Improvement on Thermostability of Pectate Lyase and Its Potential Application to Ramie Degumming
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
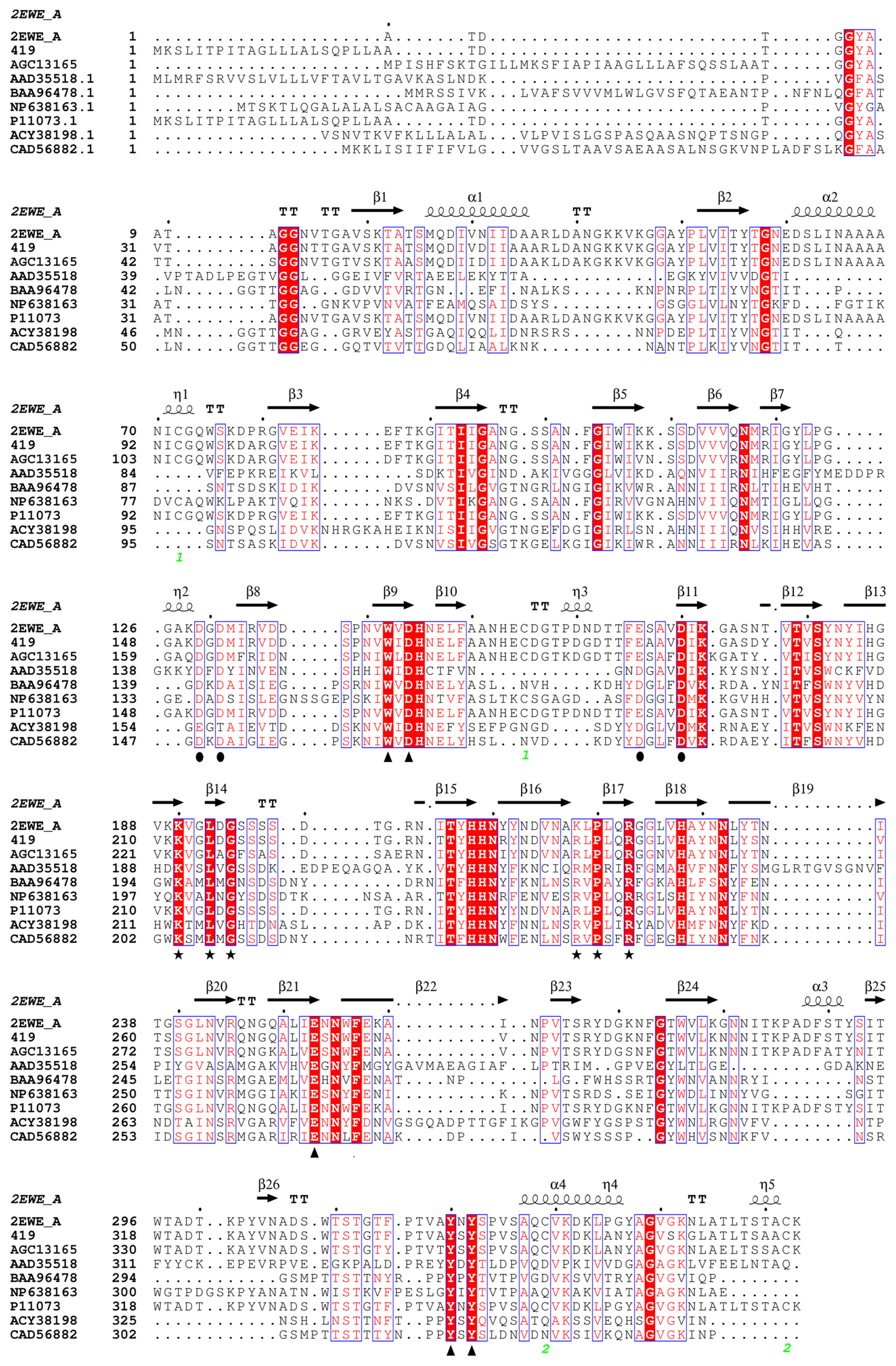
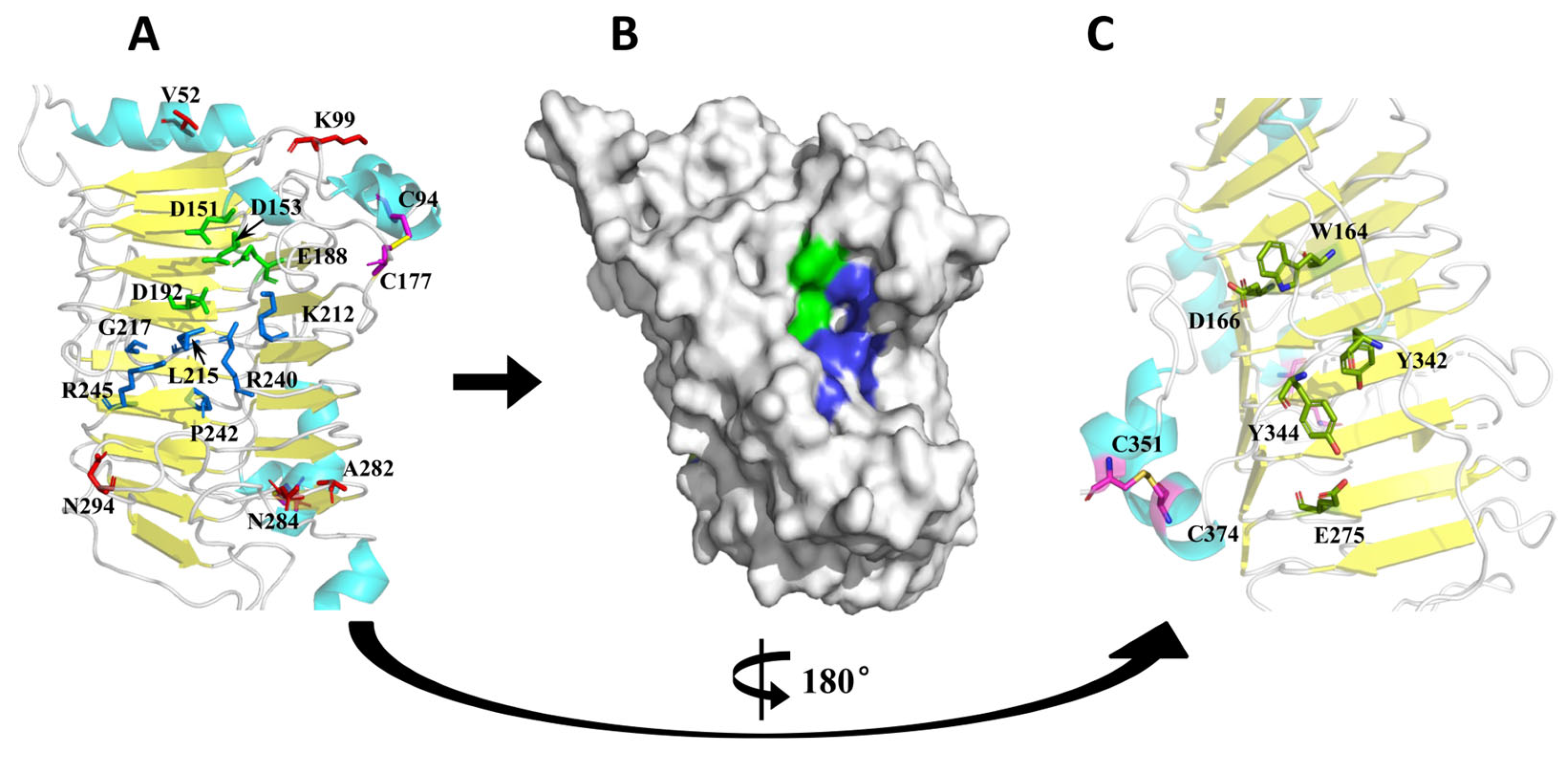
2.2.1. Prediction of Mutation Sites
2.2.2. Construction and Prokaryotic Expression of Recombinant Mutagenic Plasmid, Preparation of Cell-Free Extract
2.2.3. SDS-PAGE Analysis and Enzymatic Activity Determination
2.2.4. Thermostability of Mutants
2.2.5. Ramie Degumming through Enzymatic Method
3. Results
3.1. Selection of Mutation Sites
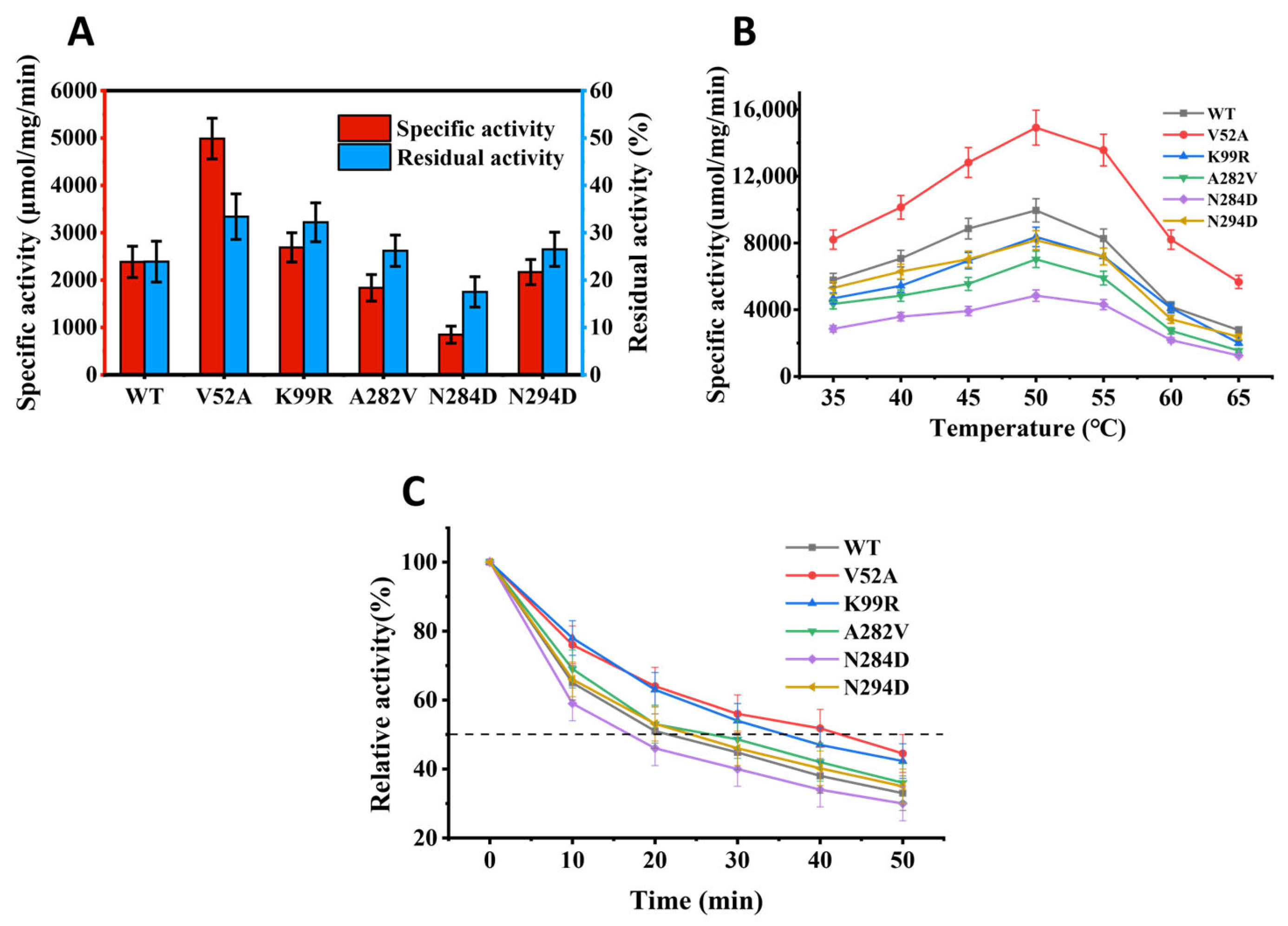
3.2. Heterologous Expression Analysis of Mutant Pectate Lyase
3.3. Determination of the Optimal Catalytic Temperature
3.4. Thermostability Test
3.5. Ramie Degumming Effect
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nicole, H.; Guy, C.; Vladimir, E.S. Bacterial pectate lyases, structural and functional diversity. Environ. Microbiol. Rep. 2014, 6, 427–440. [Google Scholar] [CrossRef]
- Pan, W.; Yang, S.; Zhan, Z.; Zhang, G. Origins and features of pectate lyases and their applications in industry. Appl. Microbiol. Biotechnol. 2020, 104, 7247–7260. [Google Scholar] [CrossRef]
- Sonia, A.; Mandhan, R.P.; Sudha, D.S.; Rakesh, K.; Jitender, S. Potential application of alkaline pectinase from Bacillus subtilis SS in pulp and paper industry. Appl. Biochem. Biotechnol. 2008, 149, 287–293. [Google Scholar] [CrossRef]
- Nwobi, A.; Cybulska, I.; Tesfai, W.; Shatilla, Y.; Rodríguez, J.; Thomsen, M.H. Simultaneous saccharification and fermentation of solid household waste following mild pretreatment using a mix of hydrolytic enzymes in combination with Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2015, 99, 929–938. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, R.; Dutta, T.; Sheikh, J. Extraction of pectinase from Candida isolated from textile mill effluent and its application in bio-scouring of cotton. Sustain. Chem. Pharm. 2020, 17, 100291. [Google Scholar] [CrossRef]
