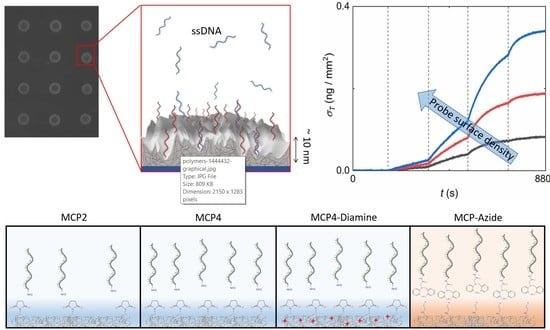
Copolymer Coatings for DNA Biosensors: Effect of Charges and Immobilization Chemistries on Yield, Strength and Kinetics of Hybridization
Abstract
1. Introduction
2. Materials and Methods
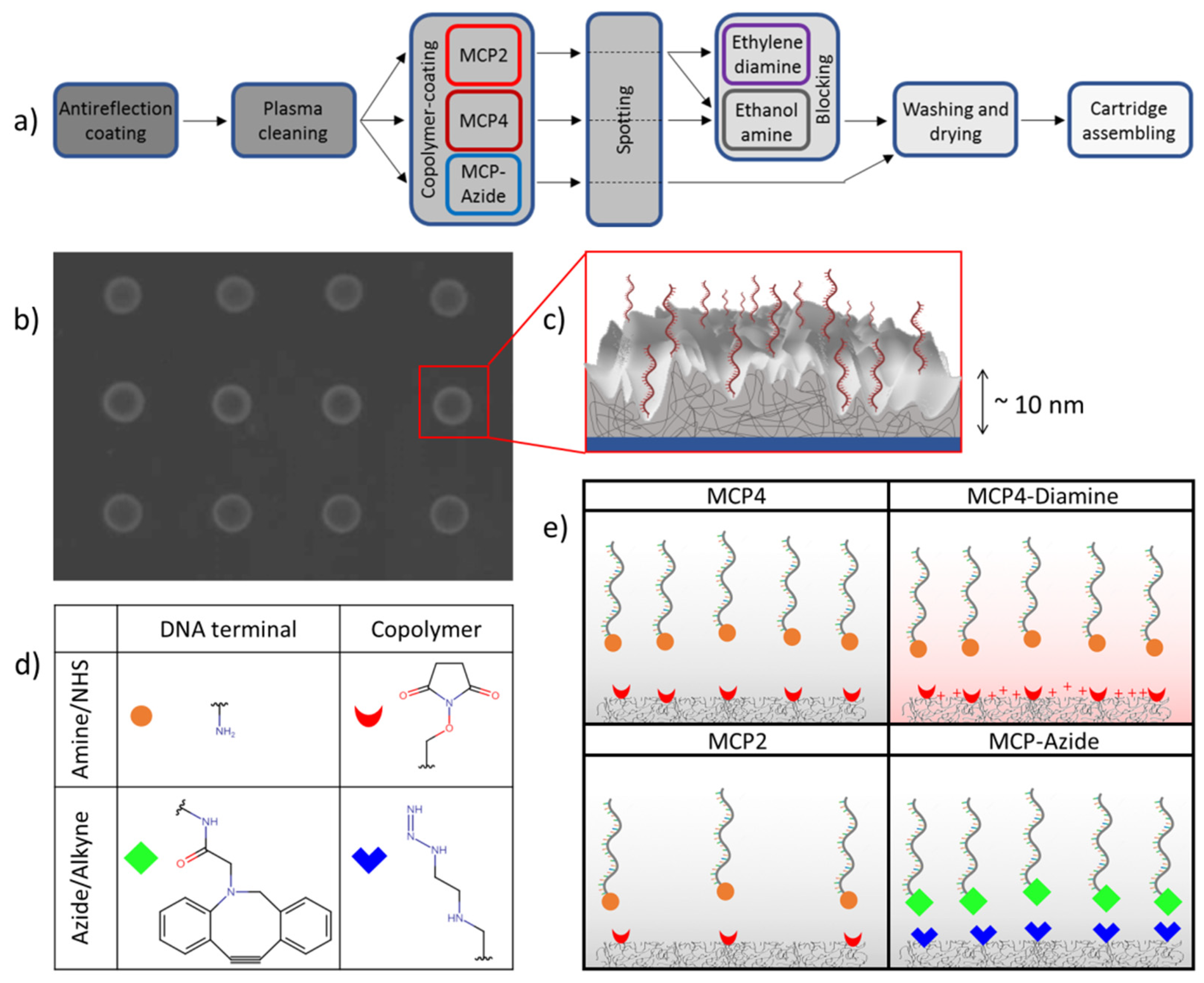
2.1. Copolymer Surface Coatings
2.2. DNA Strands and Reagents
2.3. RPI Sensor Preparation
2.4. RPI Measurement and Analysis
3. Results
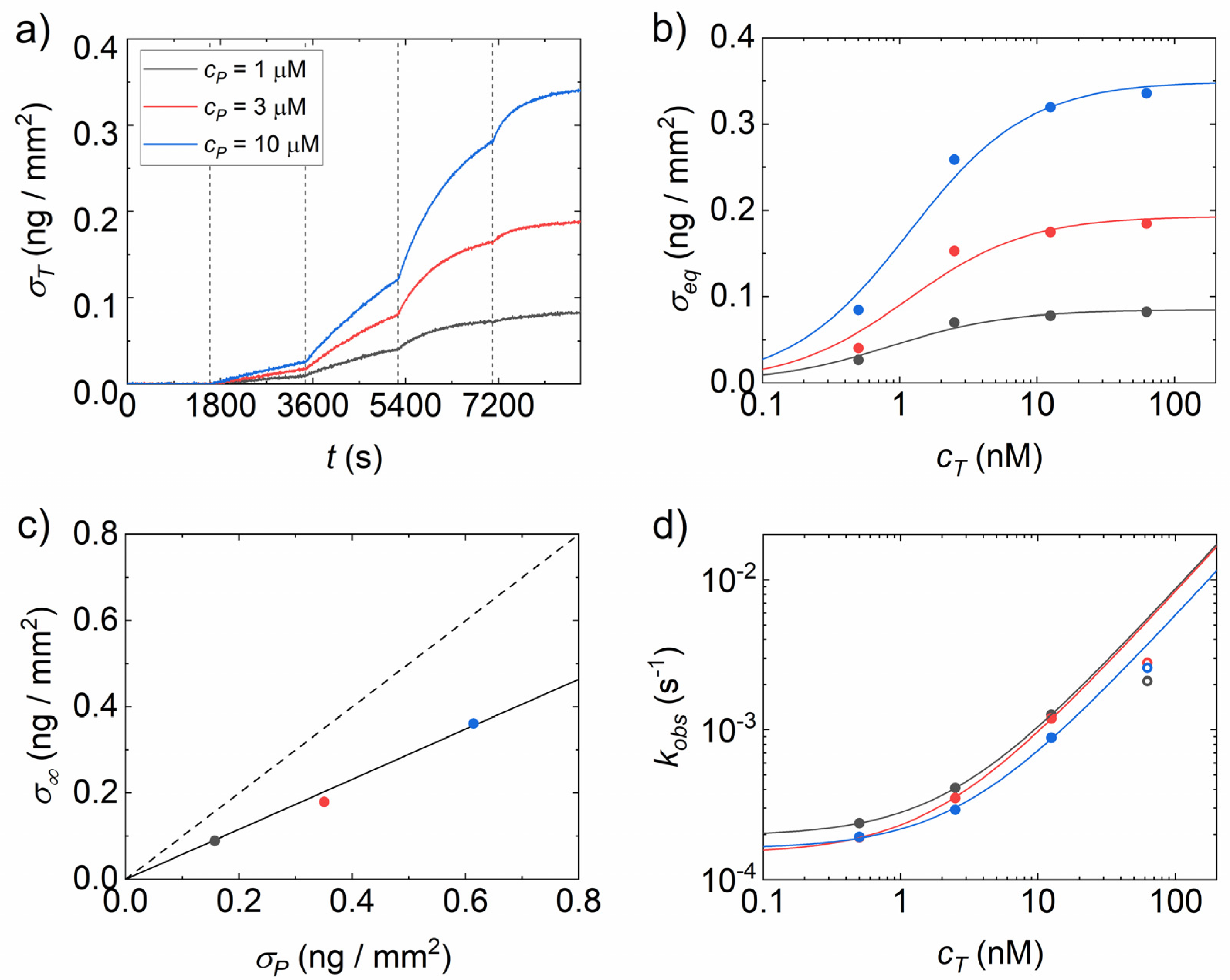
3.1. Analysis of Binding Curves
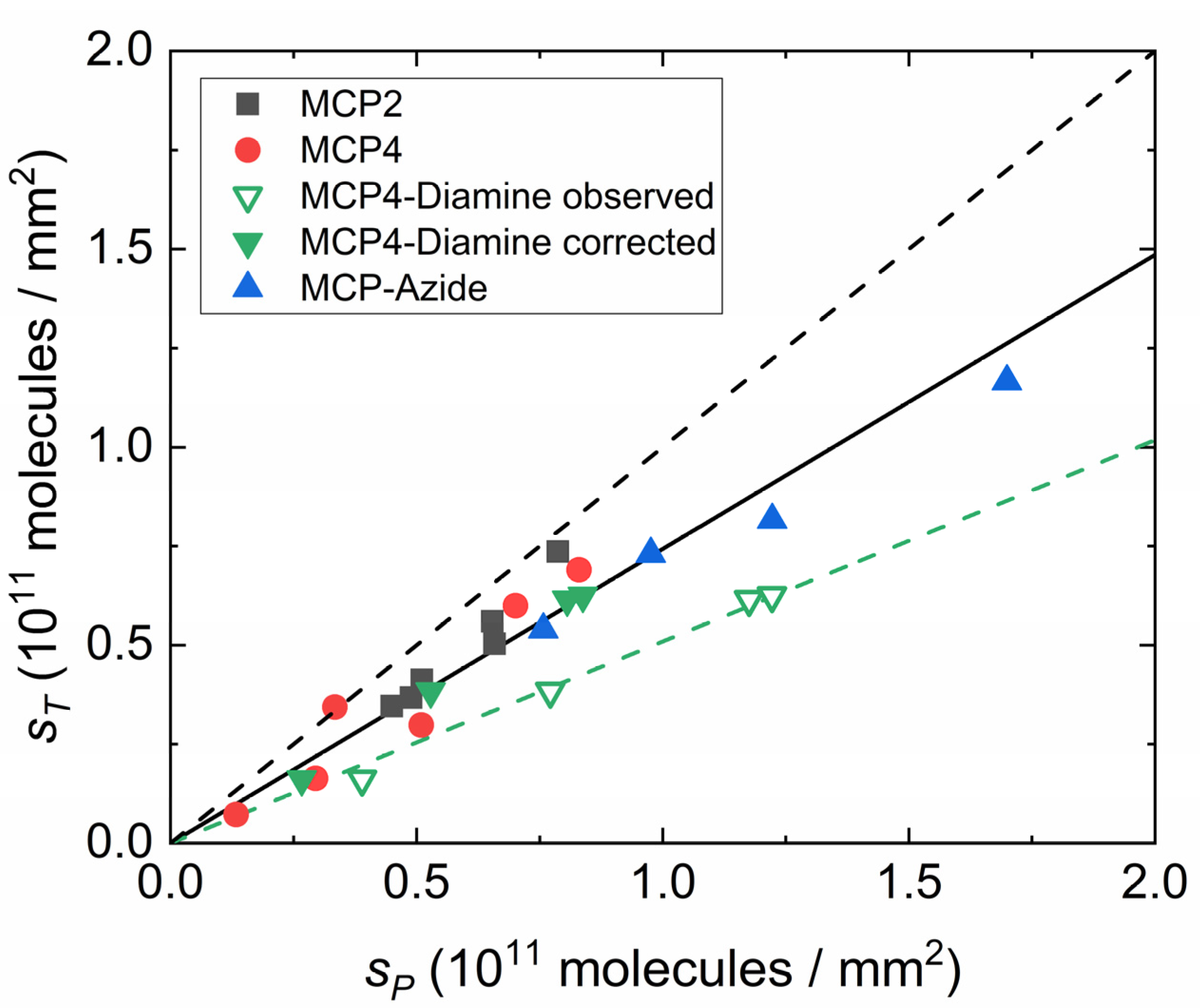
3.2. Effect of Copolymer Coatings on Hybridization Yield
3.3. Dependence of Association Rate on Probe Density
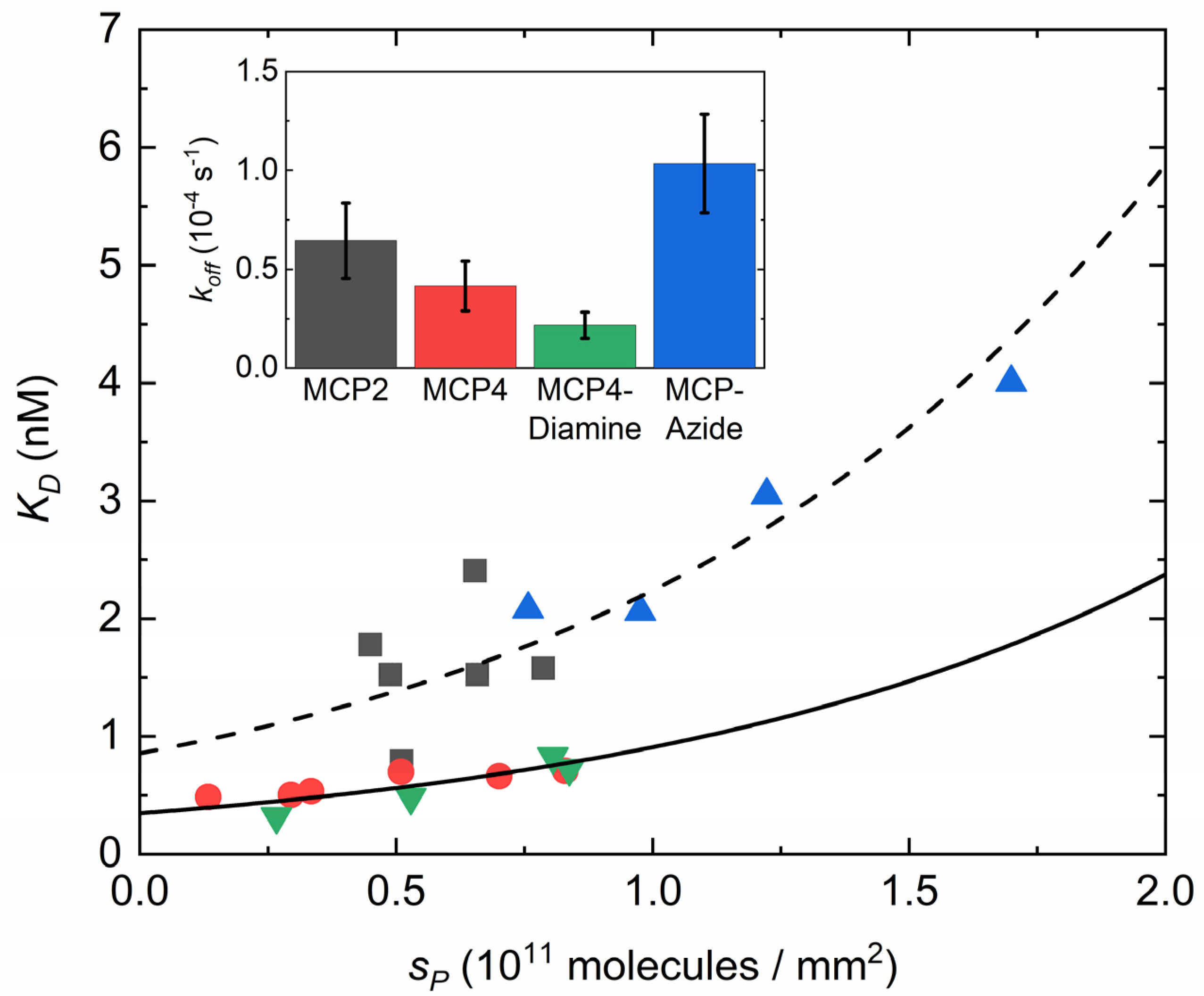
3.4. Effect of Copolymer Coatings on the Equilibrium Constant for Dissociation
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Relogio, A. Optimization of oligonucleotide-based DNA microarrays. Nucleic Acids Res. 2002, 30, e51. [Google Scholar] [CrossRef]
- Sassolas, A.; Leca-Bouvier, B.D.; Blum, L.J. DNA Biosensors and Microarrays. Chem. Rev. 2008, 108, 109–139. [Google Scholar] [CrossRef] [PubMed]
- Draghici, S.; Khatri, P.; Eklund, A.C.; Szallasi, Z. Reliability and reproducibility issues in DNA microarray measurements. Trends Genet. 2006, 22, 101–109. [Google Scholar] [CrossRef]
- Teles, F.; Fonseca, L. Trends in DNA biosensors. Talanta 2008, 77, 606–623. [Google Scholar] [CrossRef]
