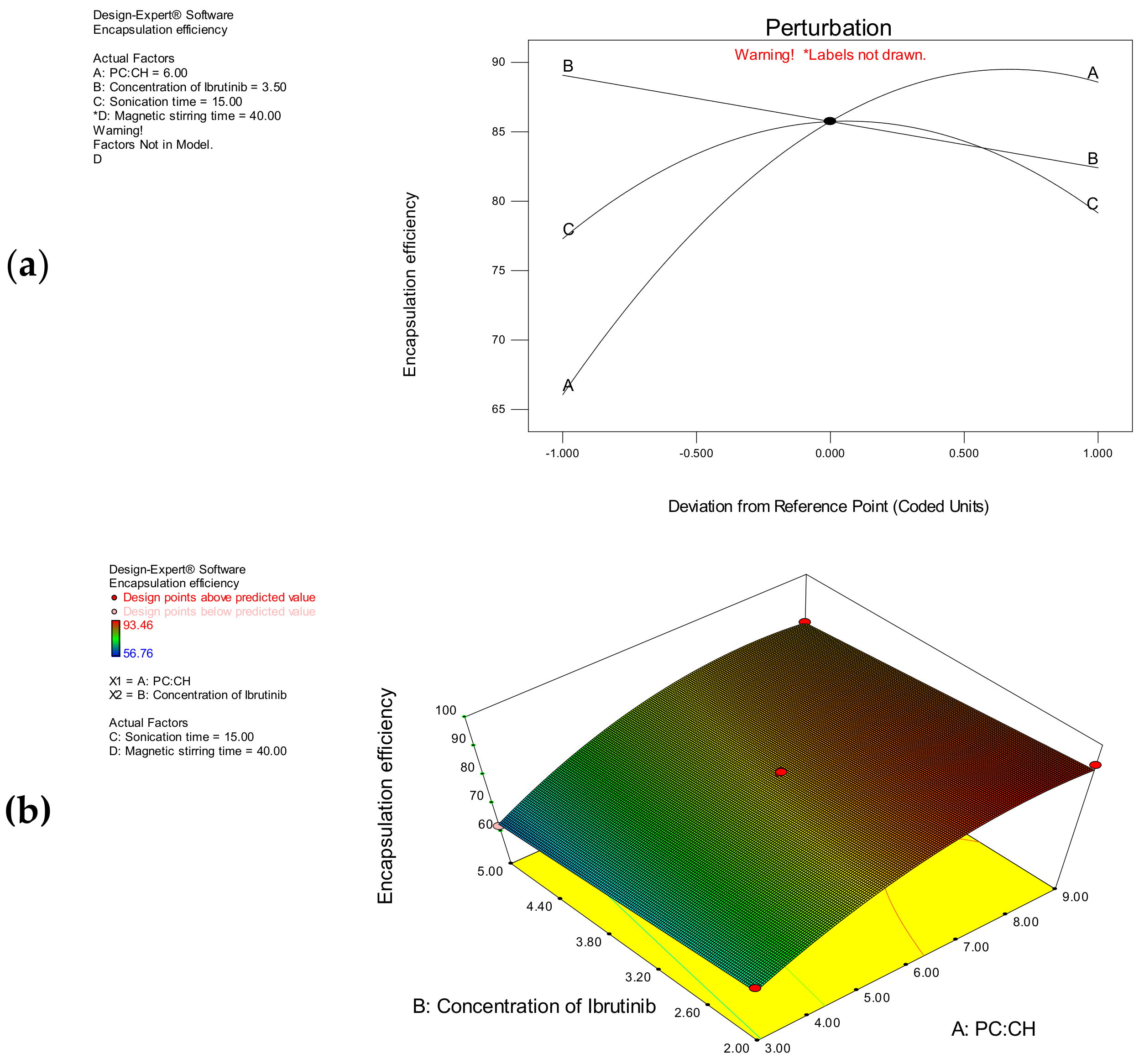
3.1.2. Encapsulation Efficiency
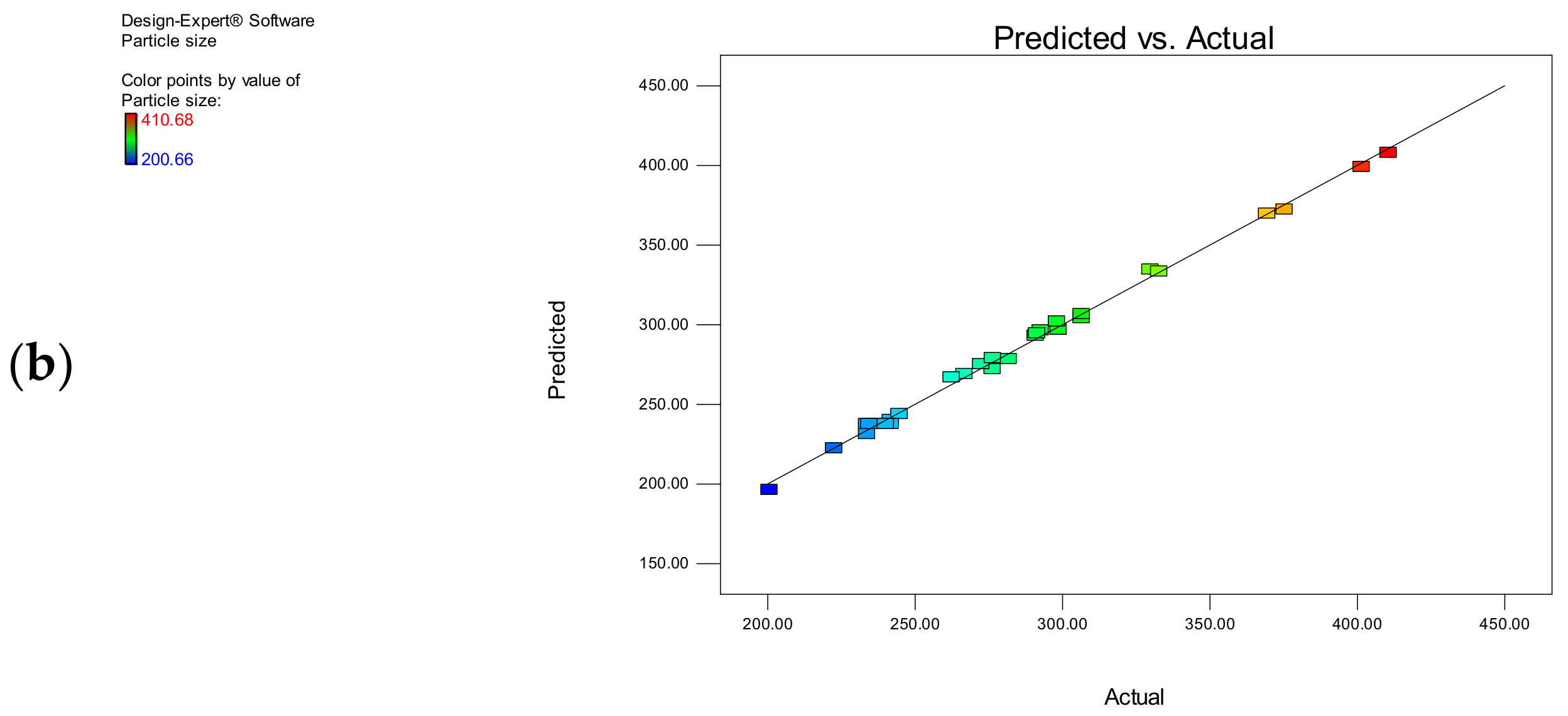
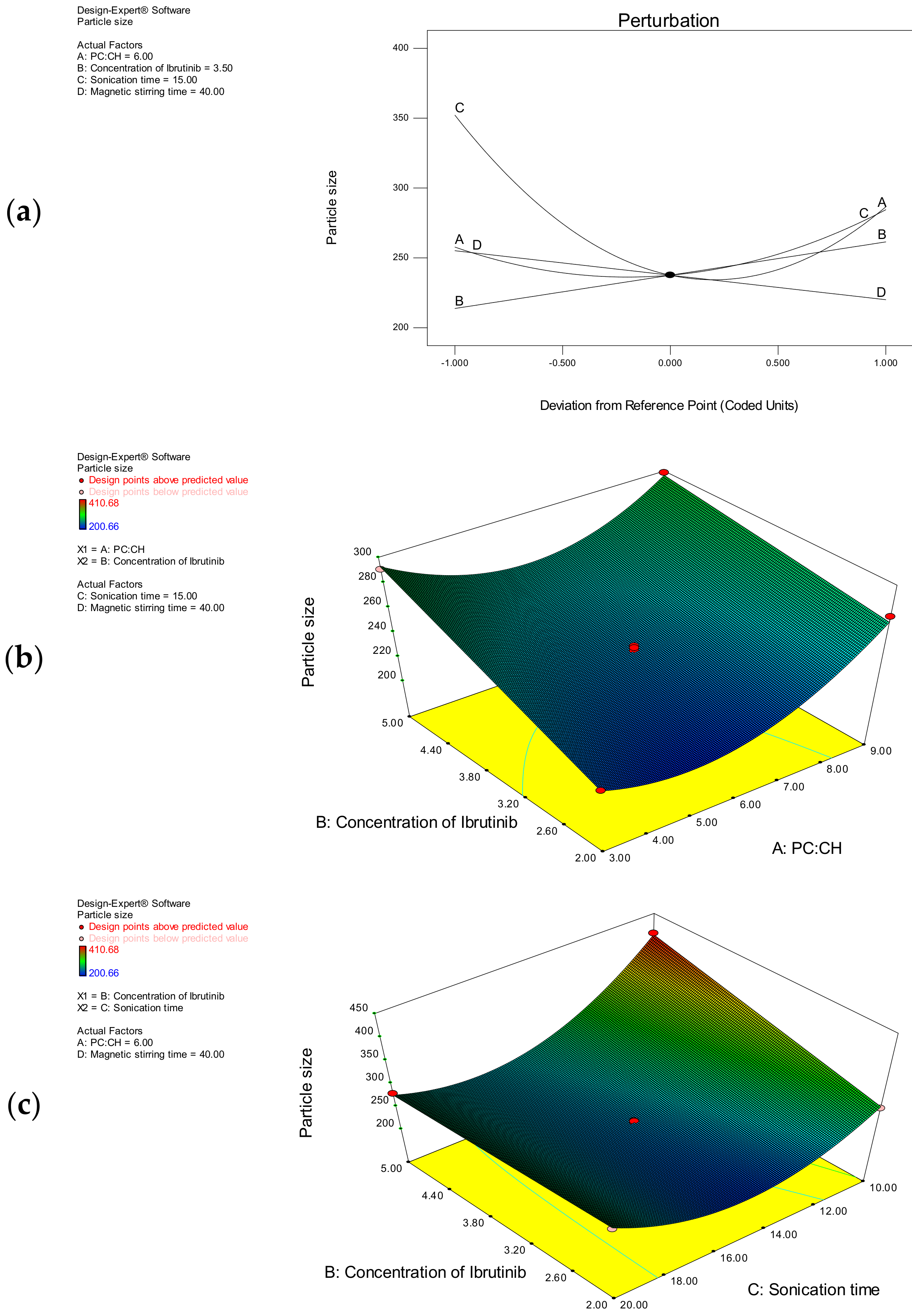
3.1.3. Particle Size
3.2. Optimization
3.3. Stability of Ibrutinib Nanoliposomes
3.3.1. pH Stability
3.3.2. Thermal Stability
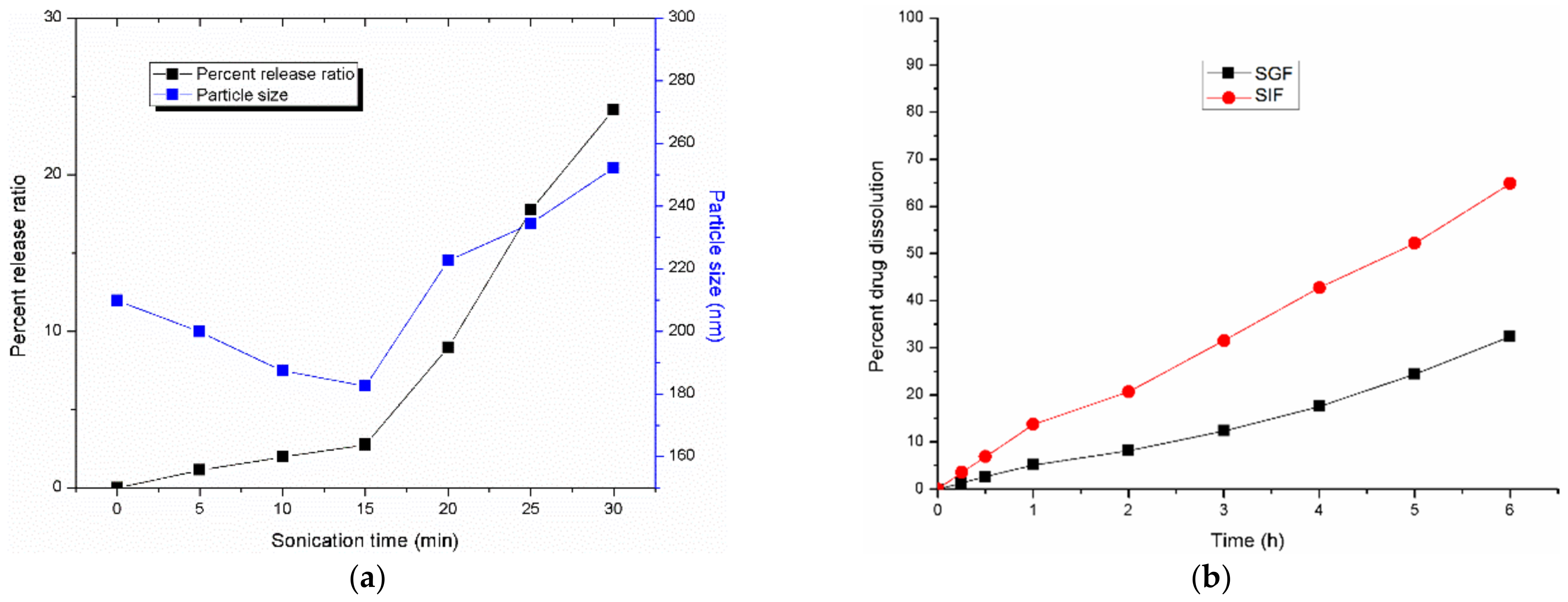
3.3.3. Effect of Ultrasound Time on the Stability of Nanoliposomes
3.4. In Vitro Dissolution Study
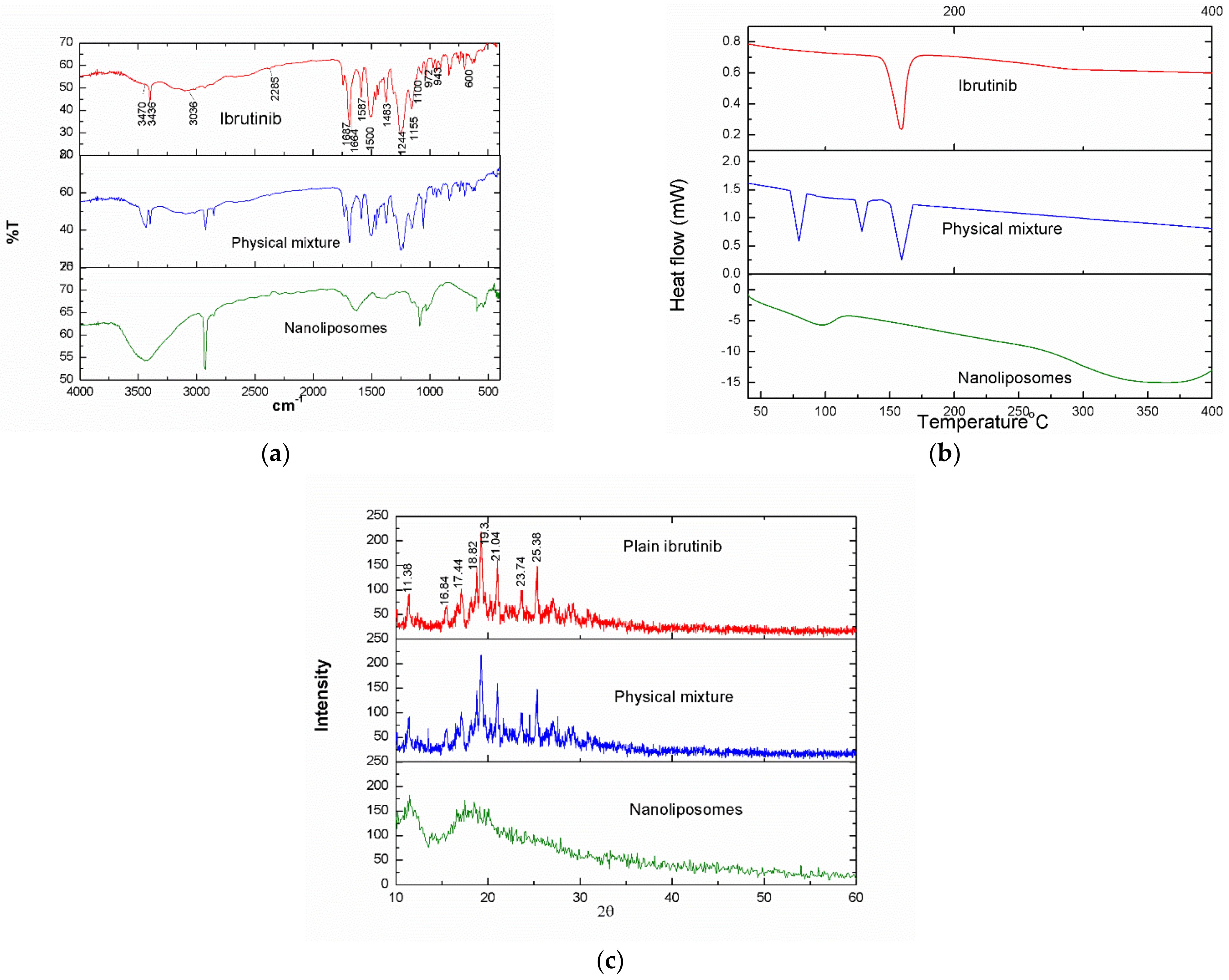
3.5. FTIR Spectra
3.6. DSC Thermograms
3.7. X-ray Diffraction Pattern
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Roskoski, R., Jr. Orally effective FDA-approved protein kinase targeted covalent inhibitors (TCIs). Pharmacol. Res. 2021, 165, 105422. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K.; Cheah, C. Non-Covalent BTK Inhibitors—The New BTKids on the Block for B-Cell Malignancies. J. Pers. Med. 2021, 11, 764. [Google Scholar] [CrossRef]
- Honigberg, L.A.; Smith, A.M.; Sirisawad, M.; Verner, E.; Loury, D.; Chang, B.; Li, S.; Pan, Z.; Thamm, D.H.; Miller, R.A.; et al. The Bruton tyrosine kinase inhibitor PCI-32765 blocks B-cell activation and is efficacious in models of autoimmune disease and B-cell malignancy. Proc. Natl. Acad. Sci. USA 2010, 107, 13075–13080. [Google Scholar] [CrossRef] [PubMed]
- Berglof, A.; Hamasy, A.; Meinke, S.; Palma, M.; Krstić, A.; Månsson, R.; Kimby, E.; Österborg, A.; Smith, C.I.E. Targets for Ibrutinib beyond B Cell Malignancies. Scand. J. Immunol. 2015, 82, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wong, J.; Sevinsky, C.J.; Kokabee, L.; Khan, F.; Sun, Y.; Conklin, D.S. Bruton’s Tyrosine Kinase Inhibitors Prevent Therapeutic Escape in Breast Cancer Cells. Mol. Cancer Ther. 2016, 15, 2198–2208. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.S.; Zhang, C.; Shokat, K.M.; Taunton, J. Structural Bioinformatics-Based Design of Selective, Irreversible Kinase Inhibitors. Science 2005, 308, 1318–1321. [Google Scholar] [CrossRef] [PubMed]
