Abstract
Recent years have witnessed a dramatic increase in the use of theoretical and computational approaches in the study and development of molecular imprinting systems. These tools are being used to either improve understanding of the mechanisms underlying the function of molecular imprinting systems or for the design of new systems. Here, we present an overview of the literature describing the application of theoretical and computational techniques to the different stages of the molecular imprinting process (pre-polymerization mixture, polymerization process and ligand–molecularly imprinted polymer rebinding), along with an analysis of trends within and the current status of this aspect of the molecular imprinting field.
1. Introduction
Molecular imprinting has been defined as: “The construction of ligand selective recognition sites in synthetic polymers where a template (atom, ion, molecule, complex, or a molecular, ionic or macromolecular assembly, including micro-organisms) is employed in order to facilitate recognition site formation during the covalent assembly of the bulk phase by a polymerization or polycondensation process, with subsequent removal of some or all of the template being necessary for recognition to occur in the spaces vacated by the templating species” [1].
The most central feature of the molecular imprinting concept [1,2,3,4,5] is the interaction between template and monomers in the pre-polymerization mixture (Figure 1b) and their effect on the structure and recognition properties of the resulting molecularly imprinted polymer (MIP), as shown in Figure 1.
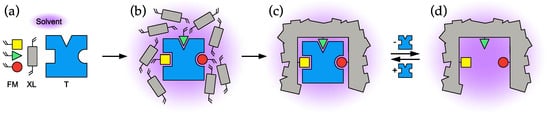
Figure 1.
Schematic description of the different stages in the molecular imprinting process. (a) The main polymer components: template (T), functional monomers (FM) and cross-linking monomer (XL). (b) Pre-polymerization mixture, (c) after polymerization, (d) after template removal.
Consequently, the molecular and physical characteristics of recognition sites in MIPs result directly from the various interactions possible in the pre-polymerization mixture, e.g., template–monomer, monomer–monomer, solvent–template/monomer, etc. An appreciation of the physical rules governing the formation of these complexes is therefore crucial for understanding the complexity of the imprinting process. If we are to achieve true rational design of molecularly imprinted systems for producing materials with predetermined recognition properties, suitable tools that can provide insight into the molecular recognition processes are needed.
Although classical thermodynamic models [6] in theory can describe the molecular events governing the synthesis and polymer–ligand recognition properties of imprinted materials, modern computational methods can be used to model the pre-polymerization mixture in much greater detail and even to characterize polymer–ligand interactions [7,8,9]. In this review, we first provide a brief background to the thermodynamic factors and theories that have been presented as a basis for explaining the recognition properties of MIPs, before reviewing the literature describing the use of computational methods for the study of the various stages of the molecular imprinting processes.
1.1. A Thermodynamic Treatment of the Molecular Imprinting Process
The physical factors underlying molecular interaction have attracted the interest of researchers for several decades. Jenck’s paradigms [10,11], the factorization of energetic contributions to molecular recognition and the intrinsic binding energy concept, are of particular note, and were employed by a number of groups. Semi-quantitative approaches, so-called back of the envelope calculations [12], were independently formulated by Andrews [12] and Williams [13,14,15], aspiring to define the physical basis for binding events. Nonetheless, as reflected in several studies [6,16,17], the thermodynamic factors controlling molecular interactions in imprinted systems are best described by Williams’ more comprehensive treatment [13,18] (Equation (1)):
where the Gibbs free energy change for complex formation (ΔGbind) is the combined energy changes associated with the loss of translational and rotational freedom (ΔGt+r), restriction of rotors upon complexation (ΔGr), hydrophobic interactions (ΔGh), residual soft vibrational modes (ΔGvib), the sum of interacting polar group contributions (∑ΔGp), adverse conformational changes (ΔGconf) and unfavorable van der Waals interactions (ΔGvdW).
ΔGbind = ΔGt+r + ΔGr + ΔGh + ΔGvib + ∑ΔGp + ΔGconf + ΔGvdW
The recognition properties of MIPs result from pre-polymerization complexation between template and functional monomer, an equilibrium process governed by the free energy of binding, ΔGbind. The position of this equilibrium dictates the number and heterogeneity of the resulting binding sites. Stronger and more regular template-functional monomer complexes are thus expected to lead to a larger number of sites with higher fidelity. The degree of template complexation by a functional monomer and the degree of heterogeneity are determined by the chemical nature of the pre-polymerization mixture and the polymerization conditions (temperature and pressure).
An investigation of changes in NMR chemical shifts and line broadening with increasing functional monomer concentration offered the first direct verification of the formation of non-covalent template-functional monomer complexes [19]. This study also indicated possible template self-association and formation of higher-order complexes, a hypothesis that was more recently supported by computational studies based on molecular dynamics (MD) [20]. Spectroscopic methods have since been used in many studies aiming to shed light on the multitude of pre-polymerization equilibria involving template-functional monomer complexation [21,22,23,24,25,26,27], self-association [27] and interactions with cross-linking monomers [28]. In such studies, it is often seen that using higher ratios of functional monomer to template, in order to increase complex formation, leads to MIPs with a higher degree of non-specific binding.
Complex formation between a template and a functional monomer carries with it an entropic penalty, ΔGt+r, associated with the loss of translational and rotational freedom. Higher-order complexes, expected to produce higher-fidelity binding sites, thus have a larger energy barrier. It follows that using a functional monomer capable of multiple simultaneous interactions should produce increased concentrations of complexed template, compared to an increased concentration of a single-point monomer. Although multi-dentate monomers are not as easily available and often require synthesis, examples of their use in MIPs have been reported [29,30,31,32].
Similar to ΔGt+r, the ΔGr term is the penalty for restricted internal bond rotation upon complexation. Thus, interactions with rigid templates are entropically favored and the resultant MIPs tend to exhibit higher selectivity than those prepared with less rigid structures. In addition, a rigid structure can adopt fewer solution conformations, which leads to a narrower site distribution. Consequently, high MIP–ligand affinities have been observed for rigid templates, such as the alkaloids morphine [33] and yohimbine [34].
MIPs have traditionally been prepared in non-polar organic media, thus relying on polar interactions to drive the equilibrium towards complexation, as reflected in the ∑ΔGp term. Examples have been reported where selectivity was enhanced by using more strongly interacting monomers [35,36,37,38], or by using crown ethers to solubilize zwitterionic template–monomer complexes at low polarity [39]. In addition, several studies have reported the use of metal ions to provide multiple coordination points between template and monomers, enabling the use of more polar solvents such as methanol or DMSO [40,41,42,43,44].
When the analyte of interest is water-soluble or otherwise incompatible with the commonly used non-polar organic solvents, other methodologies are required. For many important classes of analytes, e.g., peptides, proteins, oligonucleotides and sugars, the use of water as the porogen (solvent of polymerization) places a strong influence on the ΔGh term of Equation (1). This enables functional monomers with hydrophobic moieties to facilitate template complexation through the hydrophobic effect. Some interesting examples have been reported where polymerizable cyclodextrins were used as monomers [45,46,47,48,49]. In addition, metal ion chelation can be used as an alternative or complement.
In summary, for non-covalently imprinted polymers, the selectivity and affinity of the MIP is controlled by the various equilibria present in the pre-polymerization mixture. The positions of these equilibria are, in turn, governed by ΔGbind, as defined by the different thermodynamic terms in Equation (1). The magnitudes of the individual terms are determined by the chemical nature of all components in the mixture as well as the physical conditions during polymerization. Thus, the conceptually simple process of molecular imprinting is based on a complex series of highly interdependent equilibria, inevitably leading to polymers with heterogenous distributions of recognition sites. The same thermodynamic principles, of course, also apply to interactions between the polymer and its analyte/target in the intended application, further complicating the link from pre-polymerization conditions, over polymerization and work-up, to final use. Accordingly, there is a need for methodologies to study and understand the complexity of pre-polymerization events, to correlate these events with MIP performance and to optimize all stages of MIP synthesis.
1.2. Theoretical and Computational Strategies for MIP Development
Driven by the rapid development of molecular imprinting and its applications, several tools for in silico studies of the above-mentioned stages of MIP design and synthesis have been adopted. These tools can offer atomistic insights on aspects ranging from events in the pre-polymerization mixture to polymer–ligand interactions and even polymer morphology. Since the first applications of computational strategies to study aspects of molecular imprinting in the beginning of the 21st century, the field has grown steadily, and particularly over the last decade (Figure 2). This development has been supported through the necessary iterative interplay between these studies and experimental validation.
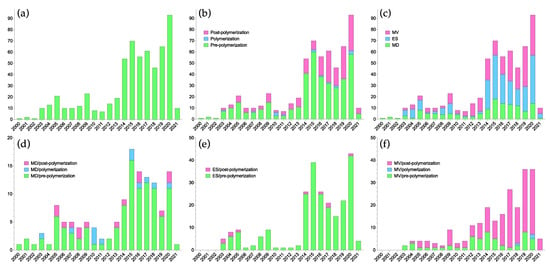
Figure 2.
Number of papers published where computational methods have been applied to some aspect of molecular imprinting. (a) Total number of papers, (b) number of papers according to stage, (c) number of papers according to method: multivariate (MV), electronic structure (ES) or MD, (d) MD-based papers according to stage, (e) ES-based papers according to stage and (f) MV-based papers according to stage. Note that a number of papers fall into more than one category.
In the past, knowledge of the molecular events underlying MIP behavior has been extracted from empirical studies of polymer–ligand interactions. Thermodynamic models [6,16,17,50,51], as discussed above, were applied in attempts to explain and understand both pre-polymerization events as well as polymer recognition characteristics. More recently, probability-based stochastic simulations of pre-polymerization monomer–template equilibria [52] contributed to this area, an example of which was the use of a stochastic algorithm [53] to simulate pre-polymerization solution heterogeneity, placing monomer-template units in a lattice matrix. Importantly, the simulated affinity distributions closely matched those measured experimentally in MIPs. Additionally, mathematical models describing pre-polymerization template–monomer complexation and subsequent template rebinding have been developed [54].
The major reason behind the recent increase in the use of computational strategies in MIP technology is likely related to increased affordability of computational power and access to appropriate software [55,56,57,58]. This has enabled the application of multivariate analyses, electronic structure calculations and full-system all-atom MD simulations to all aspects of MIP design, synthesis and evaluation. After a brief introduction of the different computational tools, focusing on methods for electronic structure calculations, MD simulations and statistics-based multivariate analyses, we review the current status of their application to the different stages of molecular imprinting.
1.2.1. Electronic Structure Calculations
The use of computational methods based on electronic structure calculations, e.g., ab initio, semi-empirical and density functional theory (DFT) strategies, for the design and evaluation of MIPs is increasing. This class of computational methods, collectively termed quantum chemistry, aims to solve the electronic Schrödinger equation based on the atomic coordinates and number of electrons of the system studied. This is impossible for systems with more than a few electrons, and therefore approximations are necessary. Ab initio methods approximate the electronic wavefunction, whereas semi-empirical and DFT methods instead approximate the Hamiltonian operator. The accuracy, and computational demand, increases from semi-empirical methods (considering only valence electrons and with some parameters derived from experiment) to DFT (calculates electron densities), and finally ab initio methods. Different strategies, basis sets and parameters are chosen to provide an acceptable approximation of the system studied within a reasonable timeframe. Typically, comparing quantum chemical calculations for isolated molecules and molecular complexes can provide information regarding interaction strength and type, and consequently, these methods are very often used for evaluation of different template–monomer combinations.
1.2.2. Molecular Dynamics
Although the development of modern MD methodology was closely intertwined with that of Monte Carlo simulations [59,60,61,62,63], it is generally considered as introduced by Alder and Wainright in a seminal study involving hard sphere simulations of gaseous argon in 1957 [64]. Other key developments include the transition to MD simulations of liquid argon by Rahman in 1964 [65], as well as liquid water simulations in 1971 by Rahman and Stillinger [66]. In MD simulations, the forces acting on and between interacting atoms and molecules are described by a set of equations and parameters, referred to as a force field [67]. Solving Newtons equations of motion allows for simulation of the motions, or dynamics, of the system. MD simulations have been applied to an increasing number of research areas, driving the development of both software and force fields. Examples include studies of surfaces [68,69], solvents [70], biomolecular interactions [71,72,73], DNA conformation [74,75], protein folding [76], phospholipid bilayers [77,78] and membrane transport of drugs [79]. Some of the more popular and commonly used force fields are AMBER [80,81,82,83], GAFF [84], CHARMM [85], OPLS [86] and GROMOS [87,88].
In comparison to electronic structure methods, MD simulations require less computational resources when treating systems of comparable size. This enables studies of much larger multimolecular systems, including MIP pre-polymerization mixtures containing thousands of molecules, with reasonable demands on both hardware resources and time. However, since electrons are not explicitly considered, MD simulations are unable to account for processes involving the movement of electrons, such as bond breaking or formation. Nevertheless, important information can be attained regarding the multitude of non-covalent interactions taking place in pre-polymerization mixtures as well as in MIP binding site models (Figure 3).
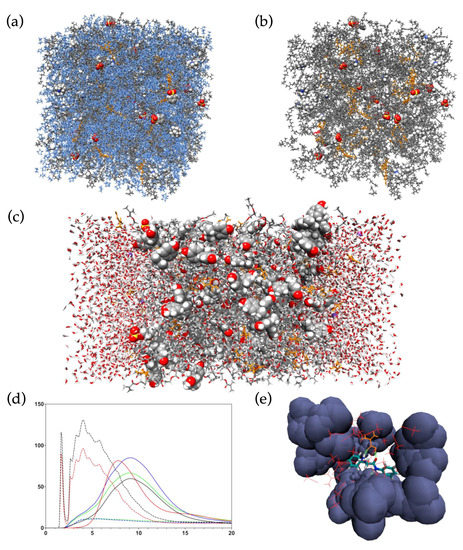
Figure 3.
Examples of MD simulation systems and MD data-derived analysis methods. (a) Full-system all-atom MD simulation of urea-based MIP anion receptors [89]. (b) The same system though excluding solvent (tetrahydrofuran) from visualization. (c) Two-phase water bisphenol A MIP emulsion polymerization simulation [90], where polymer components are flanked by aqueous phase with dissolved counter ions. (d) Radial distribution function analyses for component interactions [89]. (e) Grid density analysis plot, where local densities of interacting species are visualized [20].
1.2.3. Multivariate Analysis
Molecular imprinting and its applications, with nearly infinite combinations of pre-polymerization components, polymerization conditions, polymer workup, evaluation parameters and analytic responses, lends itself well to multivariate analysis [91,92,93]. This entails a different type of modeling than discussed above regarding molecular energies and interactions. Instead, the goal here is to produce mathematical models able to simultaneously correlate multiple experimental variables with one or more properties of a MIP and/or its application. The resultant models can then be used to optimize, e.g., the polymer recipe or analytical parameters, or to find patterns and correlations hidden in large datasets.
Application of multivariate methods usually begins with determining which parameters to study and then choosing an experimental design that allows for simultaneous evaluation of these parameters. Often, a pilot or training set of experiments is performed to determine which of the variables have the largest effect on the outcome, followed by a more focused, second experimental design in order to produce models for prediction or optimization. Common experimental designs include full or fractional factorial, Box-Behnken, Placket-Burman, Doehlert and central composite designs (Figure 4).
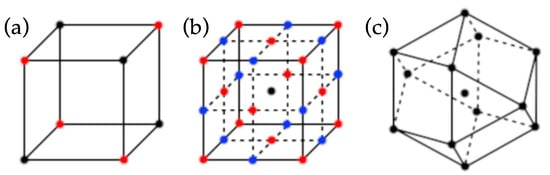
Figure 4.
Schematic illustrations of examples of experimental design concepts/methods used in the statistical evaluation of MIPs. Examples of controllable variables include, amounts of functional monomer, cross-linking monomer and porogen. (a) Black points represent experimental runs in a three-factor fractional factorial design, while the combined red and black points represent a three-factor full factorial design. (b) Red and central black points depict a three-factor central composite design, while blue and central black points represent a Box-Behnken design. (c) A three-factor Doehlert design.
The experimental data can be calibrated, or fitted, to mathematical models using a number of methods. In MIP studies, the most common are principal component analysis (PCA), partial least squares regression (PLSR), multiple linear regression (MLR) and artificial neural networks (ANN).
2. The Pre-Polymerization Stage
As seen in Figure 2b, the majority of papers employing computational treatments have focused on the pre-polymerization stage, predominantly using electronic structure methods or MD simulations.
2.1. Electronic Structure Calculations
Demands on hardware resources and simulation time both increase rapidly with the size of the system under investigation. Accordingly, the most common use of electronic structure-determining methods in MIP studies for characterization of template–monomer complexes, as discussed above, is to find the most suitable functional monomer and often also the optimal stoichiometry. Of the electronic structure-determining methods, semi-empirical strategies are less demanding on computational resources, and the two most commonly used semi-empirical methods for MIP development are AM1 [94,95,96,97,98,99,100,101,102,103,104,105,106] and PM3 [107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127], while other examples are less common [128,129,130].
The more computationally demanding ab initio and DFT methods provide higher accuracy. Investigations involving template–monomer complex studies have employed different methods, basis sets and levels of theories on several occasions. The majority of these studies employed DFT methods [124,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203,204,205,206,207,208,209,210,211,212,213,214,215,216,217,218,219,220,221,222,223,224,225,226,227,228,229,230,231,232,233,234,235,236,237,238,239,240,241,242,243,244,245,246,247,248,249,250,251,252,253,254,255,256,257,258,259,260,261,262,263,264,265,266,267,268,269,270,271,272,273,274,275,276,277,278,279,280,281,282,283,284,285,286,287,288,289,290,291,292,293,294,295,296,297], whereas ab initio-based calculations have been used in fewer instances [298,299,300,301,302,303,304,305,306,307,308,309,310,311,312,313,314,315,316,317,318,319,320,321,322,323]. The majority of these studies focus on the interaction between a single functional monomer and a single template. In one study, semi-empirical, DFT and ab initio-based calculations were compared for characterization of monomer–template interactions [324].
The growing accessibility of computational power has been accompanied by an increasing frequency of reports in the literature including quantum chemical calculations in the design and characterization of MIPs. Due to relatively high resource requirements associated with these calculations, most studies have focused primarily on subsets of pre-polymerization mixtures, restricted sets of interactions or isolated non-solvated molecular complexes in vacuo. With seemingly ever-increasing availability of computational power and emergence of novel mixed approaches combining electronic structure and MD simulations, increased use of electronic structure methods in the design and study of molecular imprinting systems is expected.
2.2. Molecular Dynamics
The nature of MD simulations makes them highly suited for studies of liquid systems, and with the assumption that MIP recognition properties originate from pre-polymerization interactions [4,51,325,326,327], MD simulations have primarily found applications in studies of this stage of MIP production. From the resulting data, or molecular trajectories, information regarding the types and strengths of all pre-polymerization interactions can be extracted and correlated with MIP recognition performance. Since the computational cost is significantly lower for force field methods than for quantum chemical calculations, MD simulations can be applied to very large systems with solvent molecules explicitly included.
In a method introduced by Piletsky et al., 20 functional monomers were initially assessed for their interaction energy with the template ephedrine in both charged and neutral states [328]. Selected monomers were then used for polymer synthesis, but also subjected to further MD simulations together with template, cross-linker and solvent, where the observed interactions could be correlated with experimental binding data. This approach has since been adapted several times in the literature [329,330,331,332,333,334,335,336,337,338,339,340,341,342,343,344,345,346,347,348,349,350,351,352,353]. In a number of reports, similar approaches have been employed to evaluate and/or characterize monomer–template interactions using MD and docking simulations as well as variations and/or combinations thereof [102,354,355,356,357,358,359,360,361,362,363,364,365,366,367,368,369,370,371,372,373,374,375,376,377,378,379,380,381,382,383,384,385,386,387,388,389,390,391].
Despite the dramatic development of computer hardware and software, multimolecular simulations involving multiple copies of monomers, template and explicit solvent are still not feasible for electronic structure methods alone. However, several examples report the combined use of quantum chemical calculations and MD simulations to study different aspects of pre-polymerization mixtures [128,143,215,216,392,393,394].
The growing number of MD studies of systems containing all MIP components and with experimental stoichiometries have highlighted the importance of these more comprehensive treatments of pre-polymerization mixtures for delineating underlying mechanisms [20,28,89,90,201,395,396,397,398,399,400,401,402,403,404,405,406,407,408,409,410,411,412,413].
MD-based investigations of other aspects of the pre-polymerization stage include studies of the structural stability of protein epitopes for template screening [414,415], mapping potential monomer interaction sites of a protein target, followed by docking of acrylamide-derived monomers and post-docking interaction energy calculations [416], studies of template interactions with Dengue virus as a support matrix to create larger binding sites [417], a series of reports attempting to correlate structural and physical properties of dummy templates and ligands with rebinding properties [418,419,420,421,422,423,424,425,426] and coarse-grained simulations studying the effect of composition on material properties and template interaction [427]. Additionally, large-scale MD simulations were performed in an attempt to mimic chromatography in a virtual capillary [428].
2.3. Multivariate Analysis
Traditionally, analysis and optimization of MIPs have been univariate in nature. This involves evaluation and optimization of one parameter with the results carried forward for optimization of the next, and so forth. This may not always be ideal as identified optima may turn out to be local or false [91]. The inherent flexibility of MIP synthesis and the interdependence of the variables makes this stage a good candidate for multivariate optimization. Consequently, a number of studies have been published applying different multivariate methods and experimental designs in order to optimize polymer composition and/or synthesis methods [210,211,212,213,305,429,430,431,432,433,434,435,436,437,438,439,440,441,442,443,444,445,446,447,448,449,450,451,452,453,454,455,456,457,458,459,460,461,462,463,464,465,466,467,468,469,470,471,472,473,474,475,476,477,478].
3. The Polymerization Stage
The polymerization reaction is the least studied aspect of molecular imprinting in general as well as in the context of computational treatment (Figure 2b). The imprinting literature is abundant with experimental correlations between pre-polymerization mixture composition and MIP recognition properties, providing support for the underlying assumption that template-functional monomer complexes are preserved in the polymer matrix. However, little direct evidence exists regarding the fate of these complexes once polymerization has been initiated, though NMR studies indicate that they are maintained during polymerization [23]. Nevertheless, there are examples of studies of the polymerization stage by means of computational methods, almost exclusively using molecular dynamics (Figure 2d). The development of reactive force fields [479,480] and other solutions enabling bond formation and breaking in force field-based simulations should further help in filling this knowledge gap.
3.1. Electronic Structure Calculations
Although this class of computational methods can accurately describe the movement of electrons and the breaking and formation of chemical bonds, the number of molecules required for a meaningful representation of the polymerization process of a MIP would lead to unreasonable computational demands. Hopefully, the technical development will eventually allow such calculations, yet to the best of our knowledge, no examples have been published.
3.2. Molecular Dynamics
A few attempts have been made to apply MD to the study of MIP polymerization. Yungerman and Srebnik used a coarse-grained Monte Carlo procedure to study the formation of binding site imperfections in MIPs [481]. Monomers were modeled as Lennard-Jones spheres and templates as rigid dumbbells made of two monomers. Consequently, the simulations only considered imprinting according to size and shape. Other similar studies have also been reported, combining Monte Carlo simulation of hard spheres with statistical mechanics [482,483,484,485,486,487] or mean field theory [488,489,490,491,492]. However, in order to replicate MIP recognition on a molecular level, it is necessary to perform atomistic simulations. Thus, Henthorn and Peppas reported all-atom MD-based simulations of the formation of glucose-imprinted polymers [493,494], where 160 template molecules, 160 functional monomers (2-hydroxyethyl methacrylate), 300 cross-linkers (ethylene glycol dimethacrylate, EGDMA), 800 water molecules and 20 initiator molecules were allowed to diffuse and relax using MD simulation, followed by a reaction step including initiation, propagation and termination. This was repeated until all radicals had been quenched. Ligand binding to the resultant polymer models was then compared with experimental data. Srebnik and co-workers combined a similar reaction scheme with lattice Monte Carlo simulations in a series of studies of protein-imprinted polymers [427,495,496,497,498,499]. Schauperl and Lewis attempted to simulate the polymerization reaction for xanthine MIPs [500]. Starting with one or more template molecules, monomers and cross-linkers were sequentially added to the system and allowed to form new bonds with the growing polymer chain. MD and energy minimization allowed for optimal host–guest interaction. The simulations were continued until a threshold density had been reached. The resultant polymer model was used to explain binding site heterogeneity. Efforts to simulate electropolymerized MIPs selective for 6-thioguanine were reported by Hyunh et al. [501]. A system with one template molecule, two functional monomers and six cross-linking monomers was subjected to MD simulation. Pre-determined “radical positions” in the monomers were allowed to form bonds if within a 3 Å distance until no additional bonds were formed. The equilibrated system was then replicated eight times, whereafter the MD simulation continued until saturation. No analysis of the resultant model was reported other than that its density was very similar to that of the polymers prepared by electropolymerization. Cowen and co-workers developed a similar algorithm for simulating the polymerization reaction during MD simulations of the pre-polymerization mixture [502,503]. Briefly, after equilibration of the system, new bonds were formed between “reactive” atoms within a suitable distance followed by another round of energy minimization. The process was repeated until no more reactions were possible.
3.3. Multivariate Analysis
The application of multivariate strategies in order to optimize polymerization conditions has been rare so far. Examples include studies of optimum polymerization temperature when comparing polymerization in bulk and surface molecular imprinting to study the role of insulin-imprinted magnetic nanoparticles [469], investigation of the influence of polymerization temperature and time on the diameter of 5-fluorouracil-imprinted MIP nanoparticles prepared via precipitation polymerization [504] and optimization of the number of cycles and scan rate in electropolymerization of ketorolac tromethamine MIPs on paper graphite electrodes [505]. It should be noted that in many of the reports discussed in Section 2.3 attempting to optimize polymer synthesis, polymerization parameters were initially included in the experimental designs. However, when it was found that variation of these parameters had no significant influence on the outcome, they were omitted from further optimization.
4. MIP Structure and Function
The bulk of the computational studies of MIP properties post-polymerization use multivariate analysis, though a handful of reports using quantum chemical calculations or MD simulations have also been presented (Figure 2d–f). This is not surprising considering the opportunities for optimization of experimental parameters at this stage, e.g., rebinding conditions.
4.1. Electronic Structure Calculations
Electronic structure methods have been used in a few instances to study aspects of MIP–template recognition. These include PM3 calculations of a binding site model for nicotinamide [110], AM1 calculations to explain recognition differences in different buffers [506], DFT studies to explain the selectivity of a phenylurea herbicide MIP [507], DFT studies to confirm the structure of a binding site in a catalytic silica MIP [508], DFT studies of the adsorption mechanism in a 5-fluorouracil MIP [294,509], DFT studies of poly-pyrrole MIP models interacting with glyphosate [510] or tryptophan [511], DFT and ab initio studies of binding site models in hydroxyzine and cetirizine MIPs [299] and ab initio studies of a binding site model for phenolic compounds [512].
4.2. Molecular Dynamics
Attempts at simulating aspects of the rebinding of a template or ligand to a MIP using force field-based methods have also been reported. In several studies, polymer models have been approximated by equilibrating templates with linear chains of functional monomers, followed by analysis of binding energies and other aspects of recognition [355,356,358,513,514,515] or docking [221,360]. Terracina et al. also used docking procedures to study selectivity in MIP models that had been optimized semi-empirically [516]. Herdes and Sarkisov created pyridine MIP models by first equilibrating systems containing pyridine, methacrylic acid, EGDMA and chloroform [517]. The template and solvent were removed, and the monomers’ positions were fixed. Monte Carlo simulations were then applied to investigate the adsorption of pyridine, benzene and toluene. Sobiech et al. constructed MIP binding site models through MD simulation of pre-formed template/functional monomer/cross-linking monomer clusters [386,518,519]. After equilibration, the template was removed, and the system was “polymerized” by replacing double bonds in the monomers with new single bonds. A similar strategy was used by Gajda et al. to mimic an aripiprazole binding site [231]. Finally, Curk et al. developed a computational approach to derive binding site models and for evaluating template rebinding. Their approach used a range of parameters, including number of template interaction points, concentrations of monomers and material properties in combination with grand canonical Monte Carlo simulations describing multiple interaction site templates [54].