- Dell’Atti, D.; Tombelli, S.; Minunni, M.; Mascini, M. Detection of clinically relevant point mutations by a novel piezoelectric biosensor. Biosens. Bioelectron. 2006, 21, 1876–1879. [Google Scholar] [CrossRef] [PubMed]
- Hai, X.; Li, Y.; Zhu, C.; Song, W.; Cao, J.; Bi, S. DNA-based label-free electrochemical biosensors: From principles to applications. TrAC Trends Anal. Chem. 2020, 133, 116098. [Google Scholar] [CrossRef]
- Zanchetta, G.; Lanfranco, R.; Giavazzi, F.; Bellini, T.; Buscaglia, M. Emerging applications of label-free optical biosensors. Nanophotonics 2017, 6, 627–645. [Google Scholar] [CrossRef]
- Oliviero, G.; Federici, S.; Colombi, P.; Bergese, P. On the difference of equilibrium constants of DNA hybridization in bulk solution and at the solid-solution interface. J. Mol. Recognit. 2011, 24, 182–187. [Google Scholar] [CrossRef]
- Qiao, W.; Chiang, H.-C.; Xie, H.; Levicky, R. Surface vs. solution hybridization: Effects of salt, temperature, and probe type. Chem. Commun. 2015, 51, 17245–17248. [Google Scholar] [CrossRef] [PubMed]
- Sonawane, M.D.; Nimse, S.B. Surface Modification Chemistries of Materials Used in Diagnostic Platforms with Biomolecules. J. Chem. 2016, 2016, 9241378. [Google Scholar] [CrossRef]
- Chiodi, E.; Marn, A.M.; Geib, M.T.; Ünlü, M.S. The Role of Surface Chemistry in the Efficacy of Protein and DNA Microarrays for Label-Free Detection: An Overview. Polymers 2021, 13, 1026. [Google Scholar] [CrossRef] [PubMed]
- Nimse, S.; Song, K.; Sonawane, M.; Sayyed, D.; Kim, T. Immobilization Techniques for Microarray: Challenges and Applications. Sensors 2014, 14, 22208–22229. [Google Scholar] [CrossRef]