- Leproult, E.; Barluenga, S.; Moras, D.; Wurtz, J.-M.; Winssinger, N. Cysteine Mapping in Conformationally Distinct Kinase Nucleotide Binding Sites: Application to the Design of Selective Covalent Inhibitors. J. Med. Chem. 2011, 54, 1347–1355. [Google Scholar] [CrossRef] [PubMed]
- Alshetaili, A.S.; Ansari, M.J.; Anwer, K.; Ganaie, M.A.; Iqbal, M.; Alshahrani, S.M.; Alalaiwe, A.S.; Alsulays, B.B.; Alshehri, S.; Sultan, A.S. Enhanced Oral Bioavailability of Ibrutinib Encapsulated Poly (Lactic-co-Glycolic Acid) Nanoparticles: Pharmacokinetic Evaluation in Rats. Curr. Pharm. Anal. 2019, 15, 661–668. [Google Scholar] [CrossRef]
- Shakeel, F.; Iqbal, M.; Ezzeldin, E. Bioavailability enhancement and pharmacokinetic profile of an anticancer drug ibrutinib by self-nanoemulsifying drug delivery system. J. Pharm. Pharmacol. 2016, 68, 772–780. [Google Scholar] [CrossRef]
- De Jong, J.; Sukbuntherng, J.; Skee, D.; Murphy, J.; O’Brien, S.; Byrd, J.C.; James, D.; Hellemans, P.; Loury, D.J.; Jiao, J.; et al. The effect of food on the pharmacokinetics of oral ibrutinib in healthy participants and patients with chronic lymphocytic leukemia. Cancer Chemother. Pharmacol. 2015, 75, 907–916. [Google Scholar] [CrossRef]
- Shono, Y.; Jantratid, E.; Kesisoglou, F.; Reppas, C.; Dressman, J.B. Forecasting in vivo oral absorption and food effect of micronized and nanosized aprepitant formulations in humans. Eur. J. Pharm. Biopharm. 2010, 76, 95–104. [Google Scholar] [CrossRef]
- Qiu, Q.; Lu, M.; Li, C.; Luo, X.; Liu, X.; Hu, L.; Liu, M.; Zheng, H.; Zhang, H.; Liu, M.; et al. Novel Self-Assembled Ibrutinib-Phospholipid Complex for Potently Peroral Delivery of Poorly Soluble Drugs with pH-Dependent Solubility. AAPS PharmSciTech 2018, 19, 3571–3583. [Google Scholar] [CrossRef]
- Massó-Vallés, D.; Jauset, T.; Soucek, L. Ibrutinib repurposing: From B-cell malignancies to solid tumors. Oncoscience 2016, 3, 147–148. [Google Scholar] [CrossRef]