4.3. Multivariate Analysis
The experimental conditions employed when evaluating or applying MIPs have a major influence on the performance of the polymer. Similar to the situation in the pre-polymerization mixture, the possible combinations of experimental parameters (e.g., analyte concentration, solvent, pH, temperature, flow rate, incubation time) are nearly endless, making this an area highly suited for multivariate optimization. Thus, different combinations of experimental designs and response surface modeling have been used for optimization of parameters when using MIPs in adsorption, separation or sensing applications [214,267,375,467,468,470,475,476,477,520,521,522,523,524,525,526,527,528,529,530,531,532,533,534,535,536,537,538,539,540,541,542,543,544,545,546,547,548,549,550,551,552,553,554,555,556,557,558,559,560,561,562,563,564,565,566,567,568,569,570,571,572,573,574,575,576,577,578,579,580,581,582,583,584,585,586,587,588,589,590,591,592,593,594,595,596,597,598,599,600,601,602,603,604,605,606,607,608,609,610,611,612,613,614,615,616,617,618,619,620,621,622,623,624,625,626,627,628,629,630,631,632,633,634]. In some studies, the optimized parameters have been improved further by using them as input for ANN models [475,621].
In other cases, multivariate methods have been used to reveal correlations hidden in the data obtained when evaluating MIPs. Different combinations of PCA and PLSR methods have been applied for interpretation of MIP binding data obtained from SERS (surface-enhanced Raman spectroscopy) [635,636,637,638,639,640,641,642,643,644], for pattern recognition in the responses from various MIP-sensor arrays [645,646,647,648,649,650,651,652,653,654,655,656,657,658,659,660], to correlate the shape of MIP-quartz crystal microbalance frequency curves to different analytes [106,661,662,663], for cyclic voltammetry measurements on binary mixtures [214,656,664,665] and for correlating the results with HPLC data [665] and to process fluorescence data [666]. PLSR and PCA have also been used to correlate bupivacaine–MIP binding with rebinding solvent properties [667,668] and with polymer morphology and pre-polymerization interactions from MD simulations [403]. Likewise, PCA has been used to examine the relation between specific analyte sorption and non-specific sorption of water in an iprodione-imprinted MIP for use in aqueous media [669].
A separate branch of multivariate analysis of chemical data is called chemometrics, in which large numbers of structure-derived properties, molecular descriptors, are generated for a set of molecules and then correlated with other properties of interest. Thus, a number of studies have been reported attempting to correlate molecular descriptors with MIP binding data using a range of multivariate tools. Rossetti et al. employed PLS models to correlate molecular descriptors with solid-phase extraction retention data for a series of biomarker pro-gastrin-releasing peptides in order to elucidate the recognition mechanism [670]. Liu et al. used MLR and PCA to couple structural and molecular parameters of a quercetin MIP to its adsorption selectivity [671]. Baggiani et al. employed PCA to correlate the chromatographic selectivity of a pentachlorophenol MIP for the template and 52 related phenols with 16 AM1-derived molecular descriptors [672]. The chromatography data from this study was later subjected to PLSR modeling using 25 descriptors, which improved the selectivity prediction capability of the model [673]. Nantasemat et al. also built ANN models for prediction of MIP selectivity using molecular descriptors for a set of templates, functional monomers and HPLC mobile phases compiled from the literature [674,675], and for bisphenol A MIPs [676]. Similar models, attempting to correlate analyte recognition with molecular descriptors, have also been reported for MIPs selective for penicillin G [677], erythromycin [678] and milk lactose [679].
5. Conclusions and Outlook
The significant growth in the number of literature reports describing computational studies of molecular imprinting systems has followed the development, availability and affordability of both hardware and software. In turn, this has enabled the use of these tools in both prognostic and diagnostic capacities in the development of molecularly imprinted materials. This development and the growing awareness of the value of the use of these tools is reinforced through validation using experimental studies. Accordingly, the combination of ready access to these computational tools and the value of the insights gained from their use should see further increases in the prevalence of their use in the molecular imprinting field.
Author Contributions
Conceptualization, I.A.N.; All authors contributed to literature collection, assessment and manuscript writing, review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by H2020-MSCA-ITN-2016, 722171-Biocapture and H2020-FETOPEN-2018-2020, 829040-MindGAP, Swedish Knowledge Foundation 2019-0114.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Wulff, G.; Sarhan, A. Über die anwendung von enzymanalog gebauten polymeren zur racemattrennung. Angew. Chem. 1972, 84, 364. [Google Scholar] [CrossRef]
- Arshady, R.; Mosbach, K. Synthesis of substrate-selective polymers by host-guest polymerization. Makromol. Chem. 1981, 182, 687–692. [Google Scholar] [CrossRef]
- Sellergren, B. (Ed.) Molecularly Imprinted Polymers: Man-Made Mimics of Antibodies and their Application in Analytical Chemistry; Elsevier: Amsterdam, The Netherlands, 2001; ISBN 978-0444828378. [Google Scholar]
- Alexander, C.; Andersson, H.S.; Andersson, L.I.; Ansell, R.J.; Kirsch, N.; Nicholls, I.A.; O’Mahony, J.; Whitcombe, M.J. Molecular imprinting science and technology: A survey of the literature for the years up to and including 2003. J. Mol. Recognit. 2006, 19, 106–180. [Google Scholar] [CrossRef]
- Whitcombe, M.J.; Kirsch, N.; Nicholls, I.A. Molecular imprinting science and technology: A survey of the literature for the years 2004-2011. J. Mol. Recognit. 2014, 27, 297–401. [Google Scholar] [CrossRef] [PubMed]
- Nicholls, I.A. Towards the rational design of molecularly imprinted polymers. J. Mol. Recognit. 1998, 11, 79–82. [Google Scholar] [CrossRef]
- Nicholls, I.A.; Andersson, H.S.; Charlton, C.; Henschel, H.; Karlsson, B.C.G.; Karlsson, J.G.; O’Mahony, J.; Rosengren, A.M.; Rosengren, K.J.; Wikman, S. Theoretical and computational strategies for rational molecularly imprinted polymer design. Biosens. Bioelectron. 2009, 25, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Nicholls, I.A.; Andersson, H.S.; Golker, K.; Henschel, H.; Karlsson, B.C.G.; Olsson, G.D.; Rosengren, A.M.; Shoravi, S.; Suriyanarayanan, S.; Wiklander, J.G.; et al. Rational design of biomimetic molecularly imprinted materials: Theoretical and computational strategies for guiding nanoscale structured polymer development. Anal. Bioanal. Chem. 2011, 400, 1771–1786. [Google Scholar] [CrossRef] [PubMed]
- Nicholls, I.A.; Karlsson, B.C.G.; Olsson, G.D.; Rosengren, A.M. Computational strategies for the design and study of molecularly imprinted materials. Ind. Eng. Chem. Res. 2013, 52, 13900–13909. [Google Scholar] [CrossRef]
- Page, M.I.; Jencks, W.P. Entropic contributions to rate accelerations in enzymic and intramolecular reactions and the chelate effect. Proc. Natl. Acad. Sci. Usa 1971, 68, 1678–1683. [Google Scholar] [CrossRef]
- Jencks, W.P. On the attribution and additivity of binding energies. Proc. Natl. Acad. Sci. Usa 1981, 78, 4046–4050. [Google Scholar] [CrossRef]
- Andrews, P.R.; Craik, D.J.; Martin, J.L. Functional group contributions to drug-receptor interactions. J. Med. Chem. 1984, 27, 1648–1657. [Google Scholar] [CrossRef]
- Williams, D.H.; Cox, J.P.L.; Doig, A.J.; Gardner, M.; Gerhard, U.; Kaye, P.T.; Lal, A.R.; Nicholls, I.A.; Salter, C.J.; Mitchell, R.C. Toward the semiquantitative estimation of binding constants. Guides for peptide-peptide binding in aqueous solution. J. Am. Chem. Soc. 1991, 113, 7020–7030. [Google Scholar] [CrossRef]
- Searle, M.S.; Williams, D.H.; Gerhard, U. Partitioning of free energy contributions in the estimation of binding constants: Residual motions and consequences for amide-amide hydrogen bond strengths. J. Am. Chem. Soc. 1992, 114, 10697–10704. [Google Scholar] [CrossRef]
- Holroyd, S.E.; Groves, P.; Searle, M.S.; Gerhard, U.; Williams, D.H. Rational design and binding of modified cell-wall peptides to vancomycin-group antibiotics: Factorising free energy contributions to binding. Tetrahedron 1993, 49, 9171–9182. [Google Scholar] [CrossRef]
- Nicholls, I.A. Thermodynamic considerations for the design of and ligand recognition by molecularly imprinted polymers. Chem. Lett. 1995, 24, 1035–1036. [Google Scholar] [CrossRef]
- Nicholls, I.A.; Adbo, K.; Andersson, H.S.; Andersson, P.O.; Ankarloo, J.; Hedin-Dahlström, J.; Jokela, P.; Karlsson, J.G.; Olofsson, L.; Rosengren, J.; et al. Can we rationally design molecularly imprinted polymers? Anal. Chim. Acta 2001, 435, 9–18. [Google Scholar] [CrossRef]
- Williams, D.H.; Stephens, E.; O’Brien, D.P.; Zhou, M. Understanding noncovalent interactions: Ligand binding energy and catalytic efficiency from ligand-induced reductions in motion within receptors and enzymes. Angew. Chem. Int. Ed. 2004, 43, 6596–6616. [Google Scholar] [CrossRef]
- Sellergren, B.; Lepistö, M.; Mosbach, K. Highly enantioselective and substrate-selective polymers obtained by molecular imprinting utilizing noncovalent interactions. NMR and chromatographic studies on the nature of recognition. J. Am. Chem. Soc. 1988, 110, 5853–5860. [Google Scholar] [CrossRef]
- Olsson, G.D.; Karlsson, B.C.G.; Shoravi, S.; Wiklander, J.G.; Nicholls, I.A. Mechanisms underlying molecularly imprinted polymer molecular memory and the role of crosslinker: Resolving debate on the nature of template recognition in phenylalanine anilide imprinted polymers. J. Mol. Recognit. 2012, 25, 69–73. [Google Scholar] [CrossRef]
- Whitcombe, M.J.; Martin, L.; Vulfson, E.N. Predicting the selectivity of imprinted polymers. Chromatographia 1998, 47, 457–464. [Google Scholar] [CrossRef]
- Svenson, J.; Andersson, H.S.; Piletsky, S.A.; Nicholls, I.A. Spectroscopic studies of the molecular imprinting self-assembly process. J. Mol. Recognit. 1998, 11, 83–86. [Google Scholar] [CrossRef]
- Svenson, J.; Karlsson, J.G.; Nicholls, I.A. Nuclear magnetic resonance study of the molecular imprinting of (−)-nicotine: Template self-association, a molecular basis for cooperative ligand binding. J. Chromatogr. A 2004, 1024, 39–44. [Google Scholar] [CrossRef]
- Ansell, R.J.; Kuah, K.L. Imprinted polymers for chiral resolution of (±)-ephedrine: Understanding the pre-polymerisation equilibrium and the action of different mobile phase modifiers. Analyst 2005, 130, 179–187. [Google Scholar] [CrossRef]
- Ansell, R.J.; Wang, D.; Kuah, J.K.L. Imprinted polymers for chiral resolution of (±)-ephedrine. Part 2: Probing pre-polymerisation equilibria in different solvents by NMR. Analyst 2008, 133, 1673–1683. [Google Scholar] [CrossRef]
- Ansell, R.J.; Wang, D. Imprinted polymers for chiral resolution of (±)-ephedrine. Part 3: NMR predictions and HPLC results with alternative functional monomers. Analyst 2009, 134, 564–576. [Google Scholar] [CrossRef] [PubMed]
- Andersson, H.S.; Nicholls, I.A. Spectroscopic evaluation of molecular imprinting polymerization systems. Bioorg. Chem. 1997, 25, 203–211. [Google Scholar] [CrossRef]
- Shoravi, S.; Olsson, G.D.; Karlsson, B.C.G.; Nicholls, I.A. On the influence of crosslinker on template complexation in molecularly imprinted polymers: A computational study of prepolymerization mixture events with correlations to template-polymer recognition behavior and NMR spectroscopic studies. Int. J. Mol. Sci. 2014, 15, 10622–10634. [Google Scholar] [CrossRef]
- Tanabe, K.; Takeuchi, T.; Matsui, J.; Ikebukuro, K.; Yano, K.; Karube, I. Recognition of barbiturates in molecularly imprinted copolymers using multiple hydrogen bonding. J. Chem. Soc. Chem. Commun. 1995, 2303–2304. [Google Scholar] [CrossRef]
- Hall, A.J.; Manesiotis, P.; Emgenbroich, M.; Quaglia, M.; De Lorenzi, E.; Sellergren, B. Urea host monomers for stoichiometric molecular imprinting of oxyanions. J. Org. Chem. 2005, 70, 1732–1736. [Google Scholar] [CrossRef] [PubMed]
- Emgenbroich, M.; Borrelli, C.; Shinde, S.; Lazraq, I.; Vilela, F.; Hall, A.J.; Oxelbark, J.; De Lorenzi, E.; Courtois, J.; Simanova, A.; et al. A phosphotyrosine-imprinted polymer receptor for the recognition of tyrosine phosphorylated peptides. Chem. Eur. J. 2008, 14, 9516–9529. [Google Scholar] [CrossRef]
- Chen, J.; Shinde, S.; Koch, M.-H.; Eisenacher, M.; Galozzi, S.; Lerari, T.; Barkovits, K.; Subedi, P.; Krüger, R.; Kuhlmann, K.; et al. Low-bias phosphopeptide enrichment from scarce samples using plastic antibodies. Sci. Rep. 2015, 5, 11438:1–11438:12. [Google Scholar] [CrossRef]
- Vlatakis, G.; Andersson, L.I.; Müller, R.; Mosbach, K. Drug assay using antibody mimics made by molecular imprinting. Nature 1993, 361, 645–647. [Google Scholar] [CrossRef] [PubMed]
- Berglund, J.; Nicholls, I.A.; Lindbladh, C.; Mosbach, K. Recognition in molecularly imprinted polymer α2-adrenoreceptor mimics. Bioorg. Med. Chem. Lett. 1996, 6, 2237–2242. [Google Scholar] [CrossRef]
- Matsui, J.; Miyoshi, Y.; Takeuchi, T. Fluoro-functionalized molecularly imprinted polymers selective for herbicides. Chem. Lett. 1995, 24, 1007–1008. [Google Scholar] [CrossRef]
- Matsui, J.; Takeuchi, T. A molecularly imprinted polymer rod as nicotine selective affinity media prepared with 2-(trifluoromethyl)acrylic acid. Anal. Commun. 1997, 34, 199–200. [Google Scholar] [CrossRef]
- Matsui, J.; Nicholls, I.A.; Takeuchi, T. Highly stereoselective molecularly imprinted polymer synthetic receptors for cinchona alkaloids. Tetrahedron Asymmetry 1996, 7, 1357–1361. [Google Scholar] [CrossRef]
- Matsui, J.; Doblhoff-Dier, O.; Takeuchi, T. 2-(trifluoromethyl)acrylic acid: A novel functional monomer in non-covalent molecular imprinting. Anal. Chim. Acta 1997, 343, 1–4. [Google Scholar] [CrossRef]
- Andersson, H.S.; Ramström, O. Crown ethers as a tool for the preparation of molecularly imprinted polymers. J. Mol. Recognit. 1998, 11, 103–106. [Google Scholar] [CrossRef]
- Vidyasankar, S.; Dhal, P.K.; Plunkett, S.D.; Arnold, F.H. Review: Selective ligand-exchange adsorbents prepared by template polymerization. Biotechnol. Bioeng. 1995, 48, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Leonhardt, A.; Mosbach, K. Enzyme-mimicking polymers exhibiting specific substrate binding and catalytic functions. React. Polym. Ion Exch. Sorbents 1987, 6, 285–290. [Google Scholar] [CrossRef]
- Matsui, J.; Nicholls, I.A.; Karube, I.; Mosbach, K. Carbon−carbon bond formation using substrate selective catalytic polymers prepared by molecular imprinting: An artificial class II aldolase. J. Org. Chem. 1996, 61, 5414–5417. [Google Scholar] [CrossRef]
- Santora, B.P.; Larsen, A.O.; Gagné, M.R. Toward the molecular imprinting of titanium lewis acids: Demonstration of Diels−Alder catalysis. Organometallics 1998, 17, 3138–3140. [Google Scholar] [CrossRef]
- Liu, J.; Wulff, G. Functional mimicry of carboxypeptidase a by a combination of transition state stabilization and a defined orientation of catalytic moieties in molecularly imprinted polymers. J. Am. Chem. Soc. 2008, 130, 8044–8054. [Google Scholar] [CrossRef]
- Lay, S.; Ni, X.; Yu, H.; Shen, S. State-of-the-art applications of cyclodextrins as functional monomers in molecular imprinting techniques: A review. J. Sep. Sci. 2016, 39, 2321–2331. [Google Scholar] [CrossRef] [PubMed]
- Asanuma, H.; Kakazu, M.; Shibata, M.; Hishiya, T. Molecularly imprinted polymer of β-cyclodextrin for the efficient recognition of cholesterol. Chem. Commun. 1997, 20, 1971–1972. [Google Scholar] [CrossRef]
- Sreenivasan, K. Synthesis and evaluation of a beta cyclodextrin-based molecularly imprinted copolymer. J. Appl. Polym. Sci. 1998, 70, 15–18. [Google Scholar] [CrossRef]
- Piletsky, S.A.; Andersson, H.S.; Nicholls, I.A. The rational use of hydrophobic effect-based recognition in molecularly imprinted polymers. J. Mol. Recognit. 1998, 11, 94–97. [Google Scholar] [CrossRef]
- Piletsky, S.A.; Andersson, H.S.; Nicholls, I.A. Combined hydrophobic and electrostatic interaction-based recognition in molecularly imprinted polymers. Macromolecules 1999, 32, 633–636. [Google Scholar] [CrossRef]
- Pande, V.S.; Grosberg, A.Y.; Tanaka, T. How to create polymers with protein-like capabilities: A theoretical suggestion. Phys. D 1997, 107, 316–321. [Google Scholar] [CrossRef]
- Piletsky, S.A.; Panasyuk, T.L.; Piletskaya, E.V.; Nicholls, I.A.; Ulbricht, M. Receptor and transport properties of imprinted polymer membranes—A review. J. Membr. Sci. 1999, 157, 263–278. [Google Scholar] [CrossRef]
- Wu, X.; Carroll, W.R.; Shimizu, K.D. Stochastic lattice model simulations of molecularly imprinted polymers. Chem. Mater. 2008, 20, 4335–4346. [Google Scholar] [CrossRef]
- Veitl, M.; Schweiger, U.; Berger, M.L. Stochastic simulation of ligand–receptor interaction. Comput. Biomed. Res. 1997, 30, 427–450. [Google Scholar] [CrossRef]
- Curk, T.; Dobnikar, J.; Frenkel, D. Rational design of molecularly imprinted polymers. Soft Matter 2016, 12, 35–44. [Google Scholar] [CrossRef]
- Nicholls, I.A.; Andersson, H.S.; Golker, K.; Henschel, H.; Karlsson, B.C.G.; Olsson, G.D.; Rosengren, A.M.; Shoravi, S.; Wiklander, J.G.; Wikman, S. Rational molecularly imprinted polymer design: Theoretical and computational strategies. In Molecular Imprinting—Principles and Applications af Micro- and Nanostructured Polymers; Lei, Y., Ed.; Pan Stanford Publishing: Singapore, 2013; pp. 71–104. ISBN 978-981-4310-99-4. [Google Scholar]
- Nicholls, I.A.; Olsson, G.D.; Karlsson, B.C.G.; Suriyanarayanan, S.; Wiklander, J.G. CHAPTER 7 Theoretical and Computational Strategies in Molecularly Imprinted Polymer Development. In Molecularly Imprinted Polymers for Analytical Chemistry Applications; The Royal Society of Chemistry: Cambridge, UK, 2018; pp. 197–226. ISBN 978-1-78262-647-3. [Google Scholar]
- Tomov, S.; McGuigan, M.; Bennett, R.; Smith, G.; Spiletic, J. Benchmarking and implementation of probability-based simulations on programmable graphics cards. Comput. Graph. 2005, 29, 71–80. [Google Scholar] [CrossRef][Green Version]
- Schadt, E.E.; Linderman, M.D.; Sorenson, J.; Lee, L.; Nolan, G.P. Computational solutions to large-scale data management and analysis. Nat. Rev. Genet. 2010, 11, 647–657. [Google Scholar] [CrossRef] [PubMed]
- Fermi, E.; Pasta, P.; Ulam, S.; Tsingou, M. Studies of Nonlinear Problems; University of California Press: Berkeley, CA, USA, 1955. [Google Scholar] [CrossRef]
- Gibson, J.B.; Goland, A.N.; Milgram, M.; Vineyard, G.H. Dynamics of radiation damage. Phys. Rev. 1960, 120, 1229–1253. [Google Scholar] [CrossRef]
- Metropolis, N.; Rosenbluth, A.W.; Rosenbluth, M.N.; Teller, A.H.; Teller, E. Equation of State Calculations by Fast Computing Machines. J. Chem. Phys. 1953, 21, 1087–1092. [Google Scholar] [CrossRef]
- Barker, J.A.; Watts, R.O. Structure of water; A Monte Carlo calculation. Chem. Phys. Lett. 1969, 3, 144–145. [Google Scholar] [CrossRef]
- McDonald, I.R.; Singer, K. Calculation of thermodynamic properties of liquid argon from Lennard-Jones parameters by a Monte Carlo method. Discuss. Faraday Soc. 1967, 43, 40–49. [Google Scholar] [CrossRef]
- Alder, B.J.; Wainwright, T.E. Phase transition for a hard sphere system. J. Chem. Phys. 1957, 27, 1208–1209. [Google Scholar] [CrossRef]
- Rahman, A. Correlations in the motion of atoms in liquid argon. Phys. Rev. 1964, 136, A405–A411. [Google Scholar] [CrossRef]
- Rahman, A.; Stillinger, F.H. Molecular dynamics study of liquid water. J. Chem. Phys. 1971, 55, 3336–3359. [Google Scholar] [CrossRef]
- Leach, A.R. Molecular dynamics simulation methods. In Molecular Modelling: Principles and Applications, 2nd ed.; Pearson: Harlow, UK, 2001; pp. 353–409. ISBN 978-0-582-38210-7. [Google Scholar]
- Hsu, Q.C.; Wu, C.D.; Fang, T.H. Studies on nanoimprint process parameters of copper by molecular dynamics analysis. Comput. Mater. Sci. 2005, 34, 314–322. [Google Scholar] [CrossRef]
- Garrison, B.J.; Delcorte, A.; Krantzman, K.D. Molecule liftoff from surfaces. Acc. Chem. Res. 2000, 33, 69–77. [Google Scholar] [CrossRef]
- van Buuren, A.R.; Marrink, S.J.; Berendsen, H.J.C. A molecular dynamics study of the decane/water interface. J. Phys. Chem. 1993, 97, 9206–9212. [Google Scholar] [CrossRef]
- Masukawa, K.M.; Kollman, P.A.; Kuntz, I.D. Investigation of neuraminidase-substrate recognition using molecular dynamics and free energy calculations. J. Med. Chem. 2003, 46, 5628–5637. [Google Scholar] [CrossRef]
- Yang, H.; Elcock, A.H. Association lifetimes of hydrophobic amino acid pairs measured directly from molecular dynamics simulations. J. Am. Chem. Soc. 2003, 125, 13968–13969. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Eriksson, L.A. Molecular dynamics study of lignin constituents in water. Holzforschung 2005, 59, 253–262. [Google Scholar] [CrossRef]
- Cheatham (III), T.E.; Kollman, P.A. Observation of the A-DNA to B-DNA transition during unrestrained molecular dynamics in aqueous solution. J. Mol. Biol. 1996, 259, 434–444. [Google Scholar] [CrossRef] [PubMed]
- Cheatham (III), T.E.; Miller, J.L.; Fox, T.; Darden, T.A.; Kollman, P.A. Molecular dynamics simulations on solvated biomolecular systems: The particle mesh ewald method leads to stable trajectories of DNA, RNA, and proteins. J. Am. Chem. Soc. 1995, 117, 4193–4194. [Google Scholar] [CrossRef]
- Duan, Y. Pathways to a protein folding intermediate observed in a 1-microsecond simulation in aqueous solution. Science 1998, 282, 740–744. [Google Scholar] [CrossRef]
- Berger, O.; Edholm, O.; Jähnig, F. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys. J. 1997, 72, 2002–2013. [Google Scholar] [CrossRef]
- van der Ploeg, P. Molecular dynamics simulation of a bilayer membrane. J. Chem. Phys. 1982, 76, 3271–3276. [Google Scholar] [CrossRef]
- de Groot, B.L.; Grubmüller, H. Water permeation across biological membranes: Mechanism and dynamics of aquaporin-1 and GlpF. Science 2001, 294, 2353–2357. [Google Scholar] [CrossRef]
- Wang, J.; Cieplak, P.; Kollman, P.A. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 2000, 21, 1049–1074. [Google Scholar] [CrossRef]
- Ponder, J.W.; Case, D.A. Force fields for protein simulations. In Protein Simulations; Daggett, V., Ed.; Elsevier: San Diego, CA, USA, 2003; Volume 66, pp. 27–85. ISBN 0-12-034266-9. [Google Scholar]
- Tian, C.; Kasavajhala, K.; Belfon, K.A.A.; Raguette, L.; Huang, H.; Migues, A.N.; Bickel, J.; Wang, Y.Z.; Pincay, J.; Wu, Q.; et al. ff19SB: Amino-acid-specific protein backbone parameters trained against quantum mechanics energy surfaces in solution. J. Chem. Theory Comput. 2020, 16, 528–552. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Brooks, B.R.; Bruccoleri, R.E.; Olafson, B.D.; States, D.J.; Swaminathan, S.; Karplus, M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983, 4, 187–217. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Maxwell, D.S.; Tirado-Rives, J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996, 118, 11225–11236. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; van der Spoel, D.; van Drunen, R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995, 91, 43–56. [Google Scholar] [CrossRef]
- Scott, W.R.P.; Hünenberger, P.H.; Tironi, I.G.; Mark, A.E.; Billeter, S.R.; Fennen, J.; Torda, A.E.; Huber, T.; Krüger, P.; van Gunsteren, W.F. The GROMOS biomolecular simulation program package. J. Phys. Chem. A 1999, 103, 3596–3607. [Google Scholar] [CrossRef]
- Shinde, S.; Incel, A.; Mansour, M.; Olsson, G.D.; Nicholls, I.A.; Esen, C.; Urraca, J.; Sellergren, B. Urea-based imprinted polymer hosts with switchable anion preference. J. Am. Chem. Soc. 2020, 142, 11404–11416. [Google Scholar] [CrossRef]