- Vanjur, L.; Carzaniga, T.; Casiraghi, L.; Chiari, M.; Zanchetta, G.; Buscaglia, M. Non-Langmuir Kinetics of DNA Surface Hybridization. Biophys. J. 2020, 119, 989–1001. [Google Scholar] [CrossRef] [PubMed]
- Cretich, M.; Pirri, G.; Damin, F.; Solinas, I.; Chiari, M. A new polymeric coating for protein microarrays. Anal. Biochem. 2004, 332, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Sola, L.; Damin, F.; Chiari, M. Array of multifunctional polymers for localized immobilization of biomolecules on microarray substrates. Anal. Chim. Acta 2019, 1047, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Özkumur, E.; Ahn, S.; Yalçın, A.; Lopez, C.A.; Çevik, E.; Irani, R.J.; DeLisi, C.; Chiari, M.; Selim Ünlü, M. Label-free microarray imaging for direct detection of DNA hybridization and single-nucleotide mismatches. Biosens. Bioelectron. 2010, 25, 1789–1795. [Google Scholar] [CrossRef] [PubMed]
- Giavazzi, F.; Salina, M.; Ceccarello, E.; Ilacqua, A.; Damin, F.; Sola, L.; Chiari, M.; Chini, B.; Cerbino, R.; Bellini, T.; et al. A fast and simple label-free immunoassay based on a smartphone. Biosens. Bioelectron. 2014, 58, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Pirri, G.; Damin, F.; Chiari, M.; Bontempi, E.; Depero, L.E. Characterization of A Polymeric Adsorbed Coating for DNA Microarray Glass Slides. Anal. Chem. 2004, 76, 1352–1358. [Google Scholar] [CrossRef]