- Haura, E.B.; Rix, U. Deploying ibrutinib to lung cancer: Another step in the quest towards drug repurposing. J. Natl. Cancer Inst. 2014, 106, dju250. [Google Scholar] [CrossRef][Green Version]
- EMA. CHMP Assessment Report: European Medicines Agency. 2014. Available online: https://www.ema.europa.eu/en/documents/assessment-report/imbruvica-epar-public-assessment-report_en.pdf (accessed on 20 July 2022).
- Ud Din, F.; Aman, W.; Ullah, I.; Qureshi, O.S.; Mustapha, O.; Shafique, S.; Zeb, A. Effective use of nanocarriers as drug delivery systems for the treatment of selected tumors. Int. J. Nanomed. 2017, 12, 7291–7309. [Google Scholar] [CrossRef]
- Gupta, S.; Kumar, P. Drug Delivery Using Nanocarriers: Indian Perspective. Proc. Natl. Acad. Sci. India Sect. B Boil. Sci. 2012, 82, 167–206. [Google Scholar] [CrossRef]
- Mozafari, M. Nanoliposomes: Preparation and analysis. In Liposomes; Methods in Molecular Biology; Springer: Berlin, Germany, 2010; Volume 605, pp. 29–50. [Google Scholar] [CrossRef]
- David, Q.; de la Luz, Z.M.; Gerardo, L.; Mendoza-Muñoz, N. Liposomal Nanotechnology in Nutraceuticals. In Handbook of Nutraceuticals and Natural Products: Biological, Medicinal, and Nutritional Properties and Applications; Wiley: Hoboken, NJ, USA, 2022; Volume 1, pp. 29–62. [Google Scholar] [CrossRef]
- Karpagam, T.; Balamuralikrishnan, B.; Varalakshmi, B.; Anand, A.V.; Sugunabai, J. An Insight on Emerging Nanomaterials for the Delivery of Various Nutraceutical Applications for the Betterment of Heath. In Emerging Nanomaterials for Advanced Technologies; Springer: Cham, Switzerland, 2022; pp. 1–27. [Google Scholar] [CrossRef]
- Jafari, S.M.; Fathi, M.; Mandala, I. Emerging product formation. In Food Waste Recovery; Academic Press: Cambridge, MA, USA, 2021; pp. 257–275. [Google Scholar] [CrossRef]
- Shahgholian, N. Encapsulation and Delivery of Nutraceuticals and Bioactive Compounds by Nanoliposomes and Tocosomes as Promising Nanocarriers. In Handbook of Nutraceuticals and Natural Products: Biological, Medicinal, and Nutritional Properties and Applications; Wiley: Hoboken, NJ, USA, 2022; Volume 1, pp. 403–439. [Google Scholar] [CrossRef]