- Shukri, N.A.; Mohd Zin, Z.; MohdMaidin, N.; Hasmadi, M.; Zainol, M.K. Ramification of pH in pectinase-assisted extraction on the antioxidant capacity of Arabica spent coffee ground (SCG) extract. J. Food Sci. 2020, 4, 1303–1311. [Google Scholar] [CrossRef]
- Neelima, M.; Kavita, S.; Mukty, S.; Archana, D.; Brajesh, P.; Hyeji, J.; Seorin, P.; Srinath, P.; Sunghun, C. Biotransformation of citrus waste-I: Production of biofuel and valuable compounds by fermentation. Processes 2021, 9, 220. [Google Scholar] [CrossRef]
- Zhang, G.; Li, S.; Xu, Y.; Wang, J.; Wang, F.; Xin, Y.; Shen, Z.; Zhang, H.; Ma, M.; Liu, H. Production of alkaline pectinase: A case study investigating the use of tobacco stalk with the newly isolated strain Bacillus tequilensis CAS-MEI-2-33. BMC Biotechnol. 2019, 19, 45. [Google Scholar] [CrossRef]
- Steven, R.H.; Robert, D.S.; Michael, G.; Margaret, L.; Frances, J. Characterization and implications of Ca2+ binding to pectate lyase C. J. Biol. Chem. 2003, 278, 12271–12277. [Google Scholar] [CrossRef] [Green Version]
- Marie-Line, G.; Miroslaw, C. Structural and mechanistic classification of uronic acid-containing polysaccharide lyases. Glycobiology 2010, 20, 1547–1573. [Google Scholar] [CrossRef] [Green Version]
- Han, C.; Li, W.; Hua, C.; Sun, F.; Bi, P.; Wang, Q. Enhancement of catalytic activity and thermostability of a thermostable cellobiohydrolase from Chaetomium thermophilum by site-directed mutagenesis. Int. J. Biol. Macromol. 2018, 116, 691–697. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Duan, S.; Zheng, K.; Feng, X.; Yang, Q.; Liu, Z.Y.; Liu, Z.; Peng, Y. An alkaline pectate lyase D from Dickeya dadantii DCE-01: Clone, expression, characterization, and potential application in ramie bio-degumming. Text. Res. J. 2019, 89, 2075–2083. [Google Scholar] [CrossRef]