- Giavazzi, F.; Salina, M.; Cerbino, R.; Bassi, M.; Prosperi, D.; Ceccarello, E.; Damin, F.; Sola, L.; Rusnati, M.; Chiari, M.; et al. Multispot, label-free biodetection at a phantom plastic–water interface. Proc. Natl. Acad. Sci. USA 2013, 110, 9350–9355. [Google Scholar] [CrossRef]
- Sola, L.; Damin, F.; Gagni, P.; Consonni, R.; Chiari, M. Synthesis of Clickable Coating Polymers by Postpolymerization Modification: Applications in Microarray Technology. Langmuir 2016, 32, 10284–10295. [Google Scholar] [CrossRef]
- Gori, A.; Sola, L.; Gagni, P.; Bruni, G.; Liprino, M.; Peri, C.; Colombo, G.; Cretich, M.; Chiari, M. Screening Complex Biological Samples with Peptide Microarrays: The Favorable Impact of Probe Orientation via Chemoselective Immobilization Strategies on Clickable Polymeric Coatings. Bioconjug. Chem. 2016, 27, 2669–2677. [Google Scholar] [CrossRef]
- Salina, M.; Giavazzi, F.; Lanfranco, R.; Ceccarello, E.; Sola, L.; Chiari, M.; Chini, B.; Cerbino, R.; Bellini, T.; Buscaglia, M. Multi-spot, label-free immunoassay on reflectionless glass. Biosens. Bioelectron. 2015, 74, 539–545. [Google Scholar] [CrossRef]
- Zanchetta, G.; Carzaniga, T.; Vanjur, L.; Casiraghi, L.; Tagliabue, G.; Morasso, C.; Bellini, T.; Buscaglia, M. Design of a rapid, multiplex, one-pot miRNA assay optimized by label-free analysis. Biosens. Bioelectron. 2021, 172, 112751. [Google Scholar] [CrossRef] [PubMed]