- Tang, B.; Peng, Y.; Yue, Q.; Pu, Y.; Li, R.; Zhao, Y.; Hai, L.; Guo, L.; Wu, Y. Design, preparation and evaluation of different branched biotin modified liposomes for targeting breast cancer. Eur. J. Med. Chem. 2020, 193, 112204. [Google Scholar] [CrossRef]
- Feuser, P.E.; Cordeiro, A.P.; Silveira, G.D.B.; Corrêa, M.E.A.B.; Silveira, P.C.L.; Sayer, C.; de Araújo, P.H.H.; Machado-De-Ávila, R.A.; Bó, A.G.D. Co-encapsulation of sodium diethyldithiocarbamate (DETC) and zinc phthalocyanine (ZnPc) in liposomes promotes increases phototoxic activity against (MDA-MB 231) human breast cancer cells. Colloids Surf. B Biointerfaces 2020, 197, 111434. [Google Scholar] [CrossRef]
- Demirci, M.; Caglar, M.; Cakir, B.; Gülseren, İ. Encapsulation by nanoliposomes. In Nanoencapsulation Technologies for the Food and Nutraceutical Industries; Elsevier: Amsterdam, The Netherlands, 2017; pp. 74–113. [Google Scholar] [CrossRef]
- Samadi, N.; Azar, P.A.; Husain, S.W.; Maibach, H.I.; Nafisi, S. Experimental design in formulation optimization of vitamin K1 oxide-loaded nanoliposomes for skin delivery. Int. J. Pharm. 2020, 579, 119136. [Google Scholar] [CrossRef]
- Tereshkina, Y.A.; Torkhovskaya, T.I.; Tikhonova, E.G.; Kostryukova, L.V.; Sanzhakov, M.A.; Korotkevich, E.I.; Khudoklinova, Y.Y.; Orlova, N.A.; Kolesanova, E.F. Nanoliposomes as drug delivery systems: Safety concerns. J. Drug Target. 2022, 30, 313–325. [Google Scholar] [CrossRef]
- Aguilar-Pérez, K.M.; Avilés-Castrillo, J.I.; Medina, D.I.; Parra-Saldivar, R.; Iqbal, H.M.N. Insight Into Nanoliposomes as Smart Nanocarriers for Greening the Twenty-First Century Biomedical Settings. Front. Bioeng. Biotechnol. 2020, 8, 579536. [Google Scholar] [CrossRef]
- Singireddy, A.; Pedireddi, S.R.; Subramanian, S. Optimization of reaction parameters for synthesis of Cyclodextrin nanosponges in controlled nanoscopic size dimensions. J. Polym. Res. 2019, 26, 93. [Google Scholar] [CrossRef]