- López-Villamizar, I.; Cabezas, A.; Pinto, R.M.; Canales, J.; Ribeiro, J.M.; Rodrigues, J.R.; Costas, M.J.; Cameselle, J.C. Molecular Dissection of Escherichia coli CpdB: Roles of the N Domain in Catalysis and Phosphate Inhibition, and of the C Domain in Substrate Specificity and Adenosine Inhibition. Int. J. Mol. Sci. 2021, 22, 1977. [Google Scholar] [CrossRef] [PubMed]
- Yury, A.D.; Alexander, V.G.; Aleksandra, M.R.; Dmitry, O.O.; Ivan, N.Z.; Veronika, Y.M.; Igor, V.U.; Arkady, P.S. Site-directed mutagenesis of GH10 xylanase A from Penicillium canescens for determining factors affecting the enzyme thermostability. Int. J. Biol. Macromol. 2017, 104, 665–671. [Google Scholar] [CrossRef]
- Chen, X.; Li, W.; Ji, P.; Zhao, Y.; Hua, C.; Han, C. Engineering the conserved and noncatalytic residues of a thermostable β-1,4-endoglucanase to improve specific activity and thermostability. Sci. Rep. 2018, 8, 2954. [Google Scholar] [CrossRef]
- Wen, Z.; Zhang, Z.; Zhong, L.; Fan, J.; Li, M.; Ma, Y.; Zhou, Y.; Zhang, W.; Guo, B.; Chen, B.; et al. Directed evolution of a plant glycosyltransferase for chemo- and regioselective glycosylation of pharmaceutically significant flavonoids. ACS Catal. 2021, 11, 14781–14790. [Google Scholar] [CrossRef]
- Chen, M.; Song, F.; Qin, Y.; Han, S.; Rao, Y.; Liang, S.; Lin, Y. Improving thermostability and catalytic activity of glycosyltransferase from panax ginseng by semi-rational design for rebaudioside D synthesis. Front. Bioeng. Biotechnol. 2022, 10, 884898. [Google Scholar] [CrossRef]
- Reetz, M.T.; Carballeira, J.D.; Vogel, A. Iterative saturation mutagenesis on the basis of B factors as a strategy for increasing protein thermostability. Angew. Chem. Int. Ed. Engl. 2006, 45, 7745–7751. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Wang, H.L.; Li, X.M.; Ma, Y.H.; Song, J.N. Characterization and high-level expression of a metagenome-derived alkaline pectate lyase in recombinant Escherichia coli. Process Biochem. 2014, 49, 69–76. [Google Scholar] [CrossRef]