- Frederix, F.; Bonroy, K.; Reekmans, G.; Laureyn, W.; Campitelli, A.; Abramov, M.A.; Dehaen, W.; Maes, G. Reduced nonspecific adsorption on covalently immobilized protein surfaces using poly(ethylene oxide) containing blocking agents. J. Biochem. Biophys. Methods 2004, 58, 67–74. [Google Scholar] [CrossRef]
- Cai, N.; Larese-Casanova, P. Application of positively-charged ethylenediamine-functionalized graphene for the sorption of anionic organic contaminants from water. J. Environ. Chem. Eng. 2016, 4, 2941–2951. [Google Scholar] [CrossRef]
- Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Farshbaf, A.; Mohtasham, N.; Zare, R.; Mohajertehran, F.; Rezaee, S.A. Potential therapeutic approaches of microRNAs for COVID-19: Challenges and opportunities. J. Oral Biol. Craniofacial Res. 2021, 11, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Vainrub, A.; Pettitt, B.M. Coulomb blockage of hybridization in two-dimensional DNA arrays. Phys. Rev. E 2002, 66, 041905. [Google Scholar] [CrossRef]
- Halperin, A.; Buhot, A.; Zhulina, E.B. Sensitivity, Specificity, and the Hybridization Isotherms of DNA Chips. Biophys. J. 2004, 86, 718–730. [Google Scholar] [CrossRef][Green Version]
- Dupuis, N.F.; Holmstrom, E.D.; Nesbitt, D.J. Single-Molecule Kinetics Reveal Cation-Promoted DNA Duplex Formation Through Ordering of Single-Stranded Helices. Biophys. J. 2013, 105, 756–766. [Google Scholar] [CrossRef]