- Shi, N.-Q.; Qi, X.-R. Preparation of Drug Liposomes by Reverse-Phase Evaporation. In Liposome-Based Drug Delivery Systems; Biomaterial Engineering; Lu, W.L., Qi, X.R., Eds.; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar] [CrossRef]
- Cohen, L.; Assaraf, Y.G.; Livney, Y.D. Novel Selectively Targeted Multifunctional Nanostructured Lipid Carriers for Prostate Cancer Treatment. Pharmaceutics 2021, 14, 88. [Google Scholar] [CrossRef]
- Anandam, S.; Selvamuthukumar, S. Optimization of microwave-assisted synthesis of cyclodextrin nanosponges using response surface methodology. J. Porous Mater. 2014, 21, 1015–1023. [Google Scholar] [CrossRef]
- Almajed, A.; Srirama, D.; Moghal, A.A.B. Response Surface Method Analysis of Chemically Stabilized Fiber-Reinforced Soil. Materials 2021, 14, 1535. [Google Scholar] [CrossRef]
- Song, H.; Chung, H.; Nam, K. Response surface modeling with Box-Behnken design for strontium removal from soil by calcium-based solution. Environ. Pollut. 2021, 274, 116577. [Google Scholar] [CrossRef]
- Wu, Z.; Guan, R.; Lyu, F.; Liu, M.; Gao, J.; Cao, G. Optimization of Preparation Conditions for Lysozyme Nanoliposomes Using Response Surface Methodology and Evaluation of Their Stability. Molecules 2016, 21, 741. [Google Scholar] [CrossRef]
- Danaei, M.; Dehghankhold, M.; Ataei, S.; Hasanzadeh Davarani, F.; Javanmard, R.; Dokhani, A.; Khorasani, S.; Mozafari, M.R. Impact of Particle Size and Polydispersity Index on the Clinical Applications of Lipidic Nanocarrier Systems. Pharmaceutics 2018, 10, 57. [Google Scholar] [CrossRef]
- Wang, J.; Yang, J.; Kopeček, J. Nanomedicines in B cell-targeting therapies. Acta Biomater. 2022, 137, 1–19. [Google Scholar] [CrossRef]
- Zhang, M.; Suo, Z.; Peng, X.; Gan, N.; Zhao, L.; Tang, P.; Wei, X.; Li, H. Microcrystalline cellulose as an effective crystal growth inhibitor for the ternary Ibrutinib formulation. Carbohydr. Polym. 2020, 229, 115476. [Google Scholar] [CrossRef]