- Olsson, G.D.; Niedergall, K.; Bach, M.; Karlsson, B.C.G.; Tovar, G.; Nicholls, I.A. Simulation of imprinted emulsion prepolymerization mixtures. Polym. J. 2015, 47, 827–830. [Google Scholar] [CrossRef]
- Esbensen, K.H. Multivariate Data Analysis—In Practice: An Introduction to Multivariate Data Analysis and Experimental Design, 5th ed.; Camo Process AS: Oslo, Norway, 2002; Volume 16, ISBN 978-8299333030. [Google Scholar]
- Carlsson, R. Design and Optimization in Organic Synthesis; Elsevier: Amsterdam, The Netherlands, 1992; ISBN 0-444-89201-X. [Google Scholar]
- Eriksson, L.; Johansson, E.; Kettaneh-Wold, N.; Wold, S. Multi- and Megavariate Data Analysis. Principles and Applications; Umetrics Academy: Umeå, Sweden, 2002; ISBN 91-973730-1-X. [Google Scholar]
- Fu, Q.; Sanbe, H.; Kagawa, C.; Kunimoto, K.-K.; Haginaka, J. Uniformly sized molecularly imprinted polymer for ( S )-nilvadipine. Comparison of chiral recognition ability with HPLC chiral stationary phases based on a protein. Anal. Chem. 2003, 75, 191–198. [Google Scholar] [CrossRef]
- Schwarz, L.; Holdsworth, C.I.; McCluskey, A.; Bowyer, M.C. Synthesis and evaluation of a molecularly imprinted polymer selective to 2,4,6-trichlorophenol. Aust. J. Chem. 2004, 57, 759–764. [Google Scholar] [CrossRef]
- Holdsworth, C.I.; Bowyer, M.C.; Lennard, C.; McCluskey, A. Formulation of cocaine-imprinted polymers utilizing molecular modelling and nmr analysis. Aust. J. Chem. 2005, 58, 315–320. [Google Scholar] [CrossRef]
- Ogawa, T.; Hoshina, K.; Haginaka, J.; Honda, C.; Tanimoto, T.; Uchida, T. Screening of bitterness-suppressing agents for quinine: The use of molecularly imprinted polymers. J. Pharm. Sci. 2005, 94, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Voshell, S.M.; Gagné, M.R. Rigidified dendritic structures for imprinting chiral information. Organometallics 2005, 24, 6338–6350. [Google Scholar] [CrossRef]
- Meng, Z.; Yamazaki, T.; Sode, K. A molecularly imprinted catalyst designed by a computational approach in catalysing a transesterification process. Biosens. Bioelectron. 2004, 20, 1068–1075. [Google Scholar] [CrossRef]
- Baggiani, C.; Anfossi, L.; Baravalle, P.; Giovannoli, C.; Tozzi, C. Selectivity features of molecularly imprinted polymers recognising the carbamate group. Anal. Chim. Acta 2005, 531, 199–207. [Google Scholar] [CrossRef]
- Zayas, H.; Holdsworth, C.I.; Bowyer, M.C.; McCluskey, A. Evaluation of 4-substituted styrenes as functional monomers for the synthesis of theophylline-specific molecularly imprinted polymers. Org. Biomol. Chem. 2014, 12, 6994–7003. [Google Scholar] [CrossRef]
- Toro, M.J.U.; Marestoni, L.D.; Sotomayor, M.D.P.T. A new biomimetic sensor based on molecularly imprinted polymers for highly sensitive and selective determination of hexazinone herbicide. Sens. Actuators B 2015, 208, 299–306. [Google Scholar] [CrossRef]
- Marestoni, L.D.; Wong, A.; Feliciano, G.T.; Marchi, M.R.R.; Tarley, C.R.T.; Sotomayor, M.D.P.T. Semi-empirical quantum chemistry method for pre-polymerization rational design of ciprofloxacin imprinted polymer and adsorption studies. J. Braz. Chem. Soc. 2016, 27, 109–118. [Google Scholar] [CrossRef]
- Tiu, B.D.B.; Pernites, R.B.; Tin, S.B.; Advincula, R.C. Detection of aspartame via microsphere-patterned and molecularly imprinted polymer arrays. Colloids Surf. A 2016, 495, 149–158. [Google Scholar] [CrossRef]
- Wong, A.; Foguel, M.V.; Khan, S.; de Oliveira, F.M.; Tarley, C.R.T.; Sotomayor, M. Development of an electrochemical sensor modified with MWCNT-COOH and MIP for detection of diuron. Electrochim. Acta 2015, 182, 122–130. [Google Scholar] [CrossRef]
- Feng, F.; Zheng, J.; Qin, P.; Han, T.; Zhao, D. A novel quartz crystal microbalance sensor array based on molecular imprinted polymers for simultaneous detection of clenbuterol and its metabolites. Talanta 2017, 167, 94–102. [Google Scholar] [CrossRef]
- Li, P.; Rong, F.; Xie, Y.; Hu, V.; Yuan, C. Study on the binding characteristic of s-naproxen imprinted polymer and the interactions between templates and monomers. J. Anal. Chem. 2004, 59, 939–944. [Google Scholar] [CrossRef]
- Luliński, P.; Maciejewska, D.; Bamburowicz-Klimkowska, M.; Szutowski, M. Dopamine-imprinted polymers: Template-monomer interactions, analysis of template removal and application to solid phase extraction. Molecules 2007, 12, 2434–2449. [Google Scholar] [CrossRef] [PubMed]
- Boysen, R.I.; Schwarz, L.J.; Li, S.; Chowdhury, J.; Hearn, M.T.W. Photo-lithographic patterning of biomimetic molecularly imprinted polymer thin films onto silicon wafers. Microsyst. Technol. 2014, 20, 2037–2043. [Google Scholar] [CrossRef]
- Wu, L.; Li, Y. Study on the recognition of templates and their analogues on molecularly imprinted polymer using computational and conformational analysis approaches. J. Mol. Recognit. 2004, 17, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Sun, B.; Li, Y.; Chang, W. Study properties of molecular imprinting polymer using a computational approach. Analyst 2003, 128, 944–949. [Google Scholar] [CrossRef]
- Rathbone, D.L.; Ali, A.; Antonaki, P.; Cheek, S. Towards a polymeric binding mimic for cytochrome CYP2D6. Biosens. Bioelectron. 2005, 20, 2353–2363. [Google Scholar] [CrossRef] [PubMed]
- Sagawa, T.; Togo, K.; Miyahara, C.; Ihara, H.; Ohkubo, K. Rate-enhancement of hydrolysis of long-chain amino acid ester by cross-linked polymers imprinted with a transition-state analogue: Evaluation of imprinting effect in kinetic analysis. Anal. Chim. Acta 2004, 504, 37–41. [Google Scholar] [CrossRef]
- Wu, L.; Li, Y. Picolinamide–Cu(Ac)2-imprinted polymer with high potential for recognition of picolinamide–copper acetate complex. Anal. Chim. Acta 2003, 482, 175–181. [Google Scholar] [CrossRef]
- Wu, L.; Li, Y. Metal ion-mediated molecular-imprinting polymer for indirect recognition of formate, acetate and propionate. Anal. Chim. Acta 2004, 517, 145–151. [Google Scholar] [CrossRef]
- Liu, B.; Ou, L.; Zhang, F.; Zhang, Z.; Li, H.; Zhu, M.; Wang, S. Validation and application of modeling algorithms for the design of molecularly imprinted polymers. J. Sep. Sci. 2014, 37, 3579–3586. [Google Scholar] [CrossRef]
- Bi, H.M.; Hu, J.P.; Liu, Y.; Chai, X.Q.; Tong, L.X.; Dong, C.H. Computer simulation study on the effect of recognized characteristics of cotinine imprinted polymer with different functional monomers. Asian J. Chem. 2014, 26, 161–163. [Google Scholar] [CrossRef]
- Luliński, P.; Dana, M.; Maciejewska, D. Synthesis and characterization of 4-(2-aminoethyl)aniline imprinted polymer as a highly effective sorbent of dopamine. Talanta 2014, 119, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Wu, X.; Wang, J.; Liang, S.; Sun, H. Highly sensitive determination of cyromazine, melamine, and their metabolites in milk by molecularly imprinted solid-phase extraction combined with ultra-performance liquid chromatography. J. Dairy Sci. 2015, 98, 2161–2171. [Google Scholar] [CrossRef] [PubMed]
- Zuo, H.G.; Zhu, J.X.; Zhan, C.R.; Shi, L.; Xing, M.; Guo, P.; Ding, Y.; Yang, H. Preparation of malathion MIP-SPE and its application in environmental analysis. Environ. Monit. Assess. 2015, 187, 394:1–394:19. [Google Scholar] [CrossRef]
- Xu, X.; Duhoranimana, E.; Zhang, X. Preparation and characterization of magnetic molecularly imprinted polymers for the extraction of hexamethylenetetramine in milk samples. Talanta 2017, 163, 31–38. [Google Scholar] [CrossRef]
- Goud, K.Y.; M, S.; Reddy, K.K.; Gobi, K.V. Development of highly selective electrochemical impedance sensor for detection of sub-micromolar concentrations of 5-Chloro-2,4-dinitrotoluene. J. Chem. Sci. 2016, 128, 763–770. [Google Scholar] [CrossRef]
- Krishnan, H.; Islam, K.M.S.; Hamzah, Z.; Ahmad, M.N. Rational computational design for the development of andrographolide molecularly imprinted polymer. In 2nd International Conference on Applied Science and Technology 2017 (ICAST’17), Kedah, Malaysia, 3–5 April 2017; Nifa, F.A.A., Lin, C.K., Hussain, A., Eds.; AIP Publishing LLC: Melville, NY, USA, 2017; Volume 1891, pp. 020083:1–020083:7. ISBN 978-0-7354-1573-7. [Google Scholar]
- Peng, M.; Li, H.; Long, R.; Shi, S.; Zhou, H.; Yang, S. Magnetic porous molecularly imprinted polymers based on surface precipitation polymerization and mesoporous SiO2 layer as sacrificial support for efficient and selective extraction and determination of chlorogenic acid in duzhong brick tea. Molecules 2018, 23, 1554. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, W.; Wu, Z.; Huang, X.; Hui, A.; He, Y.; Wang, H. Theoretical design, preparation, and evaluation of Ginkgolide B molecularly imprinted polymers. J. Sep. Sci. 2020, 43, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Nasir, A.M.; Ishak, N.H.; Said, M.S.M.; Dzahir, I.H.M. One-pot synthesis of molecular-imprinted membrane for selective extraction of caffeic acid. Polym. Bull. 2020, 77, 3953–3968. [Google Scholar] [CrossRef]
- Zuo, H.G.; Yang, H.; Zhu, J.X.; Guo, P.; Shi, L.; Zhan, C.R.; Ding, Y. Synthesis of molecularly imprinted polymer on surface of TiO2 nanowires and assessment of malathion and its metabolite in environmental water. J. Anal. Chem. 2019, 74, 1039–1055. [Google Scholar] [CrossRef]
- Terracina, J.J.; Sharfstein, S.T.; Bergkvist, M. In silico characterization of enantioselective molecularly imprinted binding sites. J. Mol. Recognit. 2018, 31, 2612:1–2612:8. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Hussain, S.; Wong, A.; Foguel, M.V.; Goncalves, L.M.; Gurgo, M.I.P.; Sotomayor, M.D.T. Synthesis and characterization of magnetic-molecularly imprinted polymers for the HPLC-UV analysis of ametryn. React. Funct. Polym. 2018, 122, 175–182. [Google Scholar] [CrossRef]
- Quinto, M.L.; Khan, S.; Picasso, G.; Sotomayor, M.D.P.T. Synthesis, characterization, and evaluation of a selective molecularly imprinted polymer for quantification of the textile dye acid violet 19 in real water samples. J. Hazard. Mater. 2020, 384, 121374:1–121374:10. [Google Scholar] [CrossRef]
- Diñeiro, Y.; Menéndez, M.I.; Blanco-López, M.C.; Lobo-Castañón, M.J.; Miranda-Ordieres, A.J.; Tuñón-Blanco, P. Computational approach to the rational design of molecularly imprinted polymers for voltammetric sensing of homovanillic acid. Anal. Chem. 2005, 77, 6741–6746. [Google Scholar] [CrossRef]
- Dong, W.; Yan, M.; Zhang, M.; Liu, Z.; Li, Y. A computational and experimental investigation of the interaction between the template molecule and the functional monomer used in the molecularly imprinted polymer. Anal. Chim. Acta 2005, 542, 186–192. [Google Scholar] [CrossRef]
- Diñeiro, Y.; Menéndez, M.I.; Blanco-López, M.C.; Lobo-Castañón, M.J.; Miranda-Ordieres, A.J.; Tuñón-Blanco, P. Computational predictions and experimental affinity distributions for a homovanillic acid molecularly imprinted polymer. Biosens. Bioelectron. 2006, 22, 364–371. [Google Scholar] [CrossRef]
- Dong, W.; Yan, M.; Liu, Z.; Wu, G.; Li, Y. Effects of solvents on the adsorption selectivity of molecularly imprinted polymers: Molecular simulation and experimental validation. Sep. Purif. Technol. 2007, 53, 183–188. [Google Scholar] [CrossRef]
- Alizadeh, T. Development of a molecularly imprinted polymer for pyridoxine using an ion-pair as template. Anal. Chim. Acta 2008, 623, 101–108. [Google Scholar] [CrossRef]
- Christoforidis, K.C.; Louloudi, M.; Rutherford, A.W.; Deligiannakis, Y. Semiquinone in molecularly imprinted hybrid amino acid−SiO2 biomimetic materials. An experimental and theoretical study. J. Phys. Chem. C 2008, 112, 12841–12852. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, F.; Tan, T.; Lei, M. Rational design and study on recognition property of paracetamol-imprinted polymer. Appl. Biochem. Biotechnol. 2008, 160, 328–342. [Google Scholar] [CrossRef]
- Campbell, S.E.; Collins, M.; Xie, L.; BelBruno, J.J. Surface morphology of spin-coated molecularly imprinted polymer films. Surf. Interface Anal. 2009, 41, 347–356. [Google Scholar] [CrossRef]
- Jacob, R.; Tate, M.; Banti, Y.; Rix, C.; Mainwaring, D.E. Synthesis, characterization, and ab initio theoretical study of a molecularly imprinted polymer selective for biosensor materials. J. Phys. Chem. A 2008, 112, 322–331. [Google Scholar] [CrossRef]
- Che, A.-F.; Wan, L.-S.; Ling, J.; Liu, Z.-M.; Xu, Z.-K. Recognition mechanism of theophylline-imprinted polymers: Two-dimensional infrared analysis and density functional theory study. J. Phys. Chem. B 2009, 113, 7053–7058. [Google Scholar] [CrossRef] [PubMed]
- Del Sole, R.; Lazzoi, M.R.; Arnone, M.; Sala, F.D.; Cannoletta, D.; Vasapollo, G. Experimental and computational studies on non-covalent imprinted microspheres as recognition system for nicotinamide molecules. Molecules 2009, 14, 2632–2649. [Google Scholar] [CrossRef]
- Demircelik, A.H.; Andac, M.; Andac, C.A.; Say, R.; Denizli, A. Molecular recognition-based detoxification of aluminum in human plasma. J. Biomater. Sci. Polym. Ed. 2009, 20, 1235–1258. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Li, X.; Guo, Z.; Qi, J. Development of a model for the rational design of molecular imprinted polymer: Computational approach for combined molecular dynamics/quantum mechanics calculations. Anal. Chim. Acta 2009, 647, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Kowalska, A.; Stobiecka, A.; Wysocki, S. A computational investigation of the interactions between harmane and the functional monomers commonly used in molecular imprinting. J. Mol. Struct. 2009, 901, 88–95. [Google Scholar] [CrossRef]
- Li, Y.; Li, X.; Dong, C.; Li, Y.; Jin, P.; Qi, J. Selective recognition and removal of chlorophenols from aqueous solution using molecularly imprinted polymer prepared by reversible addition-fragmentation chain transfer polymerization. Biosens. Bioelectron. 2009, 25, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Pietrzyk, A.; Kutner, W.; Chitta, R.; Zandler, M.E.; D’Souza, F.; Sannicolo, F.; Mussini, P.R. Melamine acoustic chemosensor based on molecularly imprinted polymer film. Anal. Chem. 2009, 81, 10061–10070. [Google Scholar] [CrossRef]
- Riahi, S.; Edris-Tabrizi, F.; Javanbakht, M.; Ganjali, M.R.; Norouzi, P. A computational approach to studying monomer selectivity towards the template in an imprinted polymer. J. Mol. Model. 2009, 15, 829–836. [Google Scholar] [CrossRef] [PubMed]
- Pardeshi, S.; Dhodapkar, R.; Kumar, A. Influence of porogens on the specific recognition of molecularly imprinted poly(acrylamide-co-ethylene glycol dimethacrylate). Compos. Interfaces 2013, 21, 13–30. [Google Scholar] [CrossRef]
- Ahmadi, F.; Karamian, E. Computational aided-molecular imprinted polymer design for solid phase extraction of metaproterenol from plasma and determination by voltammetry using modified carbon nanotube electrode. Iran. J. Pharm. Res. 2014, 13, 417–429. [Google Scholar]
- Ahmadi, F.; Yawari, E.; Nikbakht, M. Computational design of an enantioselective molecular imprinted polymer for the solid phase extraction of S-warfarin from plasma. J. Chromatogr. A 2014, 1338, 9–16. [Google Scholar] [CrossRef]
- Barros, L.A.; Pereira, L.A.; Custódio, R.; Rath, S. A novel computational approach for development of highly selective fenitrothion imprinted polymer: Theoretical predictions and experimental validations. J. Braz. Chem. Soc. 2014, 25, 619–628. [Google Scholar] [CrossRef]
- Gao, B.; He, X.P.; Jiang, Y.; Wei, J.T.; Suo, H.; Zhao, C. Computational simulation and preparation of fluorescent magnetic molecularly imprinted silica nanospheres for ciprofloxacin or norfloxacin sensing. J. Sep. Sci. 2014, 37, 3753–3759. [Google Scholar] [CrossRef]
- Liu, J.; Dai, Z.; Li, B.; Tang, S.; Jin, R. Utilization of theoretical studies of the imprinting ratio to guide experimental research into the molecular imprinted polymers formed using enrofloxacin and methacrylic acid. J. Mol. Model. 2014, 20, 2456:1–2456:10. [Google Scholar] [CrossRef] [PubMed]
- Luliński, P.; Sobiech, M.; Żołek, T.; Maciejewska, D. A separation of tyramine on a 2-(4-methoxyphenyl)ethylamine imprinted polymer: An answer from theoretical and experimental studies. Talanta 2014, 129, 155–164. [Google Scholar] [CrossRef]
- Ogunlaja, A.S.; Coombes, M.J.; Torto, N.; Tshentu, Z.R. The adsorptive extraction of oxidized sulfur-containing compounds from fuels by using molecularly imprinted chitosan materials. React. Funct. Polym. 2014, 81, 61–76. [Google Scholar] [CrossRef]
- Ogunlaja, A.S.; du Sautoy, C.; Tort, N.; Tshentu, Z.R. Design, fabrication and evaluation of intelligent sulfone-selective polybenzimidazole nanofibers. Talanta 2014, 126, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Pardo, A.; Mespouille, L.; Blankert, B.; Trouillas, P.; Surin, M.; Dubois, P.; Duez, P. Quercetin-imprinted chromatographic sorbents revisited: Optimization of synthesis and rebinding protocols for application to natural resources. J. Chromatogr. A 2014, 1364, 128–139. [Google Scholar] [CrossRef]
- Qi, P.; Wang, X.; Wang, X.; Zhang, H.; Xu, H.; Jiang, K.; Wang, Q. Computer-assisted design and synthesis of molecularly imprinted polymers for the simultaneous determination of six carbamate pesticides from environmental water. J. Sep. Sci. 2014, 37, 2955–2965. [Google Scholar] [CrossRef]
- Qin, L.; Liu, W.; Yang, Y.; Liu, X. Functional monomer screening and preparation of dibenzothiophene-imprinted polymers on the surface of carbon microsphere. Monatsh. Chem. 2014, 146, 449–458. [Google Scholar] [CrossRef]
- Roy, E.; Patra, S.; Madhuri, R.; Sharma, P.K. Gold nanoparticle mediated designing of non-hydrolytic sol-gel cross-linked metformin imprinted polymer network: A theoretical and experimental study. Talanta 2014, 120, 198–207. [Google Scholar] [CrossRef]
- Su, T.T.; Liu, J.B.; Tang, S.S.; Chang, H.B.; Jin, R.F. Theoretical study on the structures and properties of phenobarbital imprinted polymers. Chin. J. Struct. Chem. 2014, 33, 1421–1430. [Google Scholar]
- Tadi, K.K.; Motghare, R.V.; Ganesh, V. Electrochemical detection of sulfanilamide using pencil graphite electrode based on molecular imprinting technology. Electroanalysis 2014, 26, 2328–2336. [Google Scholar] [CrossRef]
- Yang, W.M.; Ma, P.F.; Fan, T.; Zhou, Z.P.; Liu, H.; Xu, W.Z. Optimal design of an imprinted preassembled system by quantum chemical calculations and preparation of a surface-imprinted material for the selective removal of quinoline. J. Appl. Polym. Sci. 2015, 132, 41730:1–41730:10. [Google Scholar] [CrossRef]
- Yilmaz, V.; Arslan, Z.; Hazer, O.; Yilmaz, H. Selective solid phase extraction of copper using a new Cu(II)-imprinted polymer and determination by inductively coupled plasma optical emission spectroscopy (ICP-OES). Microchem. J. 2014, 114, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.X.; Qu, X.; Yu, J.P.; Xu, L.C.; Zhang, Z.Q.; Hong, H.; Liu, C.S. C-13 NMR aided design of molecularly imprinted adsorbents for selectively preparative separation of erythromycin. J. Mater. Chem. B 2014, 2, 1390–1399. [Google Scholar] [CrossRef] [PubMed]
- Cao, N.L.; Zyablov, A.N.; Duvanova, O.V.; Selemenev, V.F.; Falaleev, A.V. Modeling of palmitic and oleic acids imprinted polymers based on polyamidoacid. Sorpt. Chromatogr. Process. 2015, 15, 421–428. [Google Scholar]
- Dai, Z.Q.; Liu, J.B.; Tang, S.S.; Wang, Y.; Wang, Y.M.; Jin, R.F. Optimization of enrofloxacin-imprinted polymers by computer-aided design. J. Mol. Model. 2015, 21, 290:1–290:9. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.M.; Tan, L.; Li, H.; Xu, Z.G.; Shen, G.X.; Tang, Y.W. Selective fluorescent sensing of alpha-amanitin in serum using carbon quantum dots-embedded specificity determinant imprinted polymers. Biosens. Bioelectron. 2015, 69, 265–271. [Google Scholar] [CrossRef]
- Fernandes, L.S.; Homem-de-Mello, P.; de Lima, E.C.; Honorio, K.M. Rational design of molecularly imprinted polymers for recognition of cannabinoids: A structure-property relationship study. Eur. Polym. J. 2015, 71, 364–371. [Google Scholar] [CrossRef]
- Huang, Y.J.; Zhu, Q.J. Computational modeling and theoretical calculations on the interactions between spermidine and functional monomer (methacrylic acid) in a molecularly imprinted polymer. J. Chem. 2015, 2015, 216983:1–216983:9. [Google Scholar] [CrossRef]
- Huynh, T.-P.; Chandra, B.K.C.; Sosnowska, M.; Sobczak, J.W.; Nesterov, V.N.; D’Souza, F.; Kutner, W. Nicotine molecularly imprinted polymer: Synergy of coordination and hydrogen bonding. Biosens. Bioelectron. 2015, 64, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Jun-Bo, L.; Yang, S.; Shan-Shan, T.; Rui-Fa, J. Theoretical and experimental research on the self-assembled system of molecularly imprinted polymers formed by salbutamol and methacrylic acid. J. Sep. Sci. 2015, 38, 1065–1071. [Google Scholar] [CrossRef] [PubMed]
- Karimian, N.; Gholivand, M.B.; Taherkhani, F. Computational design and development of a novel voltammetric sensor for minoxidil detection based on electropolymerized molecularly imprinted polymer. J. Electroanal. Chem. 2015, 740, 45–52. [Google Scholar] [CrossRef]
- Khan, M.S.; Pal, S.; Krupadam, R.J. Computational strategies for understanding the nature of interaction in dioxin imprinted nanoporous trappers. J. Mol. Recognit. 2015, 28, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Ktari, N.; Fourati, N.; Zerrouki, C.; Ruan, M.; Seydou, M.; Barbault, F.; Nal, F.; Yaakoubi, N.; Chehimi, M.M.; Kalfat, R. Design of a polypyrrole MIP-SAW sensor for selective detection of flumequine in aqueous media. Correlation between experimental results and DFT calculations. Rsc Adv. 2015, 5, 88666–88674. [Google Scholar] [CrossRef]
- Li, X.; He, Y.F.; Zhao, F.; Zhang, W.Y.; Ye, Z.L. Molecularly imprinted polymer-based sensors for atrazine detection by electropolymerization of o-phenylenediamine. Rsc Adv. 2015, 5, 56534–56540. [Google Scholar] [CrossRef]
- Liu, J.B.; Wang, Y.; Su, T.T.; Li, B.; Tang, S.S.; Jin, R.F. Theoretical and experimental studies on the performances of barbital-imprinted systems. J. Sep. Sci. 2015, 38, 4105–4110. [Google Scholar] [CrossRef]
- Ma, P.; Zhou, Z.; Yang, W.; Tang, B.; Liu, H.; Xu, W.; Huang, W. Preparation and application of sulfadiazine surface molecularly imprinted polymers with temperature-responsive properties. J. Appl. Polym. Sci. 2015, 132, 41769:1–41769:12. [Google Scholar] [CrossRef]
- Ma, P.F.; Yang, W.M.; Fan, T.; Liu, H.; Zhou, Z.P.; Li, J.H.; Zhang, L.; Xu, W.Z. Surface imprinted polymers for oil denitrification with the combination of computational simulation and multi-template molecular imprinting. Polym. Adv. Technol. 2015, 26, 476–486. [Google Scholar] [CrossRef]
- Maouche, N.; Ktari, N.; Bakas, I.; Fourati, N.; Zerrouki, C.; Seydou, M.; Maurel, F.; Chehimi, M.M. A surface acoustic wave sensor functionalized with a polypyrrole molecularly imprinted polymer for selective dopamine detection. J. Mol. Recognit. 2015, 28, 667–678. [Google Scholar] [CrossRef]
- Martins, N.; Carreiro, E.P.; Locati, A.; Ramalho, J.P.P.; Cabrita, M.J.; Burke, A.J.; Garcia, R. Design and development of molecularly imprinted polymers for the selective extraction of deltamethrin in olive oil: An integrated computational-assisted approach. J. Chromatogr. A 2015, 1409, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Mehamod, F.S.; KuBulat, K.; Yusof, N.F.; Othman, N.A. The development of molecular imprinting technology for caffeine extraction. Int. J. Technol. 2015, 6, 546–554. [Google Scholar] [CrossRef]
- Motghare, R.V.; Tadi, K.K.; Dhawale, P.; Deotare, S.; Kawadkar, A.K.; Chillawar, R.; Khan, S. Voltammetric determination of uric acid based on molecularly imprinted polymer modified carbon paste electrode. Electroanalysis 2015, 27, 825–832. [Google Scholar] [CrossRef]
- Muzyka, K.M. Theoretical study of energy characteristics of “artificial receptor” on melamine in pre-polymerization phase. J. Nano- Electron. Phys. 2015, 7, 01017:1–01017:5. [Google Scholar]
- Prasad, B.B.; Kumar, A. Development of molecularly imprinted polymer nanoarrays of N-acryloyl-2-mercaptobenzamide on a silver electrode for ultratrace sensing of uracil and 5-fluorouracil. J. Mater. Chem. B 2015, 3, 5864–5876. [Google Scholar] [CrossRef]