- Nava, G.; Ceccarello, E.; Giavazzi, F.; Salina, M.; Damin, F.; Chiari, M.; Buscaglia, M.; Bellini, T.; Zanchetta, G. Label-free detection of DNA single-base mismatches using a simple reflectance-based optical technique. Phys. Chem. Chem. Phys. 2016, 18, 13395–13402. [Google Scholar] [CrossRef] [PubMed]
- Peterson, A.W. The effect of surface probe density on DNA hybridization. Nucleic Acids Res. 2001, 29, 5163–5168. [Google Scholar] [CrossRef] [PubMed]
- Gong, P.; Levicky, R. DNA surface hybridization regimes. Proc. Natl. Acad. Sci. USA 2008, 105, 5301–5306. [Google Scholar] [CrossRef] [PubMed]
- Chiodi, E.; Sola, L.; Brambilla, D.; Cretich, M.; Marn, A.M.; Ünlü, M.S.; Chiari, M. Simultaneous evaluation of multiple microarray surface chemistries through real-time interferometric imaging. Anal. Bioanal. Chem. 2020, 412, 3477–3487. [Google Scholar] [CrossRef]
- Jensen, K.K.; Ørum, H.; Nielsen, P.E.; Nordén, B. Kinetics for Hybridization of Peptide Nucleic Acids (PNA) with DNA and RNA Studied with the BIAcore Technique. Biochemistry 1997, 36, 5072–5077. [Google Scholar] [CrossRef]
- Nelson, B.P.; Grimsrud, T.E.; Liles, M.R.; Goodman, R.M.; Corn, R.M. Surface Plasmon Resonance Imaging Measurements of DNA and RNA Hybridization Adsorption onto DNA Microarrays. Anal. Chem. 2001, 73, 1–7. [Google Scholar] [CrossRef]
- Peterson, A.W.; Wolf, L.K.; Georgiadis, R.M. Hybridization of Mismatched or Partially Matched DNA at Surfaces. J. Am. Chem. Soc. 2002, 124, 14601–14607. [Google Scholar] [CrossRef]
- Gao, Y. Secondary structure effects on DNA hybridization kinetics: A solution versus surface comparison. Nucleic Acids Res. 2006, 34, 3370–3377. [Google Scholar] [CrossRef]