- Zhou, Q.; Ji, P.; Zhang, J.; Li, X.; Han, C. Characterization of a novel thermostable GH45 endoglucanase from Chaetomium thermophilum and its biodegradation of pectin. J. Biosci. Bioeng. 2017, 124, 271–276. [Google Scholar] [CrossRef]
- Liu, Z.; Duan, S.; Sun, Q.; Peng, Y.; Feng, X.; Zheng, K.; Hu, Z.; Zhang, Y. A rapid process of ramie bio-degumming by Pectobacterium sp. CXJZU-120. Text. Res. J. 2012, 82, 1553–1559. [Google Scholar] [CrossRef]
- Scavetta, R.D.; Herron, S.R.; Hotchkiss, A.T.; Kita, N.; Keen, N.T.; Benen, J.A.; Kester, H.C.; Visser, J.; Jurnak, F. Structure of a plant cell wall fragment complexed to pectate lyase C. Plant Cell 1999, 11, 1081–1092. [Google Scholar] [CrossRef] [Green Version]
- Dinu, D.; Nechifor, M.T.; Stoian, G.; Costache, M.; Dinischiotu, A. Enzymes with new biochemical properties in the pectinolytic complex produced by Aspergillus niger MIUG 16. J. Biotechnol. 2007, 131, 128–137. [Google Scholar] [CrossRef]
- Wang, H.; Fu, L.; Zhang, X. Comparison of expression, purification and characterization of a new pectate lyase from Phytophthora capsici using two different methods. BMC Biotechnol. 2011, 11, 32–38. [Google Scholar] [CrossRef] [Green Version]
- Damak, N.; Abdeljalil, S.; Koubaa, A.; Trigui, S.; Ayadi, M.; Trigui-Lahiani, H.; Kallel, E.; Turki, N.; Djemal, L.; Belghith, H.; et al. Cloning and heterologous expression of a thermostable pectate lyase from Penicillium occitanis in Escherichia coli. Int. J. Biol. Macromol. 2013, 62, 549–556. [Google Scholar] [CrossRef]
- Atanasova, L.; Dubey, M.; Gruji, M.; Gudmundsson, M.; Lorenz, C.; Sandgren, M.; Kubicek, C.P.; Jensen, D.F.; Karlsson, M. Evolution and functional characterization of pectate lyase PEL12, a member of a highly expanded Clonostachys rosea polysaccharide lyase 1 family. BMC Microbial. 2018, 18, 178. [Google Scholar] [CrossRef]