- Tadi, K.K.; Motghare, R.V.; Ganesh, V. Electrochemical detection of epinephrine using a biomimic made up of hemin modified molecularly imprinted microspheres. Rsc Adv. 2015, 5, 99115–99124. [Google Scholar] [CrossRef]
- Torkashvand, M.; Gholivand, M.B.; Taherkhani, F. Fabrication of an electrochemical sensor based on computationally designed molecularly imprinted polymer for the determination of mesalamine in real samples. Mater. Sci. Eng. C 2015, 55, 209–217. [Google Scholar] [CrossRef]
- Vallo, L.L.; Laxamana, R.T.; Paredes, F.U.; Arellano, I.H.J.; Arco, S.D. Harnessing non-covalent interactions in molecular traps for probable human carcinogen butylated hydroxyanisole. Mater. Lett. 2015, 159, 317–320. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, J.-b.; Tang, S.-s.; Dai, Z.-q.; Jin, R.-f. Theoretical research on self-assembly system of molecularly imprinted polymers formed by melamine and trifluoromethacrylic acid. Struct. Chem. 2015, 27, 897–905. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, J.B.; Tang, S.S.; Jin, R.F. Preparation of melamine molecularly imprinted polymer by computer-aided design. J. Sep. Sci. 2015, 38, 2647–2654. [Google Scholar] [CrossRef]
- Yu, X.B.; Liu, J.B.; Yuan, X. Simulation and preparation of molecular imprinting system with atrazine as template. Chin. J. Struct. Chem. 2015, 34, 488–496. [Google Scholar]
- Zhang, W.; Tan, N.; Jia, X.H.; Wang, G.P.; Long, W.; Li, X.L.; Liao, S.; Hou, D. Synthesis, recognition characteristics and properties of l-3-n-butylphthalide molecularly imprinted polymers as sorbent for solid-phase extraction through precipitation polymerization. Mater. Sci. Eng. C 2015, 53, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, T.; Bagherzadeh, A.; Shamkhali, A.N. Synthesis of nano-sized stereoselective imprinted polymer by copolymerization of (S)-2-(acrylamido) propanoic acid and ethylene glycol dimethacrylate in the presence of racemic propranolol and copper ion. Mater. Sci. Eng. C 2016, 63, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Ayankojo, A.G.; Tretjakoy, A.; Reut, J.; Boroznjak, R.; Öpik, A.; Rappich, J.; Furchner, A.; Hinrichs, K.; Syritski, V. Molecularly imprinted polymer integrated with a surface acoustic wave technique for detection of sulfamethizole. Anal. Chem. 2016, 88, 1476–1484. [Google Scholar] [CrossRef] [PubMed]
- Barros, L.A.; Custodio, R.; Rath, S. Design of a new molecularly imprinted polymer selective for hydrochlorothiazide based on theoretical predictions using gibbs free energy. J. Braz. Chem. Soc. 2016, 27, 2300–2311. [Google Scholar] [CrossRef]
- Bujak, R.; Gadzala-Kopciuch, R.; Nowaczyk, A.; Raczak-Gutknecht, J.; Kordalewska, M.; Struck-Lewicka, W.; Markuszewski, M.J.; Buszewski, B. Selective determination of cocaine and its metabolite benzoylecgonine in environmental samples by newly developed sorbent materials. Talanta 2016, 146, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, M.C.; Nascimento, C.S.; Borges, K.B. Theoretical investigation on functional monomer and solvent selection for molecular imprinting of tramadol. Chem. Phys. Lett. 2016, 645, 174–179. [Google Scholar] [CrossRef]
- Li, H.; He, H.L.; Huang, J.J.; Wang, C.Z.; Gu, X.L.; Gao, Y.K.; Zhang, H.J.; Du, S.H.; Chen, L.N.; Yuan, C.S. A novel molecularly imprinted method with computational simulation for the affinity isolation and knockout of baicalein from Scutellaria baicalensis. Biomed. Chromatogr. 2016, 30, 117–125. [Google Scholar] [CrossRef]
- Liang, D.; Wang, Y.; Li, S.; Li, Y.; Zhang, M.; Li, Y.; Tian, W.; Liu, J.; Tang, S.; Li, B.; et al. Study on dicyandiamide-imprinted polymers with computer-aided design. Int. J. Mol. Sci. 2016, 17, 1750. [Google Scholar] [CrossRef]
- Luo, X.P.; Li, C.Z.; Duan, Y.Q.; Zhang, H.H.; Zhang, D.; Zhang, C.; Sun, G.B.; Sun, X.B. Molecularly imprinted polymer prepared by Pickering emulsion polymerization for removal of acephate residues from contaminated waters. J. Appl. Polym. Sci. 2016, 133, 43126:1–43126:11. [Google Scholar] [CrossRef]
- Perez, M.; Concu, R.; Ornelas, M.; Cordeiro, M.; Azenha, M.; Silva, A.F. Measurement artifacts identified in the UV-vis spectroscopic study of adduct formation within the context of molecular imprinting of naproxen. Spectrochim. ActaPart A 2016, 153, 661–668. [Google Scholar] [CrossRef]
- Prasad, B.B.; Singh, R.; Kumar, A. Development of imprinted polyneutral red/electrochemically reduced graphene oxide composite for ultra-trace sensing of 6-thioguanine. Carbon 2016, 102, 86–96. [Google Scholar] [CrossRef]
- Qiu, X.Z.; Xu, X.Y.; Liang, Y.; Hua, Y.B.; Guo, H.S. Fabrication of a molecularly imprinted polymer immobilized membrane with nanopores and its application in determination of beta(2)-agonists in pork samples. J. Chromatogr. A 2016, 1429, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.S.; Wojnarowicz, A.; Sosnowska, M.; Benincori, T.; Noworyta, K.; D’Souza, F.; Kutner, W. Potentiometric chemosensor for neopterin, a cancer biomarker, using an electrochemically synthesized molecularly imprinted polymer as the recognition unit. Biosens. Bioelectron. 2016, 77, 565–572. [Google Scholar] [CrossRef]
- Tan, L.; He, R.; Li, Y.; Liang, Y.; Li, H.; Tang, Y. Fabrication of a biomimetic adsorbent imprinted with a common specificity determinant for the removal of alpha- and beta-amanitin from plasma. J. Chromatogr. A 2016, 1459, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Tiu, B.D.B.; Krupadam, R.J.; Advincula, R.C. Pyrene-imprinted polythiophene sensors for detection of polycyclic aromatic hydrocarbons. Sens. Actuators B 2016, 228, 693–701. [Google Scholar] [CrossRef]
- Wang, Y.W.; Zhao, T.; Dai, P.; Jiang, N.; Li, F. Employment of molecularly imprinted polymers to high-throughput screen nNOS-PSD-95 interruptions: Structure and dynamics investigations on monomer-template complexation. ChemPhysChem 2016, 17, 893–901. [Google Scholar] [CrossRef]
- Wojnarowicz, A.; Sharma, P.S.; Sosnowska, M.; Lisowski, W.; Huynh, T.P.; Pszona, M.; Borowicz, P.; D’Souza, F.; Kutner, W. An electropolymerized molecularly imprinted polymer for selective carnosine sensing with impedimetric capacity. J. Mater. Chem. B 2016, 4, 1156–1165. [Google Scholar] [CrossRef]
- Zhang, Y.; Wan, J.; Cao, X. Synthesis of surface molecularly imprinting polymers for cordycepin and its application in separating cordycepin. Process Biochem. 2016, 51, 517–527. [Google Scholar] [CrossRef]
- Nezhadali, A.; Mojarrab, M. Computational study and multivariate optimization of hydrochlorothiazide analysis using molecularly imprinted polymer electrochemical sensor based on carbon nanotube/polypyrrole film. Sens. Actuators B 2014, 190, 829–837. [Google Scholar] [CrossRef]
- Nezhadali, A.; Mojarrab, M. Fabrication of an electrochemical molecularly imprinted polymer triamterene sensor based on multivariate optimization using multi-walled carbon nanotubes. J. Electroanal. Chem. 2015, 744, 85–94. [Google Scholar] [CrossRef]
- Nezhadali, A.; Mojarrab, M. Computational design and multivariate optimization of an electrochemical metoprolol sensor based on molecular imprinting in combination with carbon nanotubes. Anal. Chim. Acta 2016, 924, 86–98. [Google Scholar] [CrossRef]
- Nezhadali, A.; Senobari, S.; Mojarrab, M. 1,4-dihydroxyanthraquinone electrochemical sensor based on molecularly imprinted polymer using multi-walled carbon nanotubes and multivariate optimization method. Talanta 2016, 146, 525–532. [Google Scholar] [CrossRef]
- Gholivand, M.B.; Shariati-Rad, M.; Karimian, N.; Torkashvand, M. A chemometrics approach for simultaneous determination of cyanazine and propazine based on a carbon paste electrode modified by a molecularly imprinted polymer. Analyst 2012, 137, 1190–1198. [Google Scholar] [CrossRef]
- Mamo, S.K.; Elie, M.; Baron, M.G.; Gonzalez-Rodriguez, J. Computationally designed perrhenate ion imprinted polymers for selective trapping of rhenium ions. Acs Appl. Polym. Mater. 2020, 2, 3135–3147. [Google Scholar] [CrossRef]
- Saputra, A.; Wijaya, K.; Armunanto, R.; Tania, L.; Tahir, I. Determination of effective functional monomer and solvent for R(+)-cathinone imprinted polymer using density functional theory and molecular dynamics simulation approaches. Indones. J. Chem. 2017, 17, 516–522. [Google Scholar] [CrossRef][Green Version]
- Yu, W.L.; Liu, M.X.; Liu, R.B.; Sang, Y.X.; Wang, S.; Wang, X.H. Development of biomimetic enzyme-linked immunosorbent assay based on molecular imprinting technique for semicarbazide detection. Food Agric. Immunol. 2020, 31, 17–32. [Google Scholar] [CrossRef]
- Yu, W.L.; Tang, Y.W.; Sang, Y.X.; Liu, W.H.; Wang, S.; Wang, X.H. Preparation of a carboxylated single-walled carbon-nanotube-chitosan functional layer and its application to a molecularly imprinted electrochemical sensor to quantify semicarbazide. Food Chem. 2020, 333, 127524:1–127524:7. [Google Scholar] [CrossRef]
- Mehamod, F.S.; Othman, N.A.; Bulat, K.H.K.; Suah, F.B.M. Pyrogallol-imprinted polymers with methyl methacrylate via precipitation polymerization. In Recent Advancement on Applied Physics, Industrial Chemistry and Chemical Technology; Yatim, N.M., Harun, F.W., Ali, E.S., Eds.; AIP Publishing LLC: Melville, NY, USA, 2018; Volume 1972, pp. 020003:1–020003:10. ISBN 978-0-7354-1679-6. [Google Scholar]
- Bartold, K.; Pietrzyk-Le, A.; Huynh, T.P.; Iskierko, Z.; Sosnowska, M.; Noworyta, K.; Lisowski, W.; Sannicolo, F.; Cauteruccio, S.; Licandro, E.; et al. Programmed transfer of sequence information into a molecularly imprinted polymer for hexakis(2,2’-bithien-5-yl) DNA analogue formation toward single-nucleotide-polymorphism detection. Acs Appl. Mater. Interfaces 2017, 9, 3948–3958. [Google Scholar] [CrossRef] [PubMed]
- Cairo, P.; De Luca, G.; Tocci, E.; Drioli, E. 110th anniversary: Selective recognition of 5-fluorouracil with molecular imprinting membranes: Molecular details. Ind. Eng. Chem. Res. 2019, 58, 15497–15505. [Google Scholar] [CrossRef]
- Chittratan, P.; Phromyothin, D.; Monvisade, P. DFT-based computational investigation on functional monomer and solvent selection of molecularly imprinted polymers for recognition of chlorpyrifos organophosphate insecticide. Suranaree J. Sci. Technol. 2020, 27, 030003:1–030003:6. [Google Scholar]
- Couto, R.A.S.; Mounssef, B.; Carvalho, F.; Rodrigues, C.M.P.; Braga, A.A.C.; Aldous, L.; Goncalves, L.M.; Quinaz, M.B. Methylone screening with electropolymerized molecularly imprinted polymer on screen-printed electrodes. Sens. Actuators B 2020, 316, 128133:1–128133:10. [Google Scholar] [CrossRef]
- de Oliveira, F.M.; da Costa, M.F.; Nascentes, C.C.; Teixeira Tarley, C.R. Design of high-performance adsorption cross-linked organic functional polymers towards tricyclic antidepressants using computational simulation. J. Environ. Chem. Eng. 2019, 7, 102849:1–102849:10. [Google Scholar] [CrossRef]
- Bartold, K.; Pietrzyk-Le, A.; Lisowski, W.; Golebiewska, K.; Siklitskaya, A.; Borowicz, P.; Shao, S.; D’Souza, F.; Kutner, W. Promoting bioanalytical concepts in genetics: A TATA box molecularly imprinted polymer as a small isolated fragment of the DNA damage repairing system. Mater. Sci. Eng. C 2019, 100, 1–10. [Google Scholar] [CrossRef]
- Clarindo, J.E.S.; Viana, R.B.; Cervini, P.; Silva, A.B.F.; Cavalheiro, E.T.G. Determination of tetracycline using a graphite-polyurethane composite electrode modified with a molecularly imprinted polymer. Anal. Lett. 2020, 53, 1932–1955. [Google Scholar] [CrossRef]
- de Freitas-Marques, M.B.; Araujo, B.C.R.; da Silva, P.H.R.; Fernandes, C.; Mussel, W.D.; Sebastiao, R.D.D.; Yoshida, M.I. Multilayer perceptron network and Vyazovkin method applied to the non-isothermal kinetic study of the interaction between lumefantrine and molecularly imprinted polymer. J. Therm. Anal. Calorim. 2020, 1–9. [Google Scholar] [CrossRef]
- De Rycke, E.; Trynda, A.; Jaworowicz, M.; Dubruel, P.; De Saeger, S.; Beloglazova, N. Capacitive sensing of an amphetamine drug precursor in aqueous samples: Application of novel molecularly imprinted polymers for benzyl methyl ketone detection. Biosens. Bioelectron. 2021, 172, 112773:1–112773:8. [Google Scholar] [CrossRef]
- Ding, S.; Li, Z.; Cheng, Y.; Du, C.; Gao, J.; Zhang, Y.W.; Zhang, N.; Li, Z.; Chang, N.; Hu, X. Enhancing adsorption capacity while maintaining specific recognition performance of mesoporous silica: A novel imprinting strategy with amphiphilic ionic liquid as surfactant. Nanotechnology 2018, 29, 375604:1–375604:13. [Google Scholar] [CrossRef]
- Fahim, A.M.; Wasiniak, B.; Lukaszewicz, J.P. Molecularly imprinted polymer and computational study of (e)-4-(2-cyano-3-(dimethylamino)acryloyl)benzoic acid from poly(ethylene tereph-thalate) plastic waste. Curr. Anal. Chem. 2020, 16, 119–137. [Google Scholar] [CrossRef]
- Gajda, M.; Rybakiewicz, R.; Cieplak, M.; Zolek, T.; Maciejewska, D.; Gilant, E.; Rudzki, P.J.; Grab, K.; Kutner, A.; Borowicz, P.; et al. Low-oxidation-potential thiophene-carbazole monomers for electro-oxidative molecular imprinting: Selective chemosensing of aripiprazole. Biosens. Bioelectron. 2020, 169, 112589:1–112589:10. [Google Scholar] [CrossRef]
- Ganjavi, F.; Ansari, M.; Kazemipour, M.; Zeidabadinejad, L. Computer-aided design and synthesis of a highly selective molecularly imprinted polymer for the extraction and determination of buprenorphine in biological fluids. J. Sep. Sci. 2017, 40, 3175–3182. [Google Scholar] [CrossRef] [PubMed]
- Ganjeizadeh Rohani, F.; Mohadesi, A.; Ansari, M. A new diosgenin sensor based on molecularly imprinted polymer of para aminobenzoic acid selected by computer-aided design. J. Pharm. Biomed. Anal. 2019, 174, 552–560. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Tang, S.S.; Liu, J.B.; Jin, R.F. Computational simulation and electrochemically deposited molecularly imprinted polymer-coated carbon glass electrode for triclosan detection. Chin. J. Struct. Chem. 2020, 39, 1252–1260. [Google Scholar] [CrossRef]
- Gao, Y.; Tan, N.; Wang, J.; He, D.X.; Ji, K.; Han, J.W.; Yan, X.M. Selective recognition and preliminary separation of hepatoprotective component silybin from milk thistle seeds by the prepared core-shell magnetic molecularly imprinted polymer. J. Polym. Res. 2018, 25, 150:1–150:14. [Google Scholar] [CrossRef]
- Garcia, R.; Carreiro, E.P.; Prates Ramalho, J.P.; Burke, A.J.; Lima, J.C.; Gomes da Silva, M.D.R.; Costa Freitas, A.M.; Cabrita, M.J. A photoswitchable "host-guest" approach for the selective enrichment of dimethoate from olive oil. Anal. Chim. Acta 2018, 1035, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Garcia, R.; Carreiro, E.P.; Prates Ramalho, J.P.; Mirao, J.; Burke, A.J.; Gomes da Silva, M.D.R.; Freitas, A.M.C.; Cabrita, M.J. A magnetic controllable tool for the selective enrichment of dimethoate from olive oil samples: A responsive molecular imprinting-based approach. Food Chem. 2018, 254, 309–316. [Google Scholar] [CrossRef]
- Gu, Y.; Wang, J.; Shi, H.; Pan, M.; Liu, B.; Fang, G.; Wang, S. Electrochemiluminescence sensor based on upconversion nanoparticles and oligoaniline-crosslinked gold nanoparticles imprinting recognition sites for the determination of dopamine. Biosens. Bioelectron. 2019, 128, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Hao, Q.Q.; Lu, L.L.; Kan, X.W. Probe and analogue: Double roles of thionine for aloe-emodin selective and sensitive ratiometric detection. Sens. Actuators B 2019, 292, 247–253. [Google Scholar] [CrossRef]
- He, C.; Lay, S.; Yu, H.; Shen, S. Synthesis and application of selective adsorbent for pirimicarb pesticides in aqueous media using allyl-beta-cyclodextrin based binary functional monomers. J. Sci. Food Agric. 2018, 98, 2089–2097. [Google Scholar] [CrossRef]
- Huang, X.S.; Zhang, W.C.; Wu, Z.Y.; Li, H.H.; Yang, C.Y.; Ma, W.R.; Hui, A.L.; Zeng, Q.M.; Xiong, B.Y.; Xian, Z.J. Computer simulation aided preparation of molecularly imprinted polymers for separation of bilobalide. J. Mol. Model. 2020, 26, 198:1–198:10. [Google Scholar] [CrossRef]
- Iskierko, Z.; Checinska, A.; Sharma, P.S.; Golebiewska, K.; Noworyta, K.; Borowicz, P.; Fronc, K.; Bandi, V.; D’Souza, F.; Kutner, W. Molecularly imprinted polymer based extended-gate field-effect transistor chemosensors for phenylalanine enantioselective sensing. J. Mater. Chem. C 2017, 5, 969–977. [Google Scholar] [CrossRef]
- Khan, M.S.; Pal, S. Quantum mechanical studies on dioxin-imprinted polymer precursor composites: Fundamental insights to enhance the binding strength and selectivity of biomarkers. J. Mol. Recognit. 2018, 31, 2736:1–2736:10. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, A.; Gupta, N.; Srivastava, J.; Singh, A.K.; Singh, M. Development of highly sensitive and selective sensor for ethionamide guided by molecular modelling via electropolymerized molecularly imprinted films. Microchem. J. 2020, 152, 104355:1–104355:10. [Google Scholar] [CrossRef]
- Leepheng, P.; Limthin, D.; Homchan, W.; Suramitr, S.; Phromyothin, D. An experimental and theoretical study of molecularly imprinted electrode based on methyl methacrylate polymer for pesticide detection. Jpn. J. Appl. Phys. 2020, 59, Siij09:1–Siij09:1. [Google Scholar] [CrossRef]
- Leibl, N.; Duma, L.; Gonzato, C.; Haupt, K. Polydopamine-based molecularly imprinted thin films for electro-chemical sensing of nitro-explosives in aqueous solutions. Bioelectrochemistry 2020, 135, 107541:1–107541:9. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Niu, X.H.; Wu, H.Y.; Meng, S.C.; Zhang, W.C.; Pan, J.M.; Qiu, F.X. Impedimetric enzyme-free detection of glucose via a computation-designed molecularly imprinted electrochemical sensor fabricated on porous Ni foam. Electroanalysis 2017, 29, 1243–1251. [Google Scholar] [CrossRef]
- Linh, C.N.; Duvanova, O.V.; Yen, V.H.; Zyablov, A.N.; Nesterenko, P.N. Modeling of butyric acid recognition by molecular imprinted polyimide. J. Mol. Model. 2020, 26, 194:1–194:7. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, Z.; Yang, L.; Fan, Y.; Liu, Y. Molecular structure and spectral characteristics of hyperoside and analysis of its molecular imprinting adsorption properties based on density functional theory. J. Mol. Graph. Model. 2019, 88, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.B.; Wang, G.Y.; Tang, S.S.; Gao, Q.; Liang, D.D.; Jin, R.F. Theoretical and experimental research on self-assembly system of molecularly imprinted polymers formed via chloramphenicol and methacrylic acid. J. Sep. Sci. 2019, 42, 769–777. [Google Scholar] [CrossRef]
- Liu, J.B.; Wang, Y.; Tang, S.S.; Gao, Q.; Jin, R.F. Theoretical guidance for experimental research of the dicyandiamide and methacrylic acid molecular imprinted polymer. New J. Chem. 2017, 41, 13370–13376. [Google Scholar] [CrossRef]
- Liu, J.B.; Zhao, W.S.; Tang, S.S.; Jin, R.F. Theoretical design and adsorption properties of molecularly imprinted polymers obtained from chloramphenicol and acrylamide. Chem. Res. Chin. Univ. 2019, 36, 915–920. [Google Scholar] [CrossRef]
- Liu, J.B.; Zhao, W.S.; Wang, G.Y.; Tang, S.S.; Jin, R.F. Simulation of a phenobarbital molecularly imprinted polymerization self-assembly system and its adsorption property. Anal. Methods 2020, 12, 813–821. [Google Scholar] [CrossRef]
- Liu, Y.J.; Liu, Y.C.; Liu, Z.M.; Zhao, X.C.; Wei, J.T.; Liu, H.C.; Si, X.X.; Xu, Z.G.; Cai, Z.W. Chiral molecularly imprinted polymeric stir bar sorptive extraction for naproxen enantiomer detection in PPCPs. J. Hazard. Mater. 2020, 392, 122251:1–122251:9. [Google Scholar] [CrossRef]
- Lu, C.; Tang, Z.; Gao, X.; Ma, X.; Liu, C. Computer-aided design of magnetic dummy molecularly imprinted polymers for solid-phase extraction of ten phthalates from food prior to their determination by GC-MS/MS. Mikrochim. Acta 2018, 185, 373:1–373:11. [Google Scholar] [CrossRef]
- Manickam, P.; Arizaleta, F.; Gurusamy, M.; Bhansali, S. Theoretical studies of cortisol-imprinted prepolymerization mixtures: Structural insights into improving the selectivity of affinity sensors. J. Electrochem. Soc. 2017, 164, B3077–B3080. [Google Scholar] [CrossRef]
- Mehdipour, M.; Ansari, M.; Pournamdari, M.; Zeidabadinejad, L.; Kazemipour, M. Selective extraction of organophosphorous pesticides in plasma by magnetic molecularly imprinted polymers with the aid of computational design. Anal. Methods 2018, 10, 4136–4142. [Google Scholar] [CrossRef]
- Mehdipour, M.; Ansari, M.; Pournamdari, M.; Zeidabadinejad, L.; Kazemipour, M. Selective extraction of malathion from biological fluids by molecularly imprinted polymer coated on spinel ZnFe2O4 magnetic nanoparticles based on green synthesis. Sep. Sci. Technol. 2020, 1–11. [Google Scholar] [CrossRef]
- Mesa, R.L.; Villa, J.E.L.; Khan, S.; Peixoto, R.R.A.; Morgano, M.A.; Goncalves, L.M.; Sotomayor, M.; Picasso, G. Rational design of an ion-imprinted polymer for aqueous methylmercury sorption. Nanomaterials 2020, 10, 2541. [Google Scholar] [CrossRef]
- Nagy-Szakolczai, A.; Svab-Kovacs, A.; Krezinger, A.; Toth, B.; Nyulaszi, L.; Horvai, G. The molecular imprinting effect of propranolol and dibenzylamine as model templates: Binding strength and selectivity. Anal. Chim. Acta 2020, 1125, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, T.A.; Silva, C.F.; de Oliveira, H.L.; da Silva, R.C.S.; Nascimento, C.S.; Borges, K.B. Magnetic molecularly imprinted conducting polymer for determination of praziquantel enantiomers in milk. Analyst 2020, 145, 4245–4253. [Google Scholar] [CrossRef] [PubMed]
- Nezhadali, A.; Sadeghzadeh, S. Experimental design-artificial neural network-genetic algorithm optimization and computer-assisted design of celecoxib molecularly imprinted polymer/carbon nanotube sensor. J. Electroanal. Chem. 2017, 795, 32–40. [Google Scholar] [CrossRef]
- Pereira, T.F.D.; da Silva, A.T.M.; Borges, K.B.; Nascimento, C.S. Carvedilol-imprinted polymer: Rational design and selectivity studies. J. Mol. Struct. 2019, 1177, 101–106. [Google Scholar] [CrossRef]
- Pogany, P.; Razali, M.; Szekely, G. Experimental and theoretical investigation of the complexation of methacrylic acid and diisopropyl urea. Spectrochim. ActaPart A 2017, 170, 69–76. [Google Scholar] [CrossRef]
- Rebelo, P.; Pacheco, J.G.; Cordeiro, M.N.D.S.; Melo, A.; Delerue-Matos, C. Azithromycin electrochemical detection using a molecularly imprinted polymer prepared on a disposable screen-printed electrode. Anal. Methods 2020, 12, 1486–1494. [Google Scholar] [CrossRef]
- Ren, X.; Yang, L.; Li, Y.; Cheshari, E.C.; Li, X. The integration of molecular imprinting and surface-enhanced Raman scattering for highly sensitive detection of lysozyme biomarker aided by density functional theory. Spectrochim. ActaPart A 2020, 228, 117764:1–117764:8. [Google Scholar] [CrossRef] [PubMed]
- Rohani, F.G.; Mohadesi, A.; Ansari, M. Computational design and electropolymerization of molecularly imprinted poly(p-aminobenzoic-acid-co-dapsone) using multivariate optimization for tetradifon residue analysis. ChemistrySelect 2019, 4, 12236–12244. [Google Scholar] [CrossRef]
- Sajini, T.; Thomas, R.; Mathew, B. Computational design and fabrication of enantioselective recognition sorbents for L-phenylalanine benzyl ester on multiwalled carbon nanotubes using molecular imprinting technology. Chin. J. Polym. Sci. 2019, 37, 1305–1318. [Google Scholar] [CrossRef]
- Salajegheh, M.; Ansari, M.; Foroghi, M.M.; Kazemipour, M. Computational design as a green approach for facile preparation of molecularly imprinted polyarginine-sodium alginate-multiwalled carbon nanotubes composite film on glassy carbon electrode for theophylline sensing. J. Pharm. Biomed. Anal. 2019, 162, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Salajegheh, M.; Kazemipour, M.; Foroghi, M.M.; Ansari, M. Morphine sensing by a green modified molecularly imprinted poly L-lysine/sodium alginate-activated carbon/glassy carbon electrode based on computational design. Electroanalysis 2019, 31, 468–476. [Google Scholar] [CrossRef]
- Sales, T.A.; Ramalho, T.C. Computational design of synthetic receptors for drug detection: Interaction between molecularly imprinted polymers and MDMA (3,4-methylenedioxymethamphetamine). Theor. Chem. Acc. 2020, 139, 31:1–31:12. [Google Scholar] [CrossRef]