- Irving, D.; Gong, P.; Levicky, R. DNA Surface Hybridization: Comparison of Theory and Experiment. J. Phys. Chem. B 2010, 114, 7631–7640. [Google Scholar] [CrossRef]

| Reference | Measuring Method | DNA Length (# Bases) | Surface Coating | DNA Probe Surface Density (1010 Molecules mm−2) | Kd (nM) | kon (104 M−1 s−1) | koff (10−4·s−1) |
|---|---|---|---|---|---|---|---|
| Jensen et al., 1997 [35] | Surface plasmon resonance | 15 | Dextran and streptavidin | 25 | 1.2 | 2.9 | |
| Nelson et al., 2001 [36] | Surface plasmon resonance | 18 | 11-mercaptoundecyl amine | 1 | 55 | ||
| Peterson et al., 2002 [37] | Surface plasmon resonance | 25 | - | 1.5–3 | 16 | ||
| Gao et al., 2006 [38] | Surface plasmon resonance | 25 | - | 4.5–6.8 | 5.7 | ||
| Irving et al., 2010 [39] | cyclic voltammetry | 18 | - | 5 | 770 | ||
| Ozkumur et al., 2010 [16] | Spectral reflectance imaging biosensor | 20 | MCP4 | 4.1 | |||
| Qiao et al., 2015 [9] | Total internal reflection fluorescence | 25 | Aldehyde | 2.9 | 10−7–2 a | ||
| Nava et al., 2016 [31] | RPI (perfluoropolymer substrate) | 12 | MCP4 | 2–9 | 1–1.8 | 2–10 | 6–10 |
| Sola et al., 2019 [15] | Interferometric reflectance imaging sensor | 23 | MCP4 | 7.8 | |||
| MCP-Azide | 11.6 | ||||||
| Vanjur et al., 2020 [13] | RPI | 12 | MCP4–MCP2 | 3–9 | 1–11 | 6–40 | 1.4–9 |
| This work | RPI | 23 | MCP4 | 1.3–8.3 | 0.5–0.7 | 5.9–8.5 | 0.4 |