- Ismail, A.I. Experimental and Density Functional Theory Characteristics of Ibrutinib, a Bruton’s Kinase Inhibitor Approved for Leukemia Treatment. J. Spectrosc. 2021, 2021, 9968797. [Google Scholar] [CrossRef]

| Independent Variables | Levels | ||||
|---|---|---|---|---|---|
| Variable | Units | Low | Intermediate | High | |
| A | PC: CH | w/w | 3 | 6 | 9 |
| B | Conc. Ibrutinib | w/v | 2 | 3.5 | 5 |
| C | Sonication time | min | 10 | 15 | 20 |
| D | Stirring time | Min | 35 | 40 | 45 |
| Dependent variables | Goal | ||||
| Y1 | Encapsulation efficiecny | % | Increase | ||
| Y2 | Particle size | nm | Decrease | ||
| Exp. | A | B | C | D | Y1 | Y2 |
|---|---|---|---|---|---|---|
| 1 | 6 | 3.5 | 20 | 35 | 78.34 ± 2.78 | 306.56 ± 5.12 |
| 2 | 3 | 3.5 | 10 | 40 | 56.76 ± 0.86 | 375.38 ± 6.34 |
| 3 | 9 | 2 | 15 | 40 | 93.46 ± 0.93 | 276.34 ± 5.51 |
| 4 | 6 | 3.5 | 15 | 40 | 86.42 ± 4.08 | 234.56 ± 6.18 |
| 5 | 3 | 5 | 15 | 40 | 61.84 ± 2.87 | 290.92 ± 5.42 |
| 6 | 6 | 3.5 | 15 | 40 | 85.82 ± 2.54 | 241.72 ± 4.86 |
| 7 | 6 | 3.5 | 15 | 40 | 86.72 ± 3.32 | 237.64 ± 4.63 |
| 8 | 6 | 5 | 20 | 40 | 77.83 ± 4.08 | 281.78 ± 5.17 |
| 9 | 6 | 2 | 15 | 45 | 89.73 ± 0.78 | 200.66 ± 4.78 |
| 10 | 6 | 3.5 | 15 | 40 | 84.98 ± 2.42 | 233.78 ± 3.92 |
| 11 | 6 | 2 | 20 | 40 | 80.54± 1.28 | 291.42 ± 6.13 |
| 12 | 3 | 2 | 15 | 40 | 70.77 ± 1.44 | 222.56 ± 2.97 |
| 13 | 6 | 5 | 15 | 35 | 81.82 ± 1.56 | 276.42 ± 3.93 |
| 14 | 6 | 2 | 15 | 35 | 88.72 ± 4.18 | 233.76 ± 4.12 |
| 15 | 9 | 3.5 | 20 | 40 | 81.62 ± 3.18 | 332.88 ± 2.77 |
| 16 | 9 | 3.5 | 15 | 35 | 88.34 ± 3.76 | 298.18 ± 4.15 |
| 17 | 9 | 3.5 | 15 | 45 | 87.98 ± 4.12 | 262.46 ± 4.75 |
| 18 | 6 | 3.5 | 10 | 35 | 77.98 ± 2.12 | 369.48 ± 5.15 |
| 19 | 6 | 3.5 | 20 | 45 | 79.88 ± 3.38 | 266.82 ± 6.19 |
| 20 | 9 | 5 | 15 | 40 | 85.34 ± 1.98 | 298.72 ± 4.36 |
| 21 | 3 | 3.5 | 15 | 35 | 64.46 ± 2.24 | 272.44 ± 5.27 |
| 22 | 6 | 3.5 | 10 | 45 | 78.42 ± 1.32 | 329.92 ± 6.22 |