- Silva, C.F.; Borges, K.B.; do Nascimento, C.S. Rational design of a molecularly imprinted polymer for dinotefuran: Theoretical and experimental studies aimed at the development of an efficient adsorbent for microextraction by packed sorbent. Analyst 2017, 143, 141–149. [Google Scholar] [CrossRef] [PubMed]
- So, J.; Pang, C.; Dong, H.X.; Jang, P.; Juhyok, U.; Ri, K.; Yun, C. Adsorption of 1-naphthyl methyl carbamate in water by utilizing a surface molecularly imprinted polymer. Chem. Phys. Lett. 2018, 699, 199–207. [Google Scholar] [CrossRef]
- Su, C.L.; Li, Z.Y.; Zhang, D.; Wang, Z.M.; Zhou, X.; Liao, L.F.; Xiao, X.L. A highly sensitive sensor based on a computer-designed magnetic molecularly imprinted membrane for the determination of acetaminophen. Biosens. Bioelectron. 2020, 148, 111819:1–111819:1. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Li, Y.X.; Deng, F.F.; Pan, X.H.; Yu, H.; Marina, M.L.; Jiang, Z.J. Highly sensitive determination of amanita toxins in biological samples using beta-cyclodextrin collaborated molecularly imprinted polymers coupled with ultra-high performance liquid chromatography tandem mass spectrometry. J. Chromatogr. A 2020, 1630, 461514:1–461514:10. [Google Scholar] [CrossRef]
- Tawab, M.; Abd El-Moghny, M.G.; El Nashar, R.M. Computational design of molecularly imprinted polymer for electrochemical sensing and stability indicating study of sofosbuvir. Microchem. J. 2020, 158, 105180:1–105180:11. [Google Scholar] [CrossRef]
- Tong, Z.H.; Han, Y.; Gu, L.L.; Li, Z.Y.; Du, K.; Kong, G.H.; Liu, D.H.; Peng, J.; Shi, J.L. Preparation and application of simetryn-imprinted nanoparticles in triazine herbicide residue analysis. J. Sep. Sci. 2020, 43, 1107–1118. [Google Scholar] [CrossRef]
- Wang, F.Y.; Ling, B.P.; Li, Q.J.; Abouhany, R. Dual roles of 3-aminopropyltriethoxysilane in preparing molecularly imprinted silica particles for specific recognition of target molecules. Rsc Adv. 2020, 10, 20368–20373. [Google Scholar] [CrossRef]
- Wang, G.Y.; Liu, J.B.; Tang, S.S.; Chang, H.B.; Jin, R.F. Theoretical researches on the self-assembly system of molecularly imprinted polymers formed via phenobarbital and acrylamide. Chin. J. Struct. Chem. 2019, 38, 1069–1078. [Google Scholar] [CrossRef]
- Wang, H.; Qian, D.; Xiao, X.; Gao, S.; Cheng, J.; He, B.; Liao, L.; Deng, J. A highly sensitive and selective sensor based on a graphene-coated carbon paste electrode modified with a computationally designed boron-embedded duplex molecularly imprinted hybrid membrane for the sensing of lamotrigine. Biosens. Bioelectron. 2017, 94, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, Y.H.; Zhou, B.B.; Zhong, M. Synthesis and characterization of core-shell magnetic molecularly imprinted polymers for selective extraction of allocryptopine from the wastewater of Macleaya cordata (Willd) R. Br. J. Mol. Recognit. 2020, 33, 2844:1–2844:11. [Google Scholar] [CrossRef]
- Wang, L.L.; Yang, F.J.; Zhao, X.H.; Li, Y.Z. Screening of functional monomers and solvents for the molecular imprinting of paclitaxel separation: A theoretical study. J. Mol. Model. 2020, 26, 26:1–26:10. [Google Scholar] [CrossRef]
- Wang, S.S.; Yang, B.W.; Zhu, Q.J. Configurational simulations and theoretical calculations of molecularly imprinted polymers of histamine and 2-(trifluoromethyl)acrylic acid based on computational chemistry. J. Chin. Chem. Soc. 2017, 64, 434–439. [Google Scholar] [CrossRef]
- Wang, X.S.; Zhao, W.S.; Liu, J.B.; Tang, S.S.; Jin, R.F. Study of self-assembly system of norfloxacin molecularly imprinted polymers based on simulated design. Theor. Chem. Acc. 2021, 140, 11:1–11:8. [Google Scholar] [CrossRef]
- Wu, H.Y.; Li, X.; Meng, S.C.; Xu, J.C.; Zhang, W.C.; Jiang, Y.; Qiu, F.X. A comprehensive theoretical study of structural optimization, interaction energies calculations and solvent effects between ractopamine and functional monomers in molecular imprinting polymers. Polym. Bull. 2018, 75, 1981–1996. [Google Scholar] [CrossRef]
- Wu, H.Y.; Tian, Q.; Zheng, W.; Jiang, Y.; Xu, J.C.; Li, X.; Zhang, W.C.; Qiu, F.X. Non-enzymatic glucose sensor based on molecularly imprinted polymer: A theoretical, strategy fabrication and application. J. Solid State Electrochem. 2019, 23, 1379–1388. [Google Scholar] [CrossRef]
- Wu, M.; Fan, Y.; Li, J.; Lu, D.; Guo, Y.; Xie, L.; Wu, Y. Vinyl phosphate-functionalized, magnetic, molecularly-imprinted polymeric microspheres’ enrichment and carbon dots’ fluorescence-detection of organophosphorus pesticide residues. Polymers 2019, 11, 1770. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.W.; Liu, J.; Huang, W.B.; He, Z.K.; Zhou, J.; Li, Y.C. Recognition mechanism of molecularly imprinted polymers by aggregation-induced emission. J. Mater. Chem. C 2020, 8, 13574–13581. [Google Scholar] [CrossRef]
- Xie, L.; Xiao, N.; Li, L.; Xie, X.N.; Li, Y. Theoretical insight into the interaction between chloramphenicol and functional monomer (methacrylic acid) in molecularly imprinted polymers. Int. J. Mol. Sci. 2020, 21, 4139. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Yao, R.; Shen, S. Development of a novel assay of molecularly imprinted membrane by design-based gaussian pattern for vancomycin determination. J. Pharm. Biomed. Anal. 2019, 175, 112789:1–112789:12. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zeng, H.N.; Wan, J.F.; Cao, X.J. Computational design of a molecularly imprinted polymer compatible with an aqueous environment for solid phase extraction of chenodeoxycholic acid. J. Chromatogr. A 2020, 1609, 460490:1–460490:10. [Google Scholar] [CrossRef]
- Zeng, H.N.; Yu, X.; Wan, J.F.; Cao, X.J. Rational design and synthesis of molecularly imprinted polymers (MIP) for purifying tylosin by seeded precipitation polymerization. Process Biochem. 2020, 94, 329–339. [Google Scholar] [CrossRef]
- Zhang, B.; Fan, X.; Zhao, D. Computer-aided design of molecularly imprinted polymers for simultaneous detection of clenbuterol and its metabolites. Polymers 2018, 11, 17. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, L.; Zhang, H.; Yang, Y.Z.; Liu, X.G. Recognition of 5-fluorouracil by thermosensitive magnetic surface molecularly imprinted microspheres designed using a computational approach. J. Appl. Polym. Sci. 2017, 134, 45468:1–45468:9. [Google Scholar] [CrossRef]
- Zhang, L.H.; He, L.H.; Wang, Q.; Tang, Q.L.; Liu, F. Theoretical and experimental studies of a novel electrochemical sensor based on molecularly imprinted polymer and GQDs-PtNPs nanocomposite. Microchem. J. 2020, 158, 105196:1–105196:11. [Google Scholar] [CrossRef]
- Zhang, P.P.; Ji, X.Y.; Zhang, H.X.; Xia, B.H. Quantum investigation into intermolecular interactions between bisphenol A and 2-viny1/4-vinylpyridine: Theoretical insight into molecular imprinting complexes. Comput. Theor. Chem. 2017, 1108, 76–85. [Google Scholar] [CrossRef]
- Zhou, Q.; Wang, M.Q.; Yagi, S.; Minami, T. Extended gate-type organic transistor functionalized by molecularly imprinted polymer for taurine detection. Nanoscale 2021, 13, 100–107. [Google Scholar] [CrossRef]
- He, H.; Gu, X.; Shi, L.; Hong, J.; Zhang, H.; Gao, Y.; Du, S.; Chen, L. Molecularly imprinted polymers based on SBA-15 for selective solid-phase extraction of baicalein from plasma samples. Anal. Bioanal. Chem. 2014, 407, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Azimi, A.; Javanbakht, M. Computational prediction and experimental selectivity coefficients for hydroxyzine and cetirizine molecularly imprinted polymer based potentiometric sensors. Anal. Chim. Acta 2014, 812, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zhu, K.; Zhao, M.; Li, Y. Theoretical and experimental study of nicotinamide molecularly imprinted polymers with different porogens. Anal. Chim. Acta 2005, 549, 39–44. [Google Scholar] [CrossRef]
- Gholivand, M.B.; Khodadadian, M.; Ahmadi, F. Computer aided-molecular design and synthesis of a high selective molecularly imprinted polymer for solid-phase extraction of furosemide from human plasma. Anal. Chim. Acta 2010, 658, 225–232. [Google Scholar] [CrossRef]
- Yao, J.; Li, X.; Qin, W. Computational design and synthesis of molecular imprinted polymers with high selectivity for removal of aniline from contaminated water. Anal. Chim. Acta 2008, 610, 282–288. [Google Scholar] [CrossRef]
- Appell, M.; Jackson, M.A.; Wang, L.C.; Ho, C.-H.; Mueller, A. Determination of fusaric acid in maize using molecularly imprinted SPE clean-up. J. Sep. Sci. 2013, 37, 281–286. [Google Scholar] [CrossRef]
- Azenha, M.; Kathirvel, P.; Nogueira, P.; Fernando-Silva, A. The requisite level of theory for the computational design of molecularly imprinted silica xerogels. Biosens. Bioelectron. 2008, 23, 1843–1849. [Google Scholar] [CrossRef] [PubMed]
- Nezhadali, A.; Shadmehri, R. Neuro-genetic multi-objective optimization and computer-aided design of pantoprazole molecularly imprinted polypyrrole sensor. Sens. Actuators B 2014, 202, 240–251. [Google Scholar] [CrossRef]
- Abdel Ghani, N.T.; Mohamed El Nashar, R.; Abdel-Haleem, F.M.; Madbouly, A. Computational design, synthesis and application of a new selective molecularly imprinted polymer for electrochemical detection. Electroanalysis 2016, 28, 1530–1538. [Google Scholar] [CrossRef]
- Attia, K.A.M.; El-Abasawi, N.M.; Abdel-Azim, A.H. Experimental design of membrane sensor for selective determination of phenazopyridine hydrochloride based on computational calculations. Mater. Sci. Eng. C 2016, 61, 773–781. [Google Scholar] [CrossRef]
- El Gohary, N.A.; Madbouly, A.; El Nashar, R.M.; Mizaikoff, B. Synthesis and application of a molecularly imprinted polymer for the voltammetric determination of famciclovir. Biosens. Bioelectron. 2015, 65, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Nezhadali, A.; Rouki, Z.; Nezhadali, M. Electrochemical preparation of a molecularly imprinted polypyrrole modified pencil graphite electrode for the determination of phenothiazine in model and real biological samples. Talanta 2015, 144, 456–465. [Google Scholar] [CrossRef]
- Pace, S.J.; Nguyen, E.; Baria, M.P.; Mojica, E.R.E. Use of computational modeling in preparation and evaluation of surface imprinted xerogels for binding tetracycline. Microchim. Acta 2015, 182, 69–76. [Google Scholar] [CrossRef]
- Saad, E.M.; Madbouly, A.; Ayoub, N.; El Nashar, R.M. Preparation and application of molecularly imprinted polymer for isolation of chicoric acid from Chicorium intybus L. medicinal plant. Anal. Chim. Acta 2015, 877, 80–89. [Google Scholar] [CrossRef]
- Yang, W.M.; Liu, L.K.; Ni, X.N.; Zhou, W.; Huang, W.H.; Liu, H.; Xu, W.Z. Computer-aided design and synthesis of magnetic molecularly imprinted polymers with high selectivity for the removal of phenol from water. J. Sep. Sci. 2016, 39, 503–517. [Google Scholar] [CrossRef]
- He, H.L.; Li, H.; Gao, Y.K.; Chen, D.; Shi, L.Y.; Peng, J.; Du, S.H.; Chen, L.N. A novel molecularly imprinted polymer for the solid-phase extraction of tanshinones from serum. Anal. Lett. 2015, 48, 47–60. [Google Scholar] [CrossRef]
- El Nashar, R.M.; Abdel Ghani, N.T.; El Gohary, N.A.; Barhoum, A.; Madbouly, A. Molecularly imprinted polymers based biomimetic sensors for mosapride citrate detection in biological fluids. Mater. Sci. Eng. C 2017, 76, 123–129. [Google Scholar] [CrossRef]
- Li, X.F.; Zhong, S.A.; Chen, L.; Whittaker, A. Computer simulation and preparation of molecularly imprinted polymer membranes with chlorogenic acid as template. Polym. Int. 2011, 60, 592–598. [Google Scholar] [CrossRef]
- Gao, Q.; Zang, Y.; Zhang, Y.; Xie, J.; Li, J.Y.; Gao, J.F.; Xue, H.G. Composite polymerized molecular imprinting membrane-based electrochemical sensor for sensitive determination of curcumin by using 4-pentenoyl-aminoacyl-chitosan oligosaccharide as functional monomer oligomer. J. Electroanal. Chem. 2020, 879, 114793:1–114793:8. [Google Scholar] [CrossRef]
- Ghani, N.T.A.; Abdulla, H.; Rizk, M.S.; Dena, A.S.A.; El Nashar, R.M. Molecularly imprinted polymer/reduced graphene oxide-based carbon-paste sensor for highly sensitive determination of the anti-HCV drug daclatasvir dihydrochloride. Sens. Actuators B 2019, 283, 6–17. [Google Scholar] [CrossRef]
- Gu, X.; Huang, J.; Zhang, L.; Zhang, Y.; Wang, C.Z.; Sun, C.; Yao, D.; Li, F.; Chen, L.; Yuan, C.S. Efficient discovery and capture of new neuronal nitric oxide synthase-postsynaptic density protein-95 uncouplers from herbal medicines using magnetic molecularly imprinted polymers as artificial antibodies. J. Sep. Sci. 2017, 40, 3522–3534. [Google Scholar] [CrossRef]
- Hammam, M.A.; Abdel-Halim, M.; Madbouly, A.; Wagdy, H.A.; El Nashar, R.M. Computational design of molecularly imprinted polymer for solid phase extraction of moxifloxacin hydrochloride from Avalox (R) tablets and spiked human urine samples. Microchem. J. 2019, 148, 51–56. [Google Scholar] [CrossRef]
- Korde, B.A.; Mankar, J.S.; Phule, S.; Krupadam, R.J. Nanoporous imprinted polymers (nanoMIPs) for controlled release of cancer drug. Mater. Sci. Eng. C 2019, 99, 222–230. [Google Scholar] [CrossRef]
- Mankar, J.S.; Sharma, M.D.; Krupadam, R.J. Molecularly imprinted nanoparticles (nanoMIPs): An efficient new adsorbent for removal of arsenic from water. J. Mater. Sci. 2020, 55, 6810–6825. [Google Scholar] [CrossRef]
- Saad, E.M.; El Gohary, N.A.; Abdel-Halim, M.; Handoussa, H.; El Nashar, R.M.; Mizaikoff, B. Molecularly imprinted polymers for selective extraction of rosmarinic acid from Rosmarinus officinalis L. Food Chem. 2021, 335, 127644:1–127644:10. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Huang, W.H.; Yin, X.F.; Sarpong, K.A.; Zhang, L.M.; Li, Y.C.; Zhao, S.; Zhou, H.D.; Yang, W.M.; Xu, W.Z. Computer-aided design and synthesis of molecular imprinting polymers based on doubly oriented functional multiwalled carbon nanotubes for electrochemically sensing bisphenol A. React. Funct. Polym. 2020, 157, 104767:1–104767:11. [Google Scholar] [CrossRef]
- Chen, L.; Lee, Y.K.; Manmana, Y.; Tay, K.S.; Lee, V.S.; Rahman, N.A. Synthesis, characterization, and theoretical study of an acrylamide-based magnetic molecularly imprinted polymer for the recognition of sulfonamide drugs. e-Polymers 2015, 15, 141–150. [Google Scholar] [CrossRef]
- Mayes, A.; Whitcombe, M. Synthetic strategies for the generation of molecularly imprinted organic polymers. Adv. Drug. Del. Rev. 2005, 57, 1742–1778. [Google Scholar] [CrossRef]
- Karim, K.; Breton, F.; Rouillon, R.; Piletska, E.; Guerreiro, A.; Chianella, I.; Piletsky, S. How to find effective functional monomers for effective molecularly imprinted polymers? Adv. Drug. Del. Rev. 2005, 57, 1795–1808. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Jakusch, M.; Mizaikoff, B. Capturing molecules with templated materials—analysis and rational design of molecularly imprinted polymers. Anal. Chim. Acta 2006, 578, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Piletsky, S.A.; Karim, K.; Piletska, E.V.; Turner, A.P.F.; Day, C.J.; Freebairn, K.W.; Legge, C. Recognition of ephedrine enantiomers by molecularly imprinted polymers designed using a computational approach. Analyst 2001, 126, 1826–1830. [Google Scholar] [CrossRef]
- Piletska, E.V.; Turner, N.W.; Turner, A.P.; Piletsky, S.A. Controlled release of the herbicide simazine from computationally designed molecularly imprinted polymers. J. Control. Release 2005, 108, 132–139. [Google Scholar] [CrossRef]
- Piletska, E.V.; Romero-Guerra, M.; Guerreiro, A.R.; Karim, K.; Turner, A.P.F.; Piletsky, S.A. Adaptation of the molecular imprinted polymers towards polar environment. Anal. Chim. Acta 2005, 542, 47–51. [Google Scholar] [CrossRef]
- Piletska, E.V.; Romero-Guerra, M.; Chianella, I.; Karim, K.; Turner, A.P.F.; Piletsky, S.A. Towards the development of multisensor for drugs of abuse based on molecular imprinted polymers. Anal. Chim. Acta 2005, 542, 111–117. [Google Scholar] [CrossRef]
- Subrahmanyam, S.; Piletsky, S.A.; Piletska, E.V.; Chen, B.; Karim, K.; Turner, A.P.F. ‘Bite-and-switch’ approach using computationally designed molecularly imprinted polymers for sensing of creatinine. Biosens. Bioelectron. 2001, 16, 631–637. [Google Scholar] [CrossRef]
- Piletska, E.; Piletsky, S.; Karim, K.; Terpetschnig, E.; Turner, A. Biotin-specific synthetic receptors prepared using molecular imprinting. Anal. Chim. Acta 2004, 504, 179–183. [Google Scholar] [CrossRef]
- Chianella, I.; Lotierzo, M.; Piletsky, S.A.; Tothill, I.E.; Chen, B.; Karim, K.; Turner, A.P.F. Rational design of a polymer specific for microcystin-LR using a computational approach. Anal. Chem. 2002, 74, 1288–1293. [Google Scholar] [CrossRef] [PubMed]
- Chianella, I.; Piletsky, S.A.; Tothill, I.E.; Chen, B.; Turner, A.P.F. MIP-based solid phase extraction cartridges combined with MIP-based sensors for the detection of microcystin-LR. Biosens. Bioelectron. 2003, 18, 119–127. [Google Scholar] [CrossRef]
- Bakas, I.; Ben Oujji, N.; Istamboulié, G.; Piletsky, S.; Piletska, E.; Ait-Addi, E.; Ait-Ichou, I.; Noguer, T.; Rouillon, R. Molecularly imprinted polymer cartridges coupled to high performance liquid chromatography (HPLC-UV) for simple and rapid analysis of fenthion in olive oil. Talanta 2014, 125, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Bakas, I.; Hayat, A.; Piletsky, S.; Piletska, E.; Chehimi, M.M.; Noguer, T.; Rouillon, R. Electrochemical impedimetric sensor based on molecularly imprinted polymers/sol–gel chemistry for methidathion organophosphorous insecticide recognition. Talanta 2014, 130, 294–298. [Google Scholar] [CrossRef] [PubMed]
- Abdin, M.J.; Altintas, Z.; Tothill, I.E. In silico designed nanoMIP based optical sensor for endotoxins monitoring. Biosens. Bioelectron. 2015, 67, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Altintas, Z.; Abdin, M.J.; Tothill, A.M.; Karim, K.; Tothill, I.E. Ultrasensitive detection of endotoxins using computationally designed nanoMIPs. Anal. Chim. Acta 2016, 935, 239–248. [Google Scholar] [CrossRef]
- Altintas, Z.; France, B.; Ortiz, J.O.; Tothill, I.E. Computationally modelled receptors for drug monitoring using an optical based biomimetic SPR sensor. Sens. Actuators B 2016, 224, 726–737. [Google Scholar] [CrossRef]
- Karim, K.; Giannoudi, L.; Piletska, E.; Chianella, I.; Henry, O.Y.F.; Laitenberger, P.; Piletsky, S.A.; Cowen, T. Development of MIP sensor for monitoring propofol in clinical procedures. J. Chin. Adv. Mater. Soc. 2015, 3, 149–160. [Google Scholar] [CrossRef]
- Mistry, J.; Guerreiro, A.; Moczko, E.; Piletska, E.; Karim, K.; Piletsky, S.A. Analysis of cooperative interactions in molecularly imprinted polymer nanoparticles. Mol. Impr. 2015, 3, 55–64. [Google Scholar] [CrossRef]
- Piletska, E.V.; Abd, B.H.; Krakowiak, A.S.; Parmar, A.; Pink, D.L.; Wall, K.S.; Wharton, L.; Moczko, E.; Whitcombe, M.J.; Karim, K.; et al. Magnetic high throughput screening system for the development of nano-sized molecularly imprinted polymers for controlled delivery of curcumin. Analyst 2015, 140, 3113–3120. [Google Scholar] [CrossRef]
- Rodriguez-Dorado, R.; Carro, A.M.; Chianella, I.; Karim, K.; Concheiro, A.; Lorenzo, R.A.; Piletsky, S.; Alvarez-Lorenzo, C. Oxytetracycline recovery from aqueous media using computationally designed molecularly imprinted polymers. Anal. Bioanal. Chem. 2016, 408, 6845–6856. [Google Scholar] [CrossRef] [PubMed]
- Tsyrulneva, I.; Zaporozhets, O.; Piletska, E.; Piletsky, S. Molecular modelling and synthesis of a polymer for the extraction of amiloride and triamterene from human urine. Anal. Methods 2014, 6, 3429–3435. [Google Scholar] [CrossRef]
- Wren, S.P.; Piletsky, S.A.; Karim, K.; Gascoine, P.; Lacey, R.; Sun, T.; Grattan, K.T.V. Computational design and fabrication of optical fibre fluorescent chemical probes for the detection of cocaine. J. Lightwave Technol. 2015, 33, 2572–2579. [Google Scholar] [CrossRef]
- Xi, S.; Zhang, K.; Xiao, D.; He, H. Computational-aided design of magnetic ultra-thin dummy molecularly imprinted polymer for selective extraction and determination of morphine from urine by high-performance liquid chromatography. J. Chromatogr. A 2016, 1473, 1–9. [Google Scholar] [CrossRef]
- Breton, F.; Rouillon, R.; Piletska, E.V.; Karim, K.; Guerreiro, A.; Chianella, I.; Piletsky, S.A. Virtual imprinting as a tool to design efficient MIPs for photosynthesis-inhibiting herbicides. Biosens. Bioelectron. 2007, 22, 1948–1954. [Google Scholar] [CrossRef]
- Chianella, I.; Karim, K.; Piletska, E.V.; Preston, C.; Piletsky, S.A. Computational design and synthesis of molecularly imprinted polymers with high binding capacity for pharmaceutical applications-model case: Adsorbent for abacavir. Anal. Chim. Acta 2006, 559, 73–78. [Google Scholar] [CrossRef]
- Guerreiro, A.; Soares, A.; Piletska, E.; Mattiasson, B.; Piletsky, S. Preliminary evaluation of new polymer matrix for solid-phase extraction of nonylphenol from water samples. Anal. Chim. Acta 2008, 612, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Yañez, F.; Chianella, I.; Piletsky, S.A.; Concheiro, A.; Alvarez-Lorenzo, C. Computational modeling and molecular imprinting for the development of acrylic polymers with high affinity for bile salts. Anal. Chim. Acta 2010, 659, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Bates, F.; Busato, M.; Piletska, E.; Whitcombe, M.J.; Karim, K.; Guerreiro, A.; del Valle, M.; Giorgetti, A.; Piletsky, S. Computational design of molecularly imprinted polymer for direct detection of melamine in milk. Sep. Sci. Technol. 2017, 52, 1441–1453. [Google Scholar] [CrossRef]
- Viveiros, R.; Karim, K.; Piletsky, S.A.; Heggie, W.; Casimiro, T. Development of a molecularly imprinted polymer for a pharmaceutical impurity in supercritical CO2: Rational design using computational approach. J. Clean. Prod. 2017, 168, 1025–1031. [Google Scholar] [CrossRef]
- Wei, S.; Jakusch, M.; Mizaikoff, B. Investigating the mechanisms of 17β-estradiol imprinting by computational prediction and spectroscopic analysis. Anal. Bioanal. Chem. 2007, 389, 423–431. [Google Scholar] [CrossRef]
- Pavel, D.; Lagowski, J. Computationally designed monomers and polymers for molecular imprinting of theophylline and its derivatives. Part I. Polymer 2005, 46, 7528–7542. [Google Scholar] [CrossRef]
- Pavel, D.; Lagowski, J. Computationally designed monomers and polymers for molecular imprinting of theophylline—part II. Polymer 2005, 46, 7543–7556. [Google Scholar] [CrossRef]
- Pavel, D.; Lagowski, J.; Lepage, C.J. Computationally designed monomers for molecular imprinting of chemical warfare agents—Part V. Polymer 2006, 47, 8389–8399. [Google Scholar] [CrossRef]
- Lv, Y.; Lin, Z.; Tan, T.; Feng, W.; Qin, P.; Li, C. Application of molecular dynamics modeling for the prediction of selective adsorption properties of dimethoate imprinting polymer. Sens. Actuators B 2008, 133, 15–23. [Google Scholar] [CrossRef]
- Molinelli, A.; O’Mahony, J.; Nolan, K.; Smyth, M.R.; Jakusch, M.; Mizaikoff, B. Analyzing the mechanisms of selectivity in biomimetic self-assemblies via IR and NMR spectroscopy of prepolymerization solutions and molecular dynamics simulations. Anal. Chem. 2005, 77, 5196–5204. [Google Scholar] [CrossRef]
- Monti, S.; Cappelli, C.; Bronco, S.; Giusti, P.; Ciardelli, G. Towards the design of highly selective recognition sites into molecular imprinting polymers: A computational approach. Biosens. Bioelectron. 2006, 22, 153–163. [Google Scholar] [CrossRef]
- Yoshida, M.; Hatate, Y.; Uezu, K.; Goto, M.; Furusaki, S. Chiral-recognition polymer prepared by surface molecular imprinting technique. Colloids Surf. A 2000, 169, 259–269. [Google Scholar] [CrossRef]
- Toorisaka, E.; Uezu, K.; Goto, M.; Furusaki, S. A molecularly imprinted polymer that shows enzymatic activity. Biochem. Eng. J. 2003, 14, 85–91. [Google Scholar] [CrossRef]
- Zhang, K.; Zou, W.Y.; Zhao, H.Y.; Dramou, P.; Pham-Huy, C.; He, J.; He, H. Adsorption behavior of a computer-aid designed magnetic molecularly imprinted polymer via response surface methodology. Rsc Adv. 2015, 5, 61161–61169. [Google Scholar] [CrossRef]