| MCP4-Diamine | 2.6–8.3 | 0.3–0.8 | 2.6–6.9 | 0.2 | |||
| MCP2 | 4.5–7.9 | 0.8–2.4 | 3.4–7.6 | 0.6 | |||
| MCP-Azide | 7.5–17 | 2–4 | 2.6–5 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vanjur, L.; Carzaniga, T.; Casiraghi, L.; Zanchetta, G.; Damin, F.; Sola, L.; Chiari, M.; Buscaglia, M. Copolymer Coatings for DNA Biosensors: Effect of Charges and Immobilization Chemistries on Yield, Strength and Kinetics of Hybridization. Polymers 2021, 13, 3897. https://doi.org/10.3390/polym13223897
Vanjur L, Carzaniga T, Casiraghi L, Zanchetta G, Damin F, Sola L, Chiari M, Buscaglia M. Copolymer Coatings for DNA Biosensors: Effect of Charges and Immobilization Chemistries on Yield, Strength and Kinetics of Hybridization. Polymers. 2021; 13(22):3897. https://doi.org/10.3390/polym13223897
Chicago/Turabian StyleVanjur, Luka, Thomas Carzaniga, Luca Casiraghi, Giuliano Zanchetta, Francesco Damin, Laura Sola, Marcella Chiari, and Marco Buscaglia. 2021. "Copolymer Coatings for DNA Biosensors: Effect of Charges and Immobilization Chemistries on Yield, Strength and Kinetics of Hybridization" Polymers 13, no. 22: 3897. https://doi.org/10.3390/polym13223897
APA StyleVanjur, L., Carzaniga, T., Casiraghi, L., Zanchetta, G., Damin, F., Sola, L., Chiari, M., & Buscaglia, M. (2021). Copolymer Coatings for DNA Biosensors: Effect of Charges and Immobilization Chemistries on Yield, Strength and Kinetics of Hybridization. Polymers, 13(22), 3897. https://doi.org/10.3390/polym13223897