- Xu, Z.; Cen, Y.; Zou, S.; Xue, Y.; Zheng, Y. Recent advances in the improvement of enzyme thermostability by structure modification. Crit. Rev. Biotechnol. 2020, 40, 83–98. [Google Scholar] [CrossRef]
- Zhou, Z.; Wang, X. Rational design and structure-based engineering of alkaline pectate lyase from Paenibacillus sp. 0602 to improve thermostability. BMC Biotechnol. 2021, 21, 32. [Google Scholar] [CrossRef]
- Zhou, Z.; Liu, Y.; Chang, Z.; Wang, H.; Leier, A.; Marquez-Lago, T.T.; Ma, Y.; Li, J.; Song, J. Structure-based engineering of a pectate lyase with improved specific activity for ramie degumming. Appl. Microbiol. Biotechnol. 2017, 101, 2919–2929. [Google Scholar] [CrossRef]
- Sun, Z.; Liu, Q.; Qu, G.; Feng, Y.; Reetz, M.T. Utility of b-factors in protein science: Interpreting rigidity, flexibility, and internal motion and engineering thermostability. Chem. Rev. 2019, 119, 1626–1665. [Google Scholar] [CrossRef]
- Jochens, H.; Aerts, D.; Bornscheuer, U.T. Thermostabilization of an esterase by alignment-guided focussed directed evolution. Protein Eng. Des. Sel. 2010, 23, 903–909. [Google Scholar] [CrossRef] [Green Version]
- Adrian, B.H.; Anderson, J.L.R.; Donald, H.; Arcus, V.L.; van der Kamp, M.W.; Mulholland, A.J. Evolution of dynamical networks enhances catalysis in a designer enzyme. Nat. Chem. 2021, 13, 1017–1022. [Google Scholar] [CrossRef]
- Blanes-Mira, C.; Blanesmira, C.; Ibañez, C.; Fernándezballester, G.; Planellscases, R.; Pérezpayá, A.E. Thermal stabilization of the catalytic domain of Botulinum neurotoxin E by phosphorylation of a single tyrosine residue. Biochemistry 2001, 40, 2234–2242. [Google Scholar] [CrossRef]
- Nimpiboon, P.; Kaulpiboon, J.; Krusong, K.; Nakamura, S.; Kidokoro, S.; Pongsawasdi, P. Mutagenesis for improvement of activity and thermostability of amylomaltase from Corynebacterium glutamicum. Int. J. Biol. Macromol. 2016, 86, 820–828. [Google Scholar] [CrossRef]