- Farrington, K.; Magner, E.; Regan, F. Predicting the performance of molecularly imprinted polymers: Selective extraction of caffeine by molecularly imprinted solid phase extraction. Anal. Chim. Acta 2006, 566, 60–68. [Google Scholar] [CrossRef]
- Chen, J.; Lewis, C.; Balamurugan, D.; Yang, Z.; Ai, L.; Cai, D. Theoretical analysis of a high performance protein imprint on a nanosensor. Sens. Bio-Sens. Res. 2016, 7, 12–19. [Google Scholar] [CrossRef]
- Wang, C.L.; Hu, X.L.; Guan, P.; Qian, L.W.; Wu, D.F.; Li, J. Thymopentin magnetic molecularly imprinted polymers with room temperature ionic liquids as a functional monomer by surface-initiated ATRP. Int. J. Polym. Anal. Charact. 2014, 19, 70–82. [Google Scholar] [CrossRef]
- Wang, C.L.; Hu, X.L.; Guan, P.; Qian, L.W.; Wu, D.F.; Li, J. Superparamagnetic molecularly imprinting polymers for adsorbent and separation pentapeptides by surface ATRP. Sep. Sci. Technol. 2015, 50, 1768–1775. [Google Scholar] [CrossRef]
- Kong, Y.; Wang, N.W.; Ni, X.N.; Yu, Q.Y.; Liu, H.; Huang, W.H.; Xu, W.Z. Molecular dynamics simulations of molecularly imprinted polymer approaches to the preparation of selective materials to remove norfloxacin. J. Appl. Polym. Sci. 2016, 133, 42817:1–42817:11. [Google Scholar] [CrossRef]
- Li, J.; Hu, X.L.; Guan, P.; Song, D.M.; Qian, L.W.; Du, C.B.; Song, R.Y.; Wang, C.L. Preparation of "dummy" L-phenylalanine molecularly imprinted microspheres by using ionic liquid as a template and functional monomer. J. Sep. Sci. 2015, 38, 3279–3287. [Google Scholar] [CrossRef]
- Wang, C.L.; Hu, X.L.; Guan, P.; Wu, D.F.; Qian, L.W.; Li, J.; Song, R.Y. Separation and purification of thymopentin with molecular imprinting membrane by solid phase extraction disks. J. Pharm. Biomed. Anal. 2015, 102, 137–143. [Google Scholar] [CrossRef]
- Bitar, M.; Bou-Maroun, E.; Lerbret, A.; Ouaini, N.; Cayot, P. Binding characteristics of molecularly imprinted polymers based on fungicides in hydroalcoholic media. J. Sep. Sci. 2015, 38, 3607–3614. [Google Scholar] [CrossRef]
- Hou, S.; Wang, Y.F.; Liu, N.; Liu, J. Preparation and recognition characteristics of thymopentin molecularly imprinted polymers on SiO2. Adsorpt. Sci. Technol. 2014, 32, 833–843. [Google Scholar] [CrossRef]
- Bird, L.; Herdes, C. The porogen effect on the complexation step of trinitrotoluene-methacrylic acid: Towards efficient imprinted polymer sensors. Mol. Syst. Des. Eng. 2018, 3, 89–95. [Google Scholar] [CrossRef]
- Eroglu, B.; Dalgakiran, D.; Inan, T.; Kurkcuoglu, O.; Guner, F.S. A computational and experimental approach to develop minocycline-imprinted hydrogels and determination of their drug delivery performances. J. Polym. Res. 2018, 25, 258:1–258:10. [Google Scholar] [CrossRef]
- Fizir, M.; Wei, L.; Muchuan, N.; Itatahine, A.; Mehdi, Y.A.; He, H.; Dramou, P. QbD approach by computer aided design and response surface methodology for molecularly imprinted polymer based on magnetic halloysite nanotubes for extraction of norfloxacin from real samples. Talanta 2018, 184, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Gore, P.M.; Khurana, L.; Siddique, S.; Panicker, A.; Kandasubramanian, B. Ion-imprinted electrospun nanofibers of chitosan/1-butyl-3-methylimidazolium tetrafluoroborate for the dynamic expulsion of thorium (IV) ions from mimicked effluents. Environ. Sci. Pollut. Res. 2018, 25, 3320–3334. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Liang, J.J.; Chen, L.X.; Chen, S.L.; Zheng, H.L.; Liu, H.X.; Zhang, H.J. Removal of the environmental pollutant carbamazepine using molecular imprinted adsorbents: Molecular simulation, adsorption properties, and mechanisms. Water Res. 2020, 168, 115164:1–115164:13. [Google Scholar] [CrossRef] [PubMed]
- Li, G.Y.; Zhang, K.; Fizir, M.; Niu, M.C.; Sun, C.; Xi, S.L.; Hui, X.H.; Shi, J.R.; He, H. Rational design, preparation and adsorption study of a magnetic molecularly imprinted polymer using a dummy template and a bifunctional monomer. New J. Chem. 2017, 41, 7092–7101. [Google Scholar] [CrossRef]
- Liu, W.H.; Wang, J.; Yu, W.L.; Wang, X.H. Study on a biomimetic enzyme-linked immunosorbent assay for rapid detection of flumequine in animal foods. Food Anal. Methods 2020, 13, 403–411. [Google Scholar] [CrossRef]
- Lopez, A.S.; Ramos, M.P.; Herrero, R.; Vilarino, J.M.L. Design, synthesis and HR—MAS NMR characterization of molecular imprinted polymers with emerging contaminants templates. Sep. Purif. Technol. 2021, 257, 117860:1–117860:7. [Google Scholar] [CrossRef]
- Madikizela, L.M.; Zunngu, S.S.; Mlunguza, N.Y.; Tavengwa, N.T.; Mdluli, P.S.; Chimuka, L. Application of molecularly imprinted polymer designed for the selective extraction of ketoprofen from wastewater. Water SA 2018, 44, 406–418. [Google Scholar] [CrossRef]
- Mazouz, Z.; Mokni, M.; Fourati, N.; Zerrouki, C.; Barbault, F.; Seydou, M.; Kalfat, R.; Yaakoubi, N.; Omezzine, A.; Bouslema, A.; et al. Computational approach and electrochemical measurements for protein detection with MIP-based sensor. Biosens. Bioelectron. 2020, 151, 111978:1–111978:8. [Google Scholar] [CrossRef] [PubMed]
- Nezammahalleh, H.; Mousavizadeh, S.H.; Babaeipour, V. New potentiometric sensor based on molecularly imprinted polymer for dipicolinic acid detection in aqueous media. IEEE Sens. J. 2018, 18, 7520–7528. [Google Scholar] [CrossRef]
- Niu, M.C.; Sun, C.; Zhang, K.; Li, G.Y.; Meriem, F.; Pham-Huy, C.; Hui, X.H.; Shi, J.R.; He, H. A simple extraction method for norfloxacin from pharmaceutical wastewater with a magnetic core-shell molecularly imprinted polymer with the aid of computer simulation. New J. Chem. 2017, 41, 2614–2624. [Google Scholar] [CrossRef]
- Paredes-Ramos, M.; Sabin-Lopez, A.; Pena-Garcia, J.; Perez-Sanchez, H.; Lopez-Vilarino, J.M.; de Vicente, M.E.S. Computational aided acetaminophen—Phthalic acid molecularly imprinted polymer design for analytical determination of known and new developed recreational drugs. J. Mol. Graph. Model. 2020, 100, 107627:1–107627:8. [Google Scholar] [CrossRef] [PubMed]
- Sobiech, M.; Giebultowicz, J.; Lulinski, P. Theoretical and experimental proof for selective response of imprinted sorbent—Analysis of hordenine in human urine. J. Chromatogr. A 2020, 1613, 460677:1–460677:13. [Google Scholar] [CrossRef]
- Sobiech, M.; Lulinski, P.; Halik, P.; Maciejewska, D. The selective response of a templated polymer for the cationic drug pentamidine: Implications from molecular simulations and experimental data. Rsc Adv. 2017, 7, 46881–46893. [Google Scholar] [CrossRef]
- Wagner, S.; Zapata, C.; Wan, W.; Gawlitza, K.; Weber, M.; Rurack, K. Role of counterions in molecularly imprinted polymers for anionic species. Langmuir 2018, 34, 6963–6975. [Google Scholar] [CrossRef]
- Wang, Y.F.; Ma, Y.; Zhou, J.J.; Su, K.H.; Zhang, B.L.; Zhang, Q.Y. Thermo-sensitive surface molecularly imprinted magnetic microspheres based on bio-macromolecules and their specific recognition of bovine serum albumin. J. Sep. Sci. 2020, 43, 996–1002. [Google Scholar] [CrossRef]
- Xu, L.; Zhao, Z.X.; Huang, Y.A.; Zhu, Q.J. Preparation of chitosan molecularly imprinted polymers and the recognition mechanism for adsorption of alpha-lipoic acid. Molecules 2020, 25, 312. [Google Scholar] [CrossRef]
- Zhang, Y.Q.; Tan, X.; Liu, X.; Li, C.S.; Zeng, S.J.; Wang, H.; Zhang, S.J. Fabrication of multilayered molecularly imprinted membrane for selective recognition and separation of artemisinin. Acs Sustain. Chem. Eng. 2019, 7, 3127–3137. [Google Scholar] [CrossRef]
- Liu, R.; Li, X.; Li, Y.; Jin, P.; Qin, W.; Qi, J. Effective removal of rhodamine B from contaminated water using non-covalent imprinted microspheres designed by computational approach. Biosens. Bioelectron. 2009, 25, 629–634. [Google Scholar] [CrossRef]
- Li, Y.; Li, X.; Li, Y.; Dong, C.; Jin, P.; Qi, J. Selective recognition of veterinary drugs residues by artificial antibodies designed using a computational approach. Biomaterials 2009, 30, 3205–3211. [Google Scholar] [CrossRef] [PubMed]
- Douhaya, Y.V.; Barkaline, V.V.; Tsakalof, A. Computer-simulation-based selection of optimal monomer for imprinting of tri-O-acetyl adenosine in a polymer matrix: Calculations for benzene solution. J. Mol. Model. 2016, 22, 154:1–154:8. [Google Scholar] [CrossRef]
- O’Mahony, J.; Karlsson, B.C.G.; Mizaikoff, B.; Nicholls, I.A. Correlated theoretical, spectroscopic and X-ray crystallographic studies of a non-covalent molecularly imprinted polymerisation system. Analyst 2007, 132, 1161–1168. [Google Scholar] [CrossRef]
- Karlsson, B.C.G.; O’Mahony, J.; Karlsson, J.G.; Bengtsson, H.; Eriksson, L.A.; Nicholls, I.A. Structure and dynamics of monomer−template complexation: An explanation for molecularly imprinted polymer recognition site heterogeneity. J. Am. Chem. Soc. 2009, 131, 13297–13304. [Google Scholar] [CrossRef] [PubMed]
- O’Mahony, J.; Moloney, M.; McCormack, M.; Nicholls, I.A.; Mizaikoff, B.; Danaher, M. Design and implementation of an imprinted material for the extraction of the endocrine disruptor bisphenol A from milk. J. Chromatogr. B 2013, 931, 164–169. [Google Scholar] [CrossRef]
- Henschel, H.; Kirsch, N.; Hedin-Dahlström, J.; Whitcombe, M.J.; Wikman, S.; Nicholls, I.A. Effect of the cross-linker on the general performance and temperature dependent behaviour of a molecularly imprinted polymer catalyst of a Diels–Alder reaction. J. Mol. Catal. B Enzym. 2011, 72, 199–205. [Google Scholar] [CrossRef]
- Olsson, G.D.; Karlsson, B.C.G.; Schillinger, E.; Sellergren, B.; Nicholls, I.A. Theoretical studies of 17-β-estradiol-imprinted prepolymerization mixtures: Insights concerning the roles of cross-linking and functional monomers in template complexation and polymerization. Ind. Eng. Chem. Res. 2013, 52, 13965–13970. [Google Scholar] [CrossRef]
- Schillinger, E.; Möder, M.; Olsson, G.D.; Nicholls, I.A.; Sellergren, B. An artificial estrogen receptor through combinatorial imprinting. Chem. Eur. J. 2012, 18, 14773–14783. [Google Scholar] [CrossRef]
- Shoravi, S.; Olsson, G.D.; Karlsson, B.C.G.; Bexborn, F.; Abghoui, Y.; Hussain, J.; Wiklander, J.G.; Nicholls, I.A. In silico screening of molecular imprinting prepolymerization systems: Oseltamivir selective polymers through full-system molecular dynamics-based studies. Org. Biomol. Chem. 2016, 14, 4210–4219. [Google Scholar] [CrossRef]
- Golker, K.; Karlsson, B.C.G.; Olsson, G.D.; Rosengren, A.M.; Nicholls, I.A. Influence of composition and morphology on template recognition in molecularly imprinted polymers. Macromolecules 2013, 46, 1408–1414. [Google Scholar] [CrossRef]
- Golker, K.; Karlsson, B.C.G.; Rosengren, A.M.; Nicholls, I.A. A functional monomer is not enough: Principal component analysis of the influence of template complexation in pre-polymerization mixtures on imprinted polymer recognition and morphology. Int. J. Mol. Sci. 2014, 15, 20572–20584. [Google Scholar] [CrossRef] [PubMed]
- Cleland, D.; Olsson, G.D.; Karlsson, B.C.G.; Nicholls, I.A.; McCluskey, A. Molecular dynamics approaches to the design and synthesis of PCB targeting molecularly imprinted polymers: Interference to monomer–template interactions in imprinting of 1,2,3-trichlorobenzene. Org. Biomol. Chem. 2014, 12, 844–853. [Google Scholar] [CrossRef] [PubMed]
- Azenha, M.; Szefczyk, B.; Loureiro, D.; Kathirvel, P.; DS Cordeiro, M.N.; Fernando-Silva, A. Computational and experimental study of the effect of PEG in the preparation of damascenone-imprinted xerogels. Langmuir 2013, 29, 2024–2032. [Google Scholar] [CrossRef]
- Golker, K.; Karlsson, B.C.G.; Wiklander, J.G.; Rosengren, A.M.; Nicholls, I.A. Hydrogen bond diversity in the pre-polymerization stage contributes to morphology and MIP-template recognition—MAA versus MMA. Eur. Polym. J. 2015, 66, 558–568. [Google Scholar] [CrossRef]
- Golker, K.; Nicholls, I.A. The effect of crosslinking density on molecularly imprinted polymer morphology and recognition. Eur. Polym. J. 2016, 75, 423–430. [Google Scholar] [CrossRef]
- Qiu, C.X.; Xing, Y.H.; Yang, W.M.; Zhou, Z.P.; Wang, Y.C.; Liu, H.; Xu, W.Z. Surface molecular imprinting on hybrid SiO2-coated CdTe nanocrystals for selective optosensing of bisphenol A and its optimal design. Appl. Surf. Sci. 2015, 345, 405–417. [Google Scholar] [CrossRef]
- Qiu, C.X.; Yang, W.M.; Zhou, Z.P.; Yan, Y.S.; Xu, W.Z. Rational design and preparation of dibenzothiophene-targeting molecularly imprinted polymers with molecular dynamics approaches and surface-initiated activators regenerated by electron-transfer atom-transfer radical polymerization. J. Appl. Polym. Sci. 2015, 132, 42629:1–42629:15. [Google Scholar] [CrossRef]
- Wang, Y.C.; Wang, N.W.; Ni, X.N.; Jiang, Q.Q.; Yang, W.M.; Huang, W.H.; Xu, W.Z. A core-shell CdTe quantum dots molecularly imprinted polymer for recognizing and detecting p-nitrophenol based on computer simulation. Rsc Adv. 2015, 5, 73424–73433. [Google Scholar] [CrossRef]
- Concu, R.; Cordeiro, M.N.D.S. Molecular dynamics simulation study of the selectivity of a silica polymer for ibuprofen. Int. J. Mol. Sci. 2016, 17, 1083. [Google Scholar] [CrossRef]
- Golker, K.; Olsson, G.D.; Nicholls, I.A. The influence of a methyl substituent on molecularly imprinted polymer morphology and recognition—Acrylic acid versus methacrylic acid. Eur. Polym. J. 2017, 92, 137–149. [Google Scholar] [CrossRef]
- Xu, W.; Wang, Y.; Huang, W.; Yu, L.; Yang, Y.; Liu, H.; Yang, W. Computer-aided design and synthesis of CdTe@SiO2 core-shell molecularly imprinted polymers as a fluorescent sensor for the selective determination of sulfamethoxazole in milk and lake water. J. Sep. Sci. 2017, 40, 1091–1098. [Google Scholar] [CrossRef]
- Altintas, Z.; Takiden, A.; Utesch, T.; Mroginski, M.A.; Schmid, B.; Scheller, F.W.; Sussmuth, R.D. Integrated approaches toward high-affinity artificial protein binders obtained via computationally simulated epitopes for protein recognition. Adv. Funct. Mater. 2019, 29, 1807332:1–1807332:11. [Google Scholar] [CrossRef]
- Gomez-Arribas, L.N.; Darder, M.D.; Garcia, N.; Rodriguez, Y.; Urraca, J.L.; Moreno-Bondi, M.C. Hierarchically imprinted polymer for peptide tag recognition based on an oriented surface epitope approach. Acs Appl. Mater. Interfaces 2020, 12, 49111–49121. [Google Scholar] [CrossRef]
- Sullivan, M.V.; Dennison, S.R.; Archontis, G.; Reddy, S.M.; Hayes, J.M. Toward rational design of selective molecularly imprinted polymers (MIPs) for proteins: Computational and experimental studies of acrylamide based polymers for myoglobin. J. Phys. Chem. B 2019, 123, 5432–5443. [Google Scholar] [CrossRef] [PubMed]
- Sukjee, W.; Tancharoen, C.; Yenchitsomanus, P.T.; Gleeson, M.P.; Sangma, C. Small-molecule dengue virus co-imprinting and its application as an electrochemical sensor. ChemistryOpen 2017, 6, 340–344. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; He, X.; Cui, P.L.; Liu, J.; Li, Z.B.; Jia, B.J.; Zhang, T.; Wang, J.P.; Yuan, W.Z. Preparation of a chemiluminescence sensor for multi-detection of benzimidazoles in meat based on molecularly imprinted polymer. Food Chem. 2019, 280, 103–109. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Wang, G.N.; Liu, J.X.; Zhao, W.L.; Huang, J.J.; Xu, M.X.; Wang, J.P.; Liu, J. Dummy molecularly imprinted polymer based microplate chemiluminescence sensor for one-step detection of Sudan dyes in egg. Food Chem. 2019, 288, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.J.; Liu, J.; Liu, J.X.; Wang, J.P. A microtitre chemiluminescence sensor for detection of pyrethroids based on dual-dummy-template molecularly imprinted polymer and computational simulation. Luminescence 2020, 35, 120–128. [Google Scholar] [CrossRef]
- Li, Z.B.; Liu, J.; Liu, J.X.; Wang, Z.H.; Wang, J.P. Determination of sulfonamides in meat with dummy-template molecularly imprinted polymer-based chemiluminescence sensor. Anal. Bioanal. Chem. 2019, 411, 3179–3189. [Google Scholar] [CrossRef]
- Song, Y.P.; Li, N.; Zhang, H.C.; Wang, G.N.; Liu, J.X.; Liu, J.; Wang, J.P. Dummy template molecularly imprinted polymer for solid phase extraction of phenothiazines in meat based on computational simulation. Food Chem. 2017, 233, 422–428. [Google Scholar] [CrossRef]
- Song, Y.P.; Zhang, L.; Wang, G.N.; Liu, J.X.; Liu, J.; Wang, J.P. Dual-dummy-template molecularly imprinted polymer combining ultra performance liquid chromatography for determination of fluoroquinolones and sulfonamides in pork and chicken muscle. Food Control 2017, 82, 233–242. [Google Scholar] [CrossRef]
- Xia, W.Q.; Cui, P.L.; Wang, G.N.; Liu, J.; Wang, J.P. Application of dual-template molecularly imprinted polymer-based solid phase extraction for determination of phenothiazines and benzodiazepines in swine feed. Anal. Methods 2018, 10, 3001–3010. [Google Scholar] [CrossRef]
- Xu, M.X.; Zhao, W.L.; Liu, J.; He, T.; Huang, J.J.; Wang, J.P. Determination of beta-agonists in porcine urine by molecularly imprinted polymer based chemiluminescence. Anal. Lett. 2019, 52, 1771–1787. [Google Scholar] [CrossRef]
- Yang, K.; Wang, G.N.; Liu, H.Z.; Liu, J.; Wang, J.P. Preparation of dual-template molecularly imprinted polymer coated stir bar based on computational simulation for detection of fluoroquinolones in meat. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2017, 1046, 65–72. [Google Scholar] [CrossRef]
- Zadok, I.; Srebnik, S. Coarse-grained simulation of protein-imprinted hydrogels. J. Phys. Chem. B 2018, 122, 7091–7101. [Google Scholar] [CrossRef]
- Zink, S.; Moura, F.A.; Autreto, P.; Galvao, D.S.; Mizaikoff, B. Virtually imprinted polymers (VIPs): Understanding molecularly templated materials via molecular dynamics simulations. Phys. Chem. Chem. Phys. 2018, 20, 13145–13152. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.P.; De Biasi, V.; Perrett, D. Approaches to the rational design of molecularly imprinted polymers. Anal. Chim. Acta 2004, 504, 7–14. [Google Scholar] [CrossRef]
- Kempe, H.; Kempe, M. Novel method for the synthesis of molecularly imprinted polymer bead libraries. Macromol. Rapid Commun. 2004, 25, 315–320. [Google Scholar] [CrossRef]
- Navarro-Villoslada, F.; Vicente, B.S.; Moreno-Bondi, M.a.C. Application of multivariate analysis to the screening of molecularly imprinted polymers for bisphenol A. Anal. Chim. Acta 2004, 504, 149–162. [Google Scholar] [CrossRef]
- Navarro-Villoslada, F.; Takeuchi, T. Multivariate analysis and experimental design in the screening of combinatorial libraries of molecular imprinted polymers. Bull. Chem. Soc. Jpn. 2005, 78, 1354–1361. [Google Scholar] [CrossRef]
- Mijangos, I.; Navarro-Villoslada, F.; Guerreiro, A.; Piletska, E.; Chianella, I.; Karim, K.; Turner, A.; Piletsky, S. Influence of initiator and different polymerisation conditions on performance of molecularly imprinted polymers. Biosens. Bioelectron. 2006, 22, 381–387. [Google Scholar] [CrossRef]
- Rossi, C.; Haupt, K. Application of the Doehlert experimental design to molecularly imprinted polymers: Surface response optimization of specific template recognition as a function of the type and degree of cross-linking. Anal. Bioanal. Chem. 2007, 389, 455–460. [Google Scholar] [CrossRef]
- Koohpaei, A.R.; Shahtaheri, S.J.; Ganjali, M.R.; Forushani, A.R.; Golbabaei, F. Application of multivariate analysis to the screening of molecularly imprinted polymers (MIPs) for ametryn. Talanta 2008, 75, 978–986. [Google Scholar] [CrossRef]
- Ceolin, G.; Navarro-Villoslada, F.; Moreno-Bondi, M.C.; Horvai, G.; Horvath, V. Accelerated development procedure for molecularly imprinted polymers using membrane filterplates. J. Comb. Chem. 2009, 11, 645–652. [Google Scholar] [CrossRef]
- Salimraftar, N.; Noee, S.; Abdouss, M.; Riazi, G.; Khoshhesab, Z.M. Three-level response surface full-factorial design: Advanced chemometric approach for optimizing diclofenac sodium-imprinted polymer. Polym. Bull. 2013, 71, 19–30. [Google Scholar] [CrossRef]
- Dirion, B.; Cobb, Z.; Schillinger, E.; Andersson, L.I.; Sellergren, B. Water-compatible molecularly imprinted polymers obtained via high-throughput synthesis and experimental design. J. Am. Chem. Soc. 2003, 125, 15101–15109. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Bondi, M.C.; Benito-Pena, E.; San Vicente, B.; Navarro-Villoslada, F.; de Leon, M.E.; Orellana, G.; Aparicio, S.; Molina, J.; Kempe, M.; Fiaccabrino, G.C. Molecularly imprinted polymers as selective recognition elements for optical sensors based on fluorescent measurements. In TRANSDUCERS ’03: 12th International Conference on Solid-State Sensors, Actuators and Microsystems; IEEE: New York, NY, USA, 2003; pp. 975–978. ISBN 0-7803-7731-1. [Google Scholar]
- Ren, Y.-M.; Wei, X.-Z.; Ma, J. Preparation of Cu (II) ion imprinted magnetic composite adsorbent. Mater. Sci. Technol. 2009, 17, 680–685. [Google Scholar]
- Duan, Y.Q.; Qin, Y.; Xu, F.F.; Zhang, H.H.; Yan, Y.S.; Zhang, C.; Ma, H.L. Optimization of the process parameters of synthesis of oligomeric procyanidins imprinted polymer. Sci. Res. Essays 2010, 5, 2953–2964. [Google Scholar]
- Fernández-González, A.; Badía-Laíño, R.; Díaz-García, M.E. Improving the synthesis of a molecularly imprinted sol-gel for serine using a Plackett-Burman design. Microchim. Acta 2011, 172, 351–356. [Google Scholar] [CrossRef]
- Guardia, L.; Badía, R.; Granda-Valdés, M.; Díaz-García, M.E. Screening of a molecularly imprinted sol–gel library for nafcillin recognition. J. Sol-Gel Sci. Technol. 2012, 63, 537–545. [Google Scholar] [CrossRef]
- Jia, X.; Li, H.; Luo, J.; Lu, Q.; Peng, Y.; Shi, L.; Liu, L.; Du, S.; Zhang, G.; Chen, L. Rational design of core-shell molecularly imprinted polymer based on computational simulation and Doehlert experimental optimization: Application to the separation of tanshinone IIA from Salvia miltiorrhiza Bunge. Anal. Bioanal. Chem. 2012, 403, 2691–2703. [Google Scholar] [CrossRef] [PubMed]
- Nematollahzadeh, A.; Abdekhodaie, M.J.; Shojaei, A. Submicron nanoporous polyacrylamide beads with tunable size for verapamil imprinting. J. Appl. Polym. Sci. 2012, 125, 189–199. [Google Scholar] [CrossRef]
- Shareena, M.S.; Faizal, C.K.M. Influence of factorial design analysis on the performance of bisphenol A molecular imprinted polymer. Int. J. Chem. Environ. Eng. 2012, 3, 147–151. [Google Scholar]
- Xu, D.; Zhu, W.; Jiang, Y.; Li, X.; Li, W.; Cui, J.; Yin, J.; Li, G. Rational design of molecularly imprinted photonic films assisted by chemometrics. J. Mater. Chem. 2012, 22, 16572–16581. [Google Scholar] [CrossRef]
- Zhang, L.; Han, F.; Hu, Y.; Zheng, P.; Sheng, X.; Sun, H.; Song, W.; Lv, Y. Selective trace analysis of chloroacetamide herbicides in food samples using dummy molecularly imprinted solid phase extraction based on chemometrics and quantum chemistry. Anal. Chim. Acta 2012, 729, 36–44. [Google Scholar] [CrossRef]
- Kunath, S.; Marchyk, N.; Haupt, K.; Feller, K.H. Multi-objective optimization and design of experiments as tools to tailor molecularly imprinted polymers specific for glucuronic acid. Talanta 2013, 105, 211–218. [Google Scholar] [CrossRef]
- Mehdinia, A.; Baradaran Kayyal, T.; Jabbari, A.; Aziz-Zanjani, M.O.; Ziaei, E. Magnetic molecularly imprinted nanoparticles based on grafting polymerization for selective detection of 4-nitrophenol in aqueous samples. J. Chromatogr. A 2013, 1283, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Rostamizadeh, K.; Abdollahi, H.; Parsajoo, C. Synthesis, optimization, and characterization of molecularly imprinted nanoparticles. Int. Nano Lett. 2013, 3, 20:1–20:9. [Google Scholar] [CrossRef]
- Shekarchizadeh, H.; Ensafi, A.A.; Kadivar, M. Selective determination of sucrose based on electropolymerized molecularly imprinted polymer modified multiwall carbon nanotubes/glassy carbon electrode. Mater. Sci. Eng. C 2013, 33, 3553–3561. [Google Scholar] [CrossRef]
- Cheng, Y.; Xu, K.; Li, H.; Li, Y.; Liang, B. Preparation of urea-imprinted cross-linked chitosan and its adsorption behavior. Anal. Lett. 2014, 47, 1063–1078. [Google Scholar] [CrossRef]