| 23 | 6 | 5 | 10 | 40 | 74.78 ± 1.76 | 410.68 ± 3.18 |
| 24 | 9 | 3.5 | 10 | 40 | 79.65 ± 3.42 | 401.52 ± 4.27 |
| 25 | 3 | 3.5 | 20 | 40 | 59.84 ± 2.56 | 306.56 ± 5.16 |
| 26 | 6 | 3.5 | 15 | 40 | 85.14 ± 2.26 | 240.12 ± 3.38 |
| 27 | 3 | 3.5 | 15 | 45 | 67.66 ± 1.98 | 241.82 ± 3.92 |
| 28 | 6 | 5 | 15 | 45 | 80.96 ± 3.12 | 244.78 ± 2.96 |
| 29 | 6 | 2 | 10 | 40 | 79.34 ± 2.67 | 292.63 ± 3.18 |
| Source of Variation | Sum of Squares | Degrees of Freedom | Mean Square Value | F-Value | p-Value Prob > F |
|---|---|---|---|---|---|
| Y1—Encapsulation efficiency | |||||
| Model | 2437.769 | 5 | 487.5538 | 370.0586 | <0.0001 |
| A—PC:CH | 1520.1 | 1 | 1520.1 | 1153.773 | <0.0001 |
| B—Concentration of ibrutinib | 133.2667 | 1 | 133.2667 | 101.1509 | <0.0001 |
| C—Sonication time | 10.30453 | 1 | 10.30453 | 7.821253 | 0.0102 |
| A2 | 489.6346 | 1 | 489.6346 | 371.638 | <0.0001 |
| C2 | 389.8075 | 1 | 389.8075 | 295.8681 | <0.0001 |
| Residual | 30.3026 | 23 | 1.317504 | ||
| Lack of fit | 27.96468 | 19 | 1.471825 | 2.518179 | 0.1918 |
| Pure error | 2.33792 | 4 | 0.58448 | ||
| Total | 2468.07 | 28 | |||
| Observed R2 | 0.9877 | ||||
| Adjusted R2 | 0.9851 | ||||
| CV | 1.45 | ||||
| Y2—Particle size | |||||
| Model | 79,741.52 | 8 | 9967.69 | 817.9295 | <0.0001 |
| A—PC:CH | 2144.548 | 1 | 2144.548 | 175.9775 | <0.0001 |
| B—Concentration of ibrutinib | 6812.997 | 1 | 6812.997 | 559.0614 | <0.0001 |
| C—Sonication time | 12,909.42 | 1 | 12,909.42 | 1059.323 | <0.0001 |
| D—Stirring time | 3688.312 | 1 | 3688.312 | 302.6558 | <0.0001 |
| AB | 528.5401 | 1 | 528.5401 | 43.37098 | <0.0001 |
| AC | 4076.184 | 1 | 4076.184 | 334.4838 | <0.0001 |
| A2 | 7740.406 | 1 | 7740.406 | 635.1628 | <0.0001 |
| C2 | 46,092.1 | 1 | 46,092.1 | 3782.229 | <0.0001 |
| Residual | 243.7298 | 20 | 12.18649 | ||
| Lack of fit | 196.5759 | 16 | 12.28599 | 1.042203 | 0.5438 |
| Pure error | 47.15392 | 4 | 11.78848 | ||
| Total | 79,985.25 | 28 | |||