- Donald, J.E.; Kulp, D.W.; DeGrado, W.F. Salt bridges: Geometrically specific, designable interactions. Proteins 2011, 79, 898–915. [Google Scholar] [CrossRef] [Green Version]
- Ban, X.; Dhoble, A.S.; Li, C.; Zhang, Y.; Gu, Z.; Cheng, L.; Hong, Y.; Li, Z. Potassium and sodium ions enhance the activity and thermostability of 1,4-alphaglucan branching enzyme from Geobacillus thermoglucosidasius in the presence of glycerol. Int. J. Biol. Macromol. 2017, 102, 712–717. [Google Scholar] [CrossRef] [PubMed]
- Ban, X.; Wu, J.; Kaustubh, B.; Lahiri, P.; Dhoble, A.S.; Gu, Z.; Li, C.; Cheng, L.; Hong, Y.; Tong, Y.; et al. Additional salt bridges improve the thermostability of 1,4-α-glucan branching enzyme. Food Chem. 2020, 316, 126348. [Google Scholar] [CrossRef] [PubMed]


| Enzymes | CK | Pel419 | V52A | Novo |
|---|---|---|---|---|
| Weight loss rate (%) | 4.77 ± 0.32 | 12.89 ± 0.28 | 18.77 ± 0.23 | 19.22 ± 0.21 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, H.; Feng, X.; Yang, Q.; Zheng, K.; Yi, L.; Duan, S.; Cheng, L. Improvement on Thermostability of Pectate Lyase and Its Potential Application to Ramie Degumming. Polymers 2022, 14, 2878. https://doi.org/10.3390/polym14142878
Xu H, Feng X, Yang Q, Zheng K, Yi L, Duan S, Cheng L. Improvement on Thermostability of Pectate Lyase and Its Potential Application to Ramie Degumming. Polymers. 2022; 14(14):2878. https://doi.org/10.3390/polym14142878
Chicago/Turabian StyleXu, Huan, Xiangyuan Feng, Qi Yang, Ke Zheng, Le Yi, Shengwen Duan, and Lifeng Cheng. 2022. "Improvement on Thermostability of Pectate Lyase and Its Potential Application to Ramie Degumming" Polymers 14, no. 14: 2878. https://doi.org/10.3390/polym14142878
APA StyleXu, H., Feng, X., Yang, Q., Zheng, K., Yi, L., Duan, S., & Cheng, L. (2022). Improvement on Thermostability of Pectate Lyase and Its Potential Application to Ramie Degumming. Polymers, 14(14), 2878. https://doi.org/10.3390/polym14142878