- Mamo, S.K.; Gonzalez-Rodriguez, J. Optimisation and production of a molecular-imprinted-polymer for the electrochemical determination of triacetone triperoxide (TATP). In Proceedings of the SPIE 9253, Optics and Photonics for Counterterrorism, Crime Fighting, and Defence X; and Optical Materials and Biomaterials in Security and Defence Systems Technology XI, Amsterdam, Netherlands, 22–25 September 2014; SPIE: Bellingham, WA, USA, 2014; Volume 9253, pp. 925315:1–925315:15, ISBN 978-1-62841-316-8. [Google Scholar]
- Mamo, S.K.; Gonzalez-Rodriguez, J. Development of a molecularly imprinted polymer-based sensor for the electrochemical determination of triacetone triperoxide (TATP). Sensors 2014, 14, 23269–23282. [Google Scholar] [CrossRef]
- Mirmohseni, A.; Shojaei, M.; Pourata, R. Experimental design and multi-objective optimization of molecularly imprinted polymers for monosaccharides. Rsc Adv. 2014, 4, 20177–20184. [Google Scholar] [CrossRef]
- Muzyka, K.; Karim, K.; Guerreiro, A.; Poma, A.; Piletsky, S. Optimisation of the synthesis of vancomycin-selective molecularly imprinted polymer nanoparticles using automatic photoreactor. Nanoscale Res. Lett. 2014, 9, 154:1–154:7. [Google Scholar] [CrossRef] [PubMed]
- Zarghami, S.; Kazemimoghadam, M.; Mohammadi, T. Cu(II) removal enhancement from aqueous solutions using ion-imprinted membrane technique. Chem. Pap. 2014, 68, 809–815. [Google Scholar] [CrossRef]
- Benito-Pena, E.; Navarro-Villoslada, F.; Carrasco, S.; Jockusch, S.; Ottaviani, M.F.; Moreno-Bondi, M.C. Experimental mixture design as a tool for the synthesis of antimicrobial selective molecularly imprinted monodisperse microbeads. Acs Appl. Mater. Interfaces 2015, 7, 10966–10976. [Google Scholar] [CrossRef] [PubMed]
- Bitar, M.; Maalouly, J.; Chebib, H.; Lerbret, A.; Cayot, P.; Bou-Maroun, E. Experimental design approach in the synthesis of molecularly imprinted polymers specific for iprodione fungicide. React. Funct. Polym. 2015, 94, 17–24. [Google Scholar] [CrossRef]
- Fan, J.P.; Xu, X.K.; Xu, R.; Zhang, X.H.; Zhu, J.H. Preparation and characterization of molecular imprinted polymer functionalized with core/shell magnetic particles (Fe3O4@SiO2@MIP) for the simultaneous recognition and enrichment of four taxoids in Taxus x media. Chem. Eng. J. 2015, 279, 567–577. [Google Scholar] [CrossRef]
- Hao, Y.; Gao, R.X.; Shi, L.; Liu, D.C.; Tang, Y.H.; Guo, Z.J. Water-compatible magnetic imprinted nanoparticles served as solid-phase extraction sorbents for selective determination of trace 17beta-estradiol in environmental water samples by liquid chromatography. J. Chromatogr. A 2015, 1396, 7–16. [Google Scholar] [CrossRef]
- Atayat, A.; Mergola, L.; Mzoughi, N.; Del Sole, R. Response surface methodology approach for the preparation of a molecularly imprinted polymer for solid-phase extraction of fenoxycarb pesticide in mussels. J. Sep. Sci. 2019, 42, 3023–3032. [Google Scholar] [CrossRef]
- Biabani, M.; Nezhadali, A.; Nakhaei, A.; Nakhaei, H. Melamine recognition: Molecularly imprinted polymer for selective and sensitive determination of melamine in food samples. Int. J. Anal. Chem. 2020, 2020, 8864144:1–8864144:10. [Google Scholar] [CrossRef]
- Bitar, M.; Lafarge, C.; Sok, N.; Cayot, P.; Bou-Maroun, E. Molecularly imprinted sol-gel polymers for the analysis of iprodione fungicide in wine: Synthesis in green solvent. Food Chem. 2019, 293, 226–232. [Google Scholar] [CrossRef]
- da Silva, P.H.R.; Diniz, M.L.V.; Pianetti, G.A.; da Costa Cesar, I.; Ribeiro, E.S.M.E.S.; de Souza Freitas, R.F.; de Sousa, R.G.; Fernandes, C. Molecularly imprinted polymer for determination of lumefantrine in human plasma through chemometric-assisted solid-phase extraction and liquid chromatography. Talanta 2018, 184, 173–183. [Google Scholar] [CrossRef]
- de Oliveira, G.F.; Hudari, F.F.; Pereira, F.M.V.; Zanoni, M.V.B.; da Silva, J.L. Carbon nanotube-based molecularly imprinted voltammetric sensor for selective diuretic analysis of dialysate and hemodialysis wastewater. ChemElectroChem 2020, 7, 3006–3016. [Google Scholar] [CrossRef]
- Dorraji, P.S.; Noori, M.; Fotouhi, L. Voltammetric determination of adefovir dipivoxil by using a nanocomposite prepared from molecularly imprinted poly(o-phenylenediamine), multi-walled carbon nanotubes and carbon nitride. Mikrochim. Acta 2019, 186, 427:1–427:8. [Google Scholar] [CrossRef]
- Goudarzi, F.; Hejazi, P. Comprehensive study on the effects of total monomers’ content and polymerization temperature control on the formation of the polymer-layer in preparation of insulin-imprinted magnetic nanoparticles. Eur. Polym. J. 2020, 126, 109541:1–109541:13. [Google Scholar] [CrossRef]
- Hatamluyi, B.; Hashemzadeh, A.; Darroudi, M. A novel molecularly imprinted polymer decorated by CQDs@HBNNS nanocomposite and UiO-66-NH2 for ultra-selective electrochemical sensing of Oxaliplatin in biological samples. Sens. Actuators B 2020, 307, 127614:1–127614:10. [Google Scholar] [CrossRef]
- Li, X.T.; Wan, J.Q.; Wang, Y.; Chi, H.Y.; Yan, Z.C.; Ding, S. Selective removal and persulfate catalytic decomposition of diethyl phthalate from contaminated water on modified MIL100 through surface molecular imprinting. Chemosphere 2020, 240, 124875:1–124875:8. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.H.; Guo, X.J.; Tan, X.C.; Mai, S.F.; Chen, Z.G.; Zhai, H.Y. Molecularly imprinted monolithic column based on functionalized beta-cyclodextrin and multi-walled carbon nanotubes for selective recognition of benzimidazole residues in citrus samples. Microchem. J. 2019, 146, 1285–1294. [Google Scholar] [CrossRef]
- Liu, M.; Tran, T.M.; Abbas Elhaj, A.A.; Boen Torsetnes, S.; Jensen, O.N.; Sellergren, B.; Irgum, K. Molecularly imprinted porous monolithic materials from melamine-formaldehyde for selective trapping of phosphopeptides. Anal. Chem. 2017, 89, 9491–9501. [Google Scholar] [CrossRef] [PubMed]
- Nezhadali, A.; Bonakdar, G.A. Multivariate optimization of mebeverine analysis using molecularly imprinted polymer electrochemical sensor based on silver nanoparticles. J. Food Drug Anal. 2019, 27, 305–314. [Google Scholar] [CrossRef]
- Nezhadali, A.; Motlagh, M.O.; Sadeghzadeh, S. Spectrophotometric determination of fluoxetine by molecularly imprinted polypyrrole and optimization by experimental design, artificial neural network and genetic algorithm. Spectrochim. ActaPart A 2018, 190, 181–187. [Google Scholar] [CrossRef]
- Rohani, F.G.; Ansari, M. Electropolymerized MIP with MWCNTs on stir bar using multivariate optimization for tetradifon detection in date. Pharm. Nanotechnol. 2019, 7, 404–417. [Google Scholar] [CrossRef]
- Tavakoli, Z.; Soleimani, M.; Alavi Nikje, M.M. Characterization and performance evaluation of functional monomer effect on molecular imprinted polyurethane foam. J. Chromatogr. A 2019, 1602, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Zhai, H.; Liang, G.; Guo, X.; Chen, Z.; Yu, J.; Lin, H.; Zhou, Q. Novel coordination imprinted polymer monolithic column applied to the solid-phase extraction of flumequine from fish samples. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2019, 1118-1119, 55–62. [Google Scholar] [CrossRef]
- van Duin, A.C.T.; Dasgupta, S.; Lorant, F.; Goddard, W.A. ReaxFF: A reactive force field for hydrocarbons. J. Phys. Chem. A 2001, 105, 9396–9409. [Google Scholar] [CrossRef]
- Senftle, T.P.; Hong, S.; Islam, M.M.; Kylasa, S.B.; Zheng, Y.; Shin, Y.K.; Junkermeier, C.; Engel-Herbert, R.; Janik, M.J.; Aktulga, H.M.; et al. The ReaxFF reactive force-field: Development, applications and future directions. npj Comput. Mater. 2016, 2, 15011:1–15011:14. [Google Scholar] [CrossRef]
- Yungerman, I.; Srebnik, S. Factors contributing to binding-site imperfections in imprinted polymers. Chem. Mater. 2006, 18, 657–663. [Google Scholar] [CrossRef]
- Cheng, S.; Van Tassel, P.R. Theory and simulation of the available volume for adsorption in a chain molecule templated porous material. J. Chem. Phys. 2001, 114, 4974–4981. [Google Scholar] [CrossRef]
- Dourado, E.M.A.; Sarkisov, L. Emergence of molecular recognition phenomena in a simple model of imprinted porous materials. J. Chem. Phys. 2009, 130, 214701:1–214701:9. [Google Scholar] [CrossRef]
- Sarkisov, L.; Van Tassel, P.R. Replica Ornstein-Zernike theory of adsorption in a templated porous material: Interaction site systems. J. Chem. Phys. 2005, 123, 164706:1–164706:10. [Google Scholar] [CrossRef]
- Sarkisov, L.; Van Tassel, P.R. Integral equation theory of adsorption in templated materials: Influence of molecular attraction. J. Phys. Chem. C 2007, 111, 15726–15735. [Google Scholar] [CrossRef]
- Van Tassel, P.R. Theoretical model of adsorption in a templated porous material. Phys. Rev. E 1999, 60, R25–R28. [Google Scholar] [CrossRef]
- Zhang, L.H.; Van Tassel, P.R. Theory and simulation of adsorption in a templated porous material: Hard sphere systems. J. Chem. Phys. 2000, 112, 3006–3013. [Google Scholar] [CrossRef]
- Srebnik, S. Induced porosity in cross-linked polymer networks: Mean field theory and simulations. In Characterization of Porous Solids VI; Elsevier: Amsterdam, The Netherlands, 2002; Volume 144, pp. 43–50. ISBN 0-444-51261-6. [Google Scholar]
- Srebnik, S.; Lev, O.; Avnir, D. Pore size distribution induced by microphase separation: Effect of the leaving group during polycondensation. Chem. Mater. 2001, 13, 811–816. [Google Scholar] [CrossRef]
- Srebnik, S.; Lev, O. Toward establishing criteria for polymer imprinting using mean-field theory. J. Chem. Phys. 2002, 116, 10967–10972. [Google Scholar] [CrossRef]
- Srebnik, S. Theoretical investigation of the imprinting efficiency of molecularly imprinted polymers. Chem. Mater. 2004, 16, 883–888. [Google Scholar] [CrossRef]
- Srebnik, S.; Lev, O. Theoretical investigation of imprinted crosslinked silicates. J. Sol-Gel Sci. Technol. 2003, 26, 107–113. [Google Scholar] [CrossRef]
- Henthorn, D.B.; Peppas, N.A. Molecular simulations of recognitive polymer networks prepared by biomimetic configurational imprinting as responsive biomaterials. Mrs Online Proc. Libr. 2003, 787, 7–15. [Google Scholar] [CrossRef]
- Henthorn, D.B.; Peppas, N.A. Molecular simulations of recognitive behavior of molecularly imprinted intelligent polymeric networks. Ind. Eng. Chem. Res. 2007, 46, 6084–6091. [Google Scholar] [CrossRef]
- Levi, L.; Srebnik, S. Simulation of protein-imprinted polymers. 2. Imprinting efficiency. J. Phys. Chem. B 2010, 114, 16744–16751. [Google Scholar] [CrossRef]
- Levi, L.; Srebnik, S. Simulation of protein-imprinted polymers. 1. Imprinted pore properties. J. Phys. Chem. B 2010, 114, 107–114. [Google Scholar] [CrossRef]
- Levi, L.; Srebnik, S. Structural characterization of protein-imprinted gels using lattice Monte Carlo simulation. Macromol. Symp. 2010, 291-292, 258–270. [Google Scholar] [CrossRef]
- Levi, L.; Srebnik, S. Simulation of protein-imprinted polymers. 3. Imprinting selectivity. J. Phys. Chem. B 2011, 115, 14469–14474. [Google Scholar] [CrossRef] [PubMed]
- Yankelov, R.; Yungerman, I.; Srebnik, S. The selectivity of protein-imprinted gels and its relation to protein properties: A computer simulation study. J. Mol. Recognit. 2017, 30, 2607:1–2607:10. [Google Scholar] [CrossRef] [PubMed]
- Schauperl, M.; Lewis, D.W. Probing the structural and binding mechanism heterogeneity of molecularly imprinted polymers. J. Phys. Chem. B 2015, 119, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Huynh, T.P.; Wojnarowicz, A.; Sosnowska, M.; Srebnik, S.; Benincori, T.; Sannicolo, F.; D’Souza, F.; Kutner, W. Cytosine derivatized bis(2,2’-bithienyl)methane molecularly imprinted polymer for selective recognition of 6-thioguanine, an antitumor drug. Biosens. Bioelectron. 2015, 70, 153–160. [Google Scholar] [CrossRef]
- Cowen, T.; Busato, M.; Karim, K.; Piletsky, S.A. In silico synthesis of synthetic receptors: A polymerization algorithm. Macromol. Rapid Commun. 2016, 37, 2011–2016. [Google Scholar] [CrossRef]
- Piletska, E.V.; Guerreiro, A.; Mersiyanova, M.; Cowen, T.; Canfarotta, F.; Piletsky, S.; Karim, K.; Piletsky, S. Probing peptide sequences on their ability to generate affinity sites in molecularly imprinted polymers. Langmuir 2020, 36, 279–283. [Google Scholar] [CrossRef]
- Madadian-Bozorg, N.; Zahedi, P.; Shamsi, M.; Safarian, S. Poly (methacrylic acid)-based molecularly imprinted polymer nanoparticles containing 5-fluourouracil used in colon cancer therapy potentially. Polym. Adv. Technol. 2018, 29, 2401–2409. [Google Scholar] [CrossRef]
- Nezhadali, A.; Biabani, M. Electrochemical sensor for selective determination of ketorolac tromethamine based on molecularly imprinting polypyrrole modified with functionalized multi-wall carbon nanotubes in pharmaceutical and biological samples. Anal. Bioanal. Electrochem. 2020, 12, 48–62. [Google Scholar]
- Lai, E.P.C.; Feng, S.Y. Molecularly imprinted solid phase extraction for rapid screening of metformin. Microchem. J. 2003, 75, 159–168. [Google Scholar] [CrossRef]
- Wang, J.; Guo, R.; Chen, J.; Zhang, Q.; Liang, X. Phenylurea herbicides-selective polymer prepared by molecular imprinting using N-(4-isopropylphenyl)-N’-butyleneurea as dummy template. Anal. Chim. Acta 2005, 540, 307–315. [Google Scholar] [CrossRef]
- Tada, M.; Sasaki, T.; Iwasawa, Y. Design of a novel molecular-imprinted Rh−amine complex on SiO 2 and its shape-selective catalysis for α-methylstyrene hydrogenation. J. Phys. Chem. B 2004, 108, 2918–2930. [Google Scholar] [CrossRef]
- Li, L.F.; Chen, L.; Zhang, H.; Yang, Y.Z.; Liu, X.G.; Chen, Y.K. Temperature and magnetism bi-responsive molecularly imprinted polymers: Preparation, adsorption mechanism and properties as drug delivery system for sustained release of 5-fluorouracil. Mater. Sci. Eng. C 2016, 61, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Mazouz, Z.; Rahali, S.; Fourati, N.; Zerrouki, C.; Aloui, N.; Seydou, M.; Yaakoubi, N.; Chehimi, M.M.; Othmane, A.; Kalfat, R. Highly selective polypyrrole mip-based gravimetric and electrochemical sensors for picomolar detection of glyphosate. Sensors 2017, 17, 2586. [Google Scholar] [CrossRef]
- Bagdziunas, G. Theoretical design of molecularly imprinted polymers based on polyaniline and polypyrrole for detection of tryptophan. Mol. Syst. Des. Eng. 2020, 5, 1504–1512. [Google Scholar] [CrossRef]
- Mukawa, T.; Goto, T.; Nariai, H.; Aoki, Y.; Imamura, A.; Takeuchi, T. Novel strategy for molecular imprinting of phenolic compounds utilizing disulfide templates. J. Pharm. Biomed. Anal. 2003, 30, 1943–1947. [Google Scholar] [CrossRef]
- Lv, Y.; Lin, Z.; Feng, W.; Zhou, X.; Tan, T. Selective recognition and large enrichment of dimethoate from tea leaves by molecularly imprinted polymers. Biochem. Eng. J. 2007, 36, 221–229. [Google Scholar] [CrossRef]
- Zhao, Z.; Wang, Q.; Zhang, L.; Wu, T. Structured water and water−polymer interactions in hydrogels of molecularly imprinted polymers. J. Phys. Chem. B 2008, 112, 7515–7521. [Google Scholar] [CrossRef]
- Luo, D.; Zhao, Z.; Zhang, L.; Wang, Q.; Wang, J. On the structure of molecularly imprinted polymers by modifying charge on functional groups through molecular dynamics simulations. Mol. Simul. 2013, 40, 431–438. [Google Scholar] [CrossRef]
- Terracina, J.J.; Bergkvist, M.; Sharfstein, S.T. Computational investigation of stoichiometric effects, binding site heterogeneities, and selectivities of molecularly imprinted polymers. J. Mol. Model. 2016, 22, 139:1–139:9. [Google Scholar] [CrossRef] [PubMed]
- Herdes, C.; Sarkisov, L. Computer simulation of volatile organic compound adsorption in atomistic models of molecularly imprinted polymers. Langmuir 2009, 25, 5352–5359. [Google Scholar] [CrossRef] [PubMed]
- Sobiech, M.; Zolek, T.; Lulinski, P.; Maciejewska, D. A computational exploration of imprinted polymer affinity based on voriconazole metabolites. Analyst 2014, 139, 1779–1788. [Google Scholar] [CrossRef] [PubMed]
- Sobiech, M.; Zolek, T.; Lulinski, P.; Maciejewska, D. Separation of octopamine racemate on (R,S)-2-amino-1-phenylethanol imprinted polymer—Experimental and computational studies. Talanta 2016, 146, 556–567. [Google Scholar] [CrossRef]
- Tarley, C.R.T.; Kubota, L.T. Molecularly-imprinted solid phase extraction of catechol from aqueous effluents for its selective determination by differential pulse voltammetry. Anal. Chim. Acta 2005, 548, 11–19. [Google Scholar] [CrossRef]
- Tarley, C.R.T.; Segatelli, M.G.; Kubota, L.T. Amperometric determination of chloroguaiacol at submicromolar levels after on-line preconcentration with molecularly imprinted polymers. Talanta 2006, 69, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Koohpaei, A.R.; Shahtaheri, S.J.; Ganjali, M.R.; Forushani, A.R.; Golbabaei, F. Optimization of solid-phase extraction using developed modern sorbent for trace determination of ametryn in environmental matrices. J. Hazard. Mater. 2009, 170, 1247–1255. [Google Scholar] [CrossRef]
- Koohpaei, A.R.; Shahtaheri, S.J.; Ganjali, M.R.; Forushani, A.R.; Golbabaei, F. Molecular imprinted solid phase extraction for determination of atrazine in environmental samples. Iran. J. Environ. Health Sci. Eng. 2008, 5, 283–296. [Google Scholar]
- Alizadeh, T.; Ganjali, M.R.; Nourozi, P.; Zare, M. Multivariate optimization of molecularly imprinted polymer solid-phase extraction applied to parathion determination in different water samples. Anal. Chim. Acta 2009, 638, 154–161. [Google Scholar] [CrossRef]
- Santos, W.d.J.R.; Lima, P.R.; Tarley, C.R.T.; Kubota, L.T. A catalytically active molecularly imprinted polymer that mimics peroxidase based on hemin: Application to the determination of p-aminophenol. Anal. Bioanal. Chem. 2007, 389, 1919–1929. [Google Scholar] [CrossRef] [PubMed]
- Rahiminejad, M.; Shahtaheri, S.J.; Ganjali, M.R.; Forushani, A.R.; Golbabaei, F. Molecularly imprinted solid phase extraction for trace analysis of diazinon in drinking water. Iran. J. Environ. Health Sci. Eng. 2009, 6, 97–106. [Google Scholar]
- Santos, W.D.R.; Lima, P.R.; Tarley, C.R.T.; Hoehr, N.F.; Kubota, L.T. Synthesis and application of a peroxidase-like molecularly imprinted polymer based on hemin for selective determination of serotonin in blood serum. Anal. Chim. Acta 2009, 631, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Khajeh, M.; Sanchooli, E. Synthesis of ion-selective imprinted polymer for manganese removal from environmental water. Polym. Bull. 2010, 67, 413–425. [Google Scholar] [CrossRef]
- Alizadeh, T. An imprinted polymer for removal of Cd2+ from water samples: Optimization of adsorption and recovery steps by experimental design. Chin. J. Polym. Sci. 2011, 29, 658–669. [Google Scholar] [CrossRef]
- Khajeh, M.; Heidari, Z.S.; Sanchooli, E. Synthesis, characterization and removal of lead from water samples using lead-ion imprinted polymer. Chem. Eng. J. 2011, 166, 1158–1163. [Google Scholar] [CrossRef]
- Khajeh, M.; Sanchooli, E. A pre-concentration procedure employing a new imprinted polymer for the determination of copper in water. Int. J. Environ. Anal. Chem. 2011, 91, 1310–1319. [Google Scholar] [CrossRef]
- Alizadeh, T. Molecularly imprinted nanoparticles-based electrochemical sensor for determination of ultratrace parathion in real samples. Int. J. Environ. Anal. Chem. 2012, 92, 1742–1760. [Google Scholar] [CrossRef]
- Abedi, H.; Ebrahimzadeh, H. Imprinted polymer-based extraction for speciation analysis of inorganic tin in food and water samples. React. Funct. Polym. 2013, 73, 634–640. [Google Scholar] [CrossRef]
- Diniz, K.M.; Segatelli, M.G.; Tarley, C.R.T. Synthesis and adsorption studies of novel hybrid mesoporous copolymer functionalized with protoporphyrin for batch and on-line solid-phase extraction of Cd2+ ions. React. Funct. Polym. 2013, 73, 838–846. [Google Scholar] [CrossRef]
- Ebrahimzadeh, H.; Behbahani, M.; Yamini, Y.; Adlnasab, L.; Asgharinezhad, A.A. Optimization of Cu(II)-ion imprinted nanoparticles for trace monitoring of copper in water and fish samples using a Box–Behnken design. React. Funct. Polym. 2013, 73, 23–29. [Google Scholar] [CrossRef]
- Meng, M.; Feng, Y.; Zhang, M.; Ji, Y.; Dai, J.; Liu, Y.; Yu, P.; Yan, Y. Optimization of surface imprinted layer attached poly(vinylidene fluoride) membrane for selective separation of salicylic acid from acetylsalicylic acid using central composite design. Chem. Eng. J. 2013, 231, 132–145. [Google Scholar] [CrossRef]
- Ghorbani-Kalhor, E.; Behbahani, M.; Abolhasani, J. Application of ion-imprinted polymer nanoparticles for selective trace determination of palladium ions in food and environmental samples with the aid of experimental design methodology. Food Anal. Methods 2014, 8, 1746–1757. [Google Scholar] [CrossRef]
- Meng, M.; Feng, Y.; Liu, Y.; Wang, Y.; Yan, Y. Preparation of composite-imprinted alumina membrane for effective separation of p-hydroxybenzonic acid from its isomer using Box-Behnken design-based statistical modeling. J. Appl. Polym. Sci. 2014, 131, 40621:1–40621:11. [Google Scholar] [CrossRef]
- Meng, M.J.; Liu, Y.; Zhang, M.; Feng, Y.H.; Yan, Y.S. Introduction of an ordered porous polymer network into a ceramic alumina membrane via non-hydrolytic sol-gel methodology for targeted dynamic separation. Rsc Adv. 2014, 4, 38630–38642. [Google Scholar] [CrossRef]
- Shaikh, H.; Memon, N.; Bhanger, M.I.; Nizamani, S.M.; Denizli, A. Core-shell molecularly imprinted polymer-based solid-phase microextraction fiber for ultra trace analysis of endosulfan I and II in real aqueous matrix through gas chromatography-micro electron capture detector. J. Chromatogr. A 2014, 1337, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Meng, X.; Liu, Z.; Meng, M.; Jiang, F.; Luo, M.; Ni, L.; Qiu, J.; Liu, F.; Zhong, G. Preparation of a two-dimensional ion-imprinted polymer based on a graphene oxide/SiO2 composite for the selective adsorption of nickel ions. Langmuir 2015, 31, 8841–8851. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, G.R.; Santos, A.V.; Lima, A.S.; Soares, C.M.F.; Leite, M.S. Neural modelling in adsorption column of cholesterol-removal efficiency from milk. Lwt Food Sci. Technol. 2015, 64, 632–638. [Google Scholar] [CrossRef]
- Zare, F.; Ghaedi, M.; Daneshfar, A.; Ostovan, A. Magnetic molecularly imprinted polymer for the efficient and selective preconcentration of diazinon before its determination by high-performance liquid chromatography. J. Sep. Sci. 2015, 38, 2797–2803. [Google Scholar] [CrossRef]
- Ahamed, M.E.H.; Marjanovic, L.; Mbianda, X.Y. Statistical optimization, kinetic and isotherm studies on selective adsorption of silver and gold cyanocomplexes using aminoguanidyl-chitosan imprinted polymers. J. Adv. Chem. Eng. 2016, 6, 149:1–149:11. [Google Scholar] [CrossRef]
- Arabi, M.; Ghaedi, M.; Ostovan, A. Development of dummy molecularly imprinted based on functionalized silica nanoparticles for determination of acrylamide in processed food by matrix solid phase dispersion. Food Chem. 2016, 210, 78–84. [Google Scholar] [CrossRef]
- Arabi, M.; Ghaedi, M.; Ostovan, A.; Tashkhourian, J.; Asadallahzadeh, H. Synthesis and application of molecularly imprinted nanoparticles combined ultrasonic assisted for highly selective solid phase extraction trace amount of celecoxib from human plasma samples using design expert (DXB) software. Ultrason. Sonochem. 2016, 33, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Arabi, M.; Ghaedi, M.; Ostovan, A.; Wang, S.B. Synthesis of lab-in-a-pipette-tip extraction using hydrophilic nano-sized dummy molecularly imprinted polymer for purification and analysis of prednisolone. J. Colloid Interface Sci. 2016, 480, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Arabi, M.; Ostovan, A.; Ghaedi, M.; Purkait, M.K. Novel strategy for synthesis of magnetic dummy molecularly imprinted nanoparticles based on functionalized silica as an efficient sorbent for the determination of acrylamide in potato chips: Optimization by experimental design methodology. Talanta 2016, 154, 526–532. [Google Scholar] [CrossRef] [PubMed]