| Observed R2 | 0.9970 | ||||
| Adjusted R2 | 0.9857 | ||||
| CV | 1.22 | ||||
| Dependent Variable | Regression Equation |
|---|---|
| Encapsulation efficiency (Y1) | 85.74 + 11.25 A − 3.33B + 0.93 C − 8.42 A2 − 7.52 C2 |
| Particle size (Y2) | 237.59 + 13.37 A + 23.83 B − 32.80 C − 17.53 D − 11.49 AB − 31.92 BC + 33.49 A2 + 81.72 C2 |
| Independent Variables | Optimum Values | Predicted Values | Actual Values | |||
|---|---|---|---|---|---|---|
| Y1 | Y2 | Batch | Y1 | Y2 | ||
| A | 6.76 (w/w) | 91.39 | 204.67 | 1 | 89.94 ± 1.76 | 208.34 ± 2.42 |
| B | 2% w/v | 2 | 91.22 ± 2.12 | 211.76 ± 1.32 | ||
| C | 15.13 min | 3 | 90.86 ± 3.27 | 205.42 ± 3.14 | ||
| D | 45 min | |||||
| Batch | Particle Size (nm) | Polydispersity Index | Zeta Potential (mV) |
|---|---|---|---|
| 1 | 208.34 ± 2.42 | 0.212 ± 0.005 | 18.72 ± 3.18 |
| 2 | 211.76 ± 1.32 | 0.192 ± 0.005 | 20.43 ± 1.14 |
| 3 | 205.42 ± 3.14 | 0.234 ± 0.005 | 21.12 ± 1.18 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ashar, F.; Hani, U.; Osmani, R.A.M.; Kazim, S.M.; Selvamuthukumar, S. Preparation and Optimization of Ibrutinib-Loaded Nanoliposomes Using Response Surface Methodology. Polymers 2022, 14, 3886. https://doi.org/10.3390/polym14183886
Ashar F, Hani U, Osmani RAM, Kazim SM, Selvamuthukumar S. Preparation and Optimization of Ibrutinib-Loaded Nanoliposomes Using Response Surface Methodology. Polymers. 2022; 14(18):3886. https://doi.org/10.3390/polym14183886
Chicago/Turabian StyleAshar, Fareeaa, Umme Hani, Riyaz Ali M. Osmani, Syed Mohammed Kazim, and S. Selvamuthukumar. 2022. "Preparation and Optimization of Ibrutinib-Loaded Nanoliposomes Using Response Surface Methodology" Polymers 14, no. 18: 3886. https://doi.org/10.3390/polym14183886
APA StyleAshar, F., Hani, U., Osmani, R. A. M., Kazim, S. M., & Selvamuthukumar, S. (2022). Preparation and Optimization of Ibrutinib-Loaded Nanoliposomes Using Response Surface Methodology. Polymers, 14(18), 3886. https://doi.org/10.3390/polym14183886