- Balieiro, A.L.; Santos, R.A.; Pereira, M.M.; Figueiredo, R.T.; Freitas, L.S.; de Alsina, O.L.S.; Lima, A.S.; Soares, C.M.F. Adsorption process of molecularly imprinted silica for extraction of lactose from milk. Braz. J. Chem. Eng. 2016, 33, 361–372. [Google Scholar] [CrossRef]
- Jafary Omid, N.; Morovati, H.; Amini, M.; Dehpour, A.R.; Partoazar, A.; Rafiee-Tehrani, M.; Dorkoosh, F. Development of molecularly imprinted olanzapine nano-particles: In vitro characterization and in vivo evaluation. Aaps Pharmscitech 2016, 17, 1457–1467. [Google Scholar] [CrossRef]
- Khajeh, M.; Moghaddam, S.A.; Bohlooli, M.; Ghaffari-Moghaddam, M. Application of the artificial neural network and imperialist competitive algorithm for optimization of molecularly imprinted solid phase extraction of methylene blue. e-Polymers 2016, 16, 243–253. [Google Scholar] [CrossRef]
- Kolaei, M.; Dashtian, K.; Rafiee, Z.; Ghaedi, M. Ultrasonic-assisted magnetic solid phase extraction of morphine in urine samples by new imprinted polymer-supported on MWCNT-Fe3O4-NPs: Central composite design optimization. Ultrason. Sonochem. 2016, 33, 240–248. [Google Scholar] [CrossRef]
- Li, G.Z.; Wang, W.; Wang, Q.; Zhu, T. Deep eutectic solvents modified molecular imprinted polymers for optimized purification of chlorogenic acid from honeysuckle. J. Chromatogr. Sci. 2016, 54, 271–279. [Google Scholar] [CrossRef]
- Terzopoulou, Z.; Papageorgiou, M.; Kyzas, G.Z.; Bikiaris, D.N.; Lambropoulou, D.A. Preparation of molecularly imprinted solid-phase microextraction fiber for the selective removal and extraction of the antiviral drug abacavir in environmental and biological matrices. Anal. Chim. Acta 2016, 913, 63–75. [Google Scholar] [CrossRef]
- Asfaram, A.; Ghaedi, M.; Dashtian, K. Rapid ultrasound-assisted magnetic microextraction of gallic acid from urine, plasma and water samples by HKUST-1-MOE-Fe3O4-GA-MIP-NPs: UV-vis detection and optimization study. Ultrason. Sonochem. 2017, 34, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Asfaram, A.; Ghaedi, M.; Dashtian, K. Ultrasound assisted combined molecularly imprinted polymer for selective extraction of nicotinamide in human urine and milk samples: Spectrophotometric determination and optimization study. Ultrason. Sonochem. 2017, 34, 640–650. [Google Scholar] [CrossRef]
- Bahrani, S.; Ghaedi, M.; Khoshnood Mansoorkhani, M.J.; Ostovan, A. A highly selective nanocomposite based on MIP for curcumin trace levels quantification in food samples and human plasma following optimization by central composite design. J. Chromatogr. B 2017, 1040, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Davarani, S.S.; Rezayati Zad, Z.; Taheri, A.R.; Rahmatian, N. Highly selective solid phase extraction and preconcentration of Azathioprine with nano-sized imprinted polymer based on multivariate optimization and its trace determination in biological and pharmaceutical samples. Mater. Sci. Eng. C 2017, 71, 572–583. [Google Scholar] [CrossRef]
- Arvand, M.; Alirezanejad, F. New sensing material of molecularly imprinted polymer for the selective recognition of sulfamethoxazole in foods and plasma and employing the Taguchi optimization methodology to optimize the carbon paste electrode. J. Iran. Chem. Soc. 2012, 10, 93–105. [Google Scholar] [CrossRef]
- Arvand, M.; Fallahi, P. Man-tailored biomimetic sensor of molecularly imprinted materials for the potentiometric measurement of rivastigmine in tablets and biological fluids and employing the taguchi optimization methodology to optimize the mip-based membranes. Electroanalysis 2012, 24, 1852–1863. [Google Scholar] [CrossRef]
- Alizadeh, T.; Rezaloo, F. Toluene chemiresistor sensor based on nano-porous toluene-imprinted polymer. Int. J. Environ. Anal. Chem. 2013, 93, 919–934. [Google Scholar] [CrossRef]
- Alizadeh, T.; Hamedsoltani, L. Graphene/graphite/molecularly imprinted polymer nanocomposite as the highly selective gas sensor for nitrobenzene vapor recognition. J. Environ. Chem. Eng. 2014, 2, 1514–1526. [Google Scholar] [CrossRef]
- Shojaei, S.; Nasirizadeh, N.; Entezam, M.; Koosha, M.; Azimzadeh, M. An electrochemical nanosensor based on molecularly imprinted polymer (MIP) for detection of gallic acid in fruit juices. Food Anal. Methods 2016, 9, 2721–2731. [Google Scholar] [CrossRef]
- Abidi, H.; Ghaedi, M.; Rafiei, A.; Jelowdar, A.; Arabi, M.; Ostovan, A.; Asfaram, A. A molecularly imprinted polymer coupled with high-performance liquid chromatography-UV for the determination of albendazole in plasma and urine samples: CCD-RSM design. New J. Chem. 2018, 42, 15937–15945. [Google Scholar] [CrossRef]
- Dil, E.A.; Ghaedi, M.; Asfaram, A.; Mehrabi, F.; Shokrollahi, A.; Matin, A.A.; Tayebi, L. Magnetic dual-template molecularly imprinted polymer based on syringe-to-syringe magnetic solid-phase microextraction for selective enrichment of p-Coumaric acid and ferulic acid from pomegranate, grape, and orange samples. Food Chem. 2020, 325, 126902:1–126902:7. [Google Scholar] [CrossRef]
- Ansari, S. Application of hollow porous molecularly imprinted polymers using K2Ti4O9 coupled with SPE-HPLC for the determination of celecoxib in human urine samples: Optimization by central composite design (CCD). Anal. Methods 2017, 9, 3200–3212. [Google Scholar] [CrossRef]
- Ansari, S.; Karimi, M. Synthesis and application of molecularly imprinted polymer for highly selective solid phase extraction trace amount of sotalol from human urine samples: Optimization by central composite design (CCD). Med. Chem. Res. 2017, 26, 2477–2490. [Google Scholar] [CrossRef]
- Ansari, S.; Masoum, S. A multi-walled carbon nanotube-based magnetic molecularly imprinted polymer as a highly selective sorbent for ultrasonic-assisted dispersive solid-phase microextraction of sotalol in biological fluids. Analyst 2018, 143, 2862–2875. [Google Scholar] [CrossRef]
- Ansari, S.; Masoum, S. Ultrasound-assisted dispersive solid-phase microextraction of capecitabine by multi-stimuli responsive molecularly imprinted polymer modified with chitosan nanoparticles followed by HPLC analysis. Microchim. Acta 2020, 187, 366:1–366:11. [Google Scholar] [CrossRef]
- Afsharipour, R.; Dadfarnia, S.; Shabani, A.M.H.; Kazemi, E. Design of a pseudo stir bar sorptive extraction using graphenized pencil lead as the base of the molecularly imprinted polymer for extraction of nabumetone. Spectrochim. ActaPart A 2020, 238, 118427:1–118427:12. [Google Scholar] [CrossRef] [PubMed]
- Arabi, M.; Ghaedi, M.; Ostovan, A. Development of a lower toxic approach based on green synthesis of water-compatible molecularly imprinted nanoparticles for the extraction of hydrochlorothiazide from human urine. Acs Sustain. Chem. Eng. 2017, 5, 3775–3785. [Google Scholar] [CrossRef]
- Arabi, M.; Ghaedi, M.; Ostovan, A. Synthesis and application of in-situ molecularly imprinted silica monolithic in pipette-tip solid-phase microextraction for the separation and determination of gallic acid in orange juice samples. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2017, 1048, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Arabi, M.; Ghaedi, M.; Ostovan, A. Water compatible molecularly imprinted nanoparticles as a restricted access material for extraction of hippuric acid, a biological indicator of toluene exposure, from human urine. Microchim. Acta 2017, 184, 879–887. [Google Scholar] [CrossRef]
- Arabi, M.; Ostovan, A.; Bagheri, A.R.; Guo, X.; Li, J.; Ma, J.; Chen, L. Hydrophilic molecularly imprinted nanospheres for the extraction of rhodamine B followed by HPLC analysis: A green approach and hazardous waste elimination. Talanta 2020, 215, 120933:1–120933:8. [Google Scholar] [CrossRef]
- Arabzadeh, N.; Mohammadi, A.; Darwish, M.; Abuzerr, S. Construction of a TiO2-Fe3O4-decorated molecularly imprinted polymer nanocomposite for tartrazine degradation: Response surface methodology modeling and optimization. J. Chin. Chem. Soc. 2019, 66, 474–483. [Google Scholar] [CrossRef]
- Asfaram, A.; Arabi, M.; Ostovan, A.; Sadeghi, H.; Ghaedi, M. Simple and selective detection of quercetin in extracts of plants and food samples by dispersive-micro-solid phase extraction based on core-shell magnetic molecularly imprinted polymers. New J. Chem. 2018, 42, 16144–16153. [Google Scholar] [CrossRef]
- Asgari, S.; Bagheri, H.; Es-Haghi, A.; AminiTabrizi, R. An imprinted interpenetrating polymer network for microextraction in packed syringe of carbamazepine. J. Chromatogr. A 2017, 1491, 1–8. [Google Scholar] [CrossRef]
- Atarodi, H.; Faghihian, H. Selective photodegradation of atrazine by a novel molecularly imprinted nanophotocatalyst prepared on the basis of chitosan. J. Photochem. Photobiol. A 2019, 382, 111892:1–111892:11. [Google Scholar] [CrossRef]
- Azizi, A.; Shahhoseini, F.; Bottaro, C.S. Magnetic molecularly imprinted polymers prepared by reversible addition fragmentation chain transfer polymerization for dispersive solid phase extraction of polycyclic aromatic hydrocarbons in water. J. Chromatogr. A 2020, 1610, 460534:1–460534:15. [Google Scholar] [CrossRef]
- Bagheri, A.R.; Arabi, M.; Ghaedi, M.; Ostovan, A.; Wang, X.; Li, J.; Chen, L. Dummy molecularly imprinted polymers based on a green synthesis strategy for magnetic solid-phase extraction of acrylamide in food samples. Talanta 2019, 195, 390–400. [Google Scholar] [CrossRef] [PubMed]
- Bagheri, A.R.; Ghaedi, M. Synthesis of chitosan based molecularly imprinted polymer for pipette-tip solid phase extraction of Rhodamine B from chili powder samples. Int. J. Biol. Macromol. 2019, 139, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Bahrani, S.; Ghaedi, M.; Arabi, M. Construction of molecularly imprinted nanoparticles by employing ultrasound waves for selective determination of doxepin from human plasma samples: Modeling and optimization. Biomed. Chromatogr. 2019, 33, 4675:1–4675:9. [Google Scholar] [CrossRef] [PubMed]
- Bazrafshan, A.A.; Ghaedi, M.; Rafiee, Z.; Hajati, S.; Ostovan, A. Nano-sized molecularly imprinted polymer for selective ultrasound-assisted microextraction of pesticide carbaryl from water samples: Spectrophotometric determination. J. Colloid Interface Sci. 2017, 498, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Beigzadeh, Z.; Golbabaei, F.; Khadem, M.; Omidi, F.; Someah, M.S.; Shahtaheri, S.J. Development of molecularly imprinted membranes for selective determination of urinary ultra-trace 5-fluorouracil as antineoplastic drug used in chemotherapy. Macromol. Res. 2020, 28, 390–399. [Google Scholar] [CrossRef]
- Benedetti, B.; Di Carro, M.; Magi, E. Multivariate optimization of an extraction procedure based on magnetic molecular imprinted polymer for the determination of polycyclic aromatic hydrocarbons in sea water. Microchem. J. 2019, 145, 1199–1206. [Google Scholar] [CrossRef]
- da Silva, J.L.; Buffon, E.; Beluomini, M.A.; Pradela, L.A.; Araujo, D.A.G.; Santos, A.L.; Takeuchi, R.M.; Stradiotto, N.R. Non-enzymatic lactose molecularly imprinted sensor based on disposable graphite paper electrode. Anal. Chim. Acta 2021, 1143, 53–64. [Google Scholar] [CrossRef]
- Dahaghin, Z.; Mousavi, H.Z.; Sajjadi, S.M. A novel magnetic ion imprinted polymer as a selective magnetic solid phase for separation of trace lead(II) ions from agricultural products, and optimization using a Box-Behnken design. Food Chem. 2017, 237, 275–281. [Google Scholar] [CrossRef]
- Dil, E.A.; Asfaram, A.; Javadian, H. A new approach for microextraction of trace albendazole sulfoxide drug from the samples of human plasma and urine, and water by the molecularly imprinted polymer nanoparticles combined with HPLC. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2020, 1158, 122249:1–122249:12. [Google Scholar] [CrossRef]
- Dil, E.A.; Doustimotlagh, A.H.; Javadian, H.; Asfaram, A.; Ghaedi, M. Nano-sized Fe3O4@SiO2-molecular imprinted polymer as a sorbent for dispersive solid-phase microextraction of melatonin in the methanolic extract of Portulaca oleracea, biological, and water samples. Talanta 2021, 221, 121620:1–121620:10. [Google Scholar] [CrossRef] [PubMed]
- Dramou, P.; Itatahine, A.; Fizir, M.; Ait Mehdi, Y.; Kutoka, P.T.; He, H. Preparation of novel molecularly imprinted magnetic graphene oxide and their application for quercetin determination. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2019, 1124, 273–283. [Google Scholar] [CrossRef]
- Ganjavi, F.; Ansari, M.; Kazemipour, M.; Zeidabadinejad, L. Computational design, synthesis and utilization of a magnetic molecularly imprinted polymer on graphene oxide nanosheets for highly selective extraction and determination of buprenorphine in biological fluids and tablets. Anal. Methods 2018, 10, 5214–5226. [Google Scholar] [CrossRef]
- Garcia, Y.; Smolinska-Kempisty, K.; Pereira, E.; Piletska, E.; Piletsky, S. Development of competitive ’pseudo’-ELISA assay for measurement of cocaine and its metabolites using molecularly imprinted polymer nanoparticles. Anal. Methods 2017, 9, 4592–4598. [Google Scholar] [CrossRef]
- Ghani, S.M.; Rezaei, B.; Jamei, H.R.; Ensafi, A.A. Novel synthesis of a dual fluorimetric sensor for the simultaneous analysis of levodopa and pyridoxine. Anal. Bioanal. Chem. 2021, 413, 377–387. [Google Scholar] [CrossRef]
- Gholami, H.; Arabi, M.; Ghaedi, M.; Ostovan, A.; Bagheri, A.R. Column packing elimination in matrix solid phase dispersion by using water compatible magnetic molecularly imprinted polymer for recognition of melamine from milk samples. J. Chromatogr. A 2019, 1594, 13–22. [Google Scholar] [CrossRef]
- Gholami, H.; Ghaedi, M.; Arabi, M.; Ostovan, A.; Bagheri, A.R.; Mohamedian, H. Application of molecularly imprinted biomembrane for advancement of matrix solid-phase dispersion for clean enrichment of parabens from powder sunscreen samples: Optimization of chromatographic conditions and green approach. Acs Omega 2019, 4, 3839–3849. [Google Scholar] [CrossRef]
- Gholami, H.; Ghaedi, M.; Ostovan, A.; Arabi, M.; Bagheri, A.R. Preparation of hollow porous molecularly imprinted and aluminum(III) doped silica nanospheres for extraction of the drugs valsartan and losartan prior to their quantitation by HPLC. Mikrochim. Acta 2019, 186, 702:1–702:9. [Google Scholar] [CrossRef] [PubMed]
- Ghorbani, A.; Ansari, S.; Masoum, S. Ultrasonic-assisted solid-phase extraction of sotalol in human urine samples using molecularly imprinted nanoparticles: Experimental design and adsorption study. Sep. Sci. Technol. 2018, 53, 2782–2796. [Google Scholar] [CrossRef]
- Gonzalez, A.; Cerda, V. Development of an automatic sequential injection analysis-lab on valve system exploiting molecularly imprinted polymers coupled with high performance liquid chromatography for the determination of estrogens in wastewater samples. Talanta 2020, 209, 120564:1–120564:6. [Google Scholar] [CrossRef] [PubMed]
- Hashemi, S.H.; Kaykhaii, M.; Keikha, A.J.; Naruie, N. Application of molecularly imprinted polymer pipette tip micro-solid phase extraction of nalidixic acid and acetaminophen from pills and seawater samples and their determination by spectrophotometry. Chem. Pap. 2020, 74, 4009–4023. [Google Scholar] [CrossRef]
- Hashemi, S.H.; Kaykhaii, M.; Keikha, A.J.; Parkaz, A. Application of response surface methodology to optimize pipette tip micro-solid phase extraction of dyes from seawater by molecularly imprinted polymer and their determination by HPLC. J. Iran. Chem. Soc. 2019, 16, 2613–2627. [Google Scholar] [CrossRef]
- Hashemi, S.H.; Kaykhaii, M.; Keikha, A.J.; Sajjadi, Z. Application of Box-Behnken design in response surface methodology for the molecularly imprinted polymer pipette-tip solid phase extraction of methyl red from seawater samples and its determination by spectrophotometery. Mar. Pollut. Bull. 2018, 137, 306–314. [Google Scholar] [CrossRef]
- Hashemi, S.H.; Keykha, F. Application of the response surface methodology in the optimization of modified molecularly imprinted polymer based pipette-tip micro-solid phase extraction for spectrophotometric determination of nicotine in seawater and human plasma. Anal. Methods 2019, 11, 5405–5412. [Google Scholar] [CrossRef]
- Hashemi, S.H.; Najari, F. Response surface methodology of pre-concentration of chorophenols from seawater samples by molecularly imprinted stir bar sorptive extraction combined with HPLC: Box-Behnken design. J. Chromatogr. Sci. 2019, 57, 279–289. [Google Scholar] [CrossRef]
- Hashemi, S.H.; Ziyaadini, M.; Kaykhaii, M.; Jamali Keikha, A.; Naruie, N. Separation and determination of ciprofloxacin in seawater, human blood plasma and tablet samples using molecularly imprinted polymer pipette-tip solid phase extraction and its optimization by response surface methodology. J. Sep. Sci. 2020, 43, 505–513. [Google Scholar] [CrossRef]
- Heravizadeh, O.R.; Khadem, M.; Dehghani, F.; Shahtaheri, S.J. Determination of fenthion in urine samples using molecularly imprinted nanoparticles: Modelling and optimisation by response surface methodology. Int. J. Environ. Anal. Chem. 2020, 1–15. [Google Scholar] [CrossRef]
- Heravizadeh, O.R.; Khadem, M.; Nabizadeh, R.; Shahtaheri, S.J. Synthesis of molecular imprinted polymer nanoparticles followed by application of response surface methodology for optimization of metribuzin extraction from urine samples. Chem. Pap. 2018, 72, 3057–3068. [Google Scholar] [CrossRef]
- Heravizadeh, O.R.; Khadem, M.; Nabizadeh, R.; Shahtaheri, S.J. Synthesis of molecularly imprinted nanoparticles for selective exposure assessment of permethrin: Optimization by response surface methodology. J. Environ. Health Sci. Eng. 2019, 17, 393–406. [Google Scholar] [CrossRef]
- Jafari, S.; Nasirizadeh, N.; Dehghani, M. Developing a highly sensitive electrochemical sensor using thiourea-imprinted polymers based on an MWCNT modified carbon ceramic electrode. J. Electroanal. Chem. 2017, 802, 139–146. [Google Scholar] [CrossRef]
- Kaabipour, M.; Khodadoust, S.; Zeraatpisheh, F. Preparation of magnetic molecularly imprinted polymer for dispersive solid-phase extraction of valsartan and its determination by high-performance liquid chromatography: Box-Behnken design. J. Sep. Sci. 2020, 43, 912–919. [Google Scholar] [CrossRef] [PubMed]
- Kalogiouri, N.P.; Tsalbouris, A.; Kabir, A.; Furton, K.G.; Samanidou, V.F. Synthesis and application of molecularly imprinted polymers using sol-gel matrix imprinting technology for the efficient solid-phase extraction of BPA from water. Microchem. J. 2020, 157, 104965:1–104965:9. [Google Scholar] [CrossRef]
- Kamari, K.; Taheri, A. Preparation and evaluation of magnetic core-shell mesoporous molecularly imprinted polymers for selective adsorption of amitriptyline in biological samples. J. Taiwan Inst. Chem. Eng. 2018, 86, 230–239. [Google Scholar] [CrossRef]
- Khadem, M.; Faridbod, F.; Norouzi, P.; Foroushani, A.R.; Ganjali, M.R.; Yarahmadi, R.; Shahtaheri, S.J. Voltammetric determination of carbofuran pesticide in biological and environmental samples using a molecularly imprinted polymer sensor, a multivariate optimization. J. Anal. Chem. 2020, 75, 669–678. [Google Scholar] [CrossRef]
- Khodadoust, S.; Nasiriani, T.; Zeraatpisheh, F. Preparation of a magnetic molecularly imprinted polymer for the selective adsorption of chlordiazepoxide and its determination by central composite design optimized HPLC. New J. Chem. 2018, 42, 14444–14452. [Google Scholar] [CrossRef]
- Li, G.; Row, K.H. Selective extraction of 3,4-dihydroxybenzoic acid in Ilex chinensis Sims by meticulous mini-solid-phase microextraction using ternary deep eutectic solvent-based molecularly imprinted polymers. Anal. Bioanal. Chem. 2018, 410, 7849–7858. [Google Scholar] [CrossRef]
- Li, G.; Row, K.H. Hydrophilic molecularly imprinted chitosan based on deep eutectic solvents for the enrichment of gallic acid in red ginseng tea. Polymers 2019, 11, 1434. [Google Scholar] [CrossRef]
- Li, G.; Row, K.H. Deep eutectic solvents skeleton typed molecularly imprinted chitosan microsphere coated magnetic graphene oxide for solid-phase microextraction of chlorophenols from environmental water. J. Sep. Sci. 2020, 43, 1063–1070. [Google Scholar] [CrossRef]
- Li, G.; Wang, X.; Row, K.H. Magnetic solid-phase extraction with Fe3O4/molecularly imprinted polymers modified by deep eutectic solvents and ionic liquids for the rapid purification of alkaloid isomers (theobromine and theophylline) from green tea. Molecules 2017, 22, 1061. [Google Scholar] [CrossRef]
- Li, G.Z.; Row, K.H. Deep eutectic solvents cross-linked molecularly imprinted chitosan microsphere for the micro-solid phase extraction of p-hydroxybenzoic acid from pear rind. J. Sep. Sci. 2020, 44, 549–556. [Google Scholar] [CrossRef]
- Mirzajani, R.; Kardani, F.; Ramezani, Z. A nanocomposite consisting of graphene oxide, zeolite imidazolate framework 8, and a molecularly imprinted polymer for (multiple) fiber solid phase microextraction of sterol and steroid hormones prior to their quantitation by HPLC. Mikrochim. Acta 2019, 186, 129:1–129:14. [Google Scholar] [CrossRef]
- Nezhadali, A.; Mehrdadian, Z.; Mojarrab, M. Nanogram/mL detection of nortriptyline: Preparation of a molecularly imprinted polymer for spectrophotometric determination of nortriptyline based on multivariate optimization methods. J. Sep. Sci. 2019, 42, 3479–3486. [Google Scholar] [CrossRef]
- Nezhadali, A.; Shadmehri, R.; Rajabzadeh, F.; Sadeghzadeh, S. Selective determination of closantel by artificial neural network-genetic algorithm optimized molecularly imprinted polypyrrole using UV-visible spectrophotometry. Spectrochim. ActaPart A 2020, 243, 118779:1–118779:7. [Google Scholar] [CrossRef]
- Ostovan, A.; Ghaedi, M.; Arabi, M. Fabrication of water-compatible superparamagnetic molecularly imprinted biopolymer for clean separation of baclofen from bio-fluid samples: A mild and green approach. Talanta 2018, 179, 760–768. [Google Scholar] [CrossRef] [PubMed]
- Ostovan, A.; Ghaedi, M.; Arabi, M.; Asfaram, A. Hollow porous molecularly imprinted polymer for highly selective clean-up followed by influential preconcentration of ultra-trace glibenclamide from bio-fluid. J. Chromatogr. A 2017, 1520, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Parvinizadeh, F.; Daneshfar, A. Fabrication of a magnetic metal-organic framework molecularly imprinted polymer for extraction of anti-malaria agent hydroxychloroquine. New J. Chem. 2019, 43, 8508–8516. [Google Scholar] [CrossRef]
- Ostovan, A.; Ghaedi, M.; Arabi, M.; Yang, Q.; Li, J.; Chen, L. Hydrophilic multitemplate molecularly imprinted biopolymers based on a green synthesis strategy for determination of B-family vitamins. Acs Appl. Mater. Interfaces 2018, 10, 4140–4150. [Google Scholar] [CrossRef] [PubMed]
- Pesavento, M.; Merli, D.; Biesuz, R.; Alberti, G.; Marchetti, S.; Milanese, C. A MIP-based low-cost electrochemical sensor for 2-furaldehyde detection in beverages. Anal. Chim. Acta 2021, 1142, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Rahmani, M.E.; Ansari, M.; Nateghi, M.; Kazemipour, M. Computation-assisted molecularly imprinted polymer synthesis for extraction of naltrexone from urine using experimental design and determination by UPLC-DAD. J. Aoac Int. 2017, 100, 700–711. [Google Scholar] [CrossRef] [PubMed]
- Tavakoli, Z.; Soleimani, M.; Nikje, M.M.A. Evaluation of an experimentally designed molecular imprinted polyurethane foam performance for extraction of alprazolam. J. Chromatogr. Sci. 2019, 57, 662–670. [Google Scholar] [CrossRef] [PubMed]
- Toudeshki, R.M.; Dadfarnia, S.; Haji Shabani, A.M. Surface molecularly imprinted polymer on magnetic multi-walled carbon nanotubes for selective recognition and preconcentration of metformin in biological fluids prior to its sensitive chemiluminescence determination: Central composite design optimization. Anal. Chim. Acta 2019, 1089, 78–89. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.Y.; Wu, P.; Cui, Y.R.; Fizir, M.; Shi, J.R.; He, H. Selective recognition and enrichment of sterigmatocystin in wheat by thermo-responsive imprinted polymer based on magnetic halloysite nanotubes. J. Chromatogr. A 2020, 1619, 460952:1–460952:12. [Google Scholar] [CrossRef]
- Yuan, X.C.; Gao, X.; Yuan, Y.; Ji, Y.H.; Xiong, Z.L.; Zhao, L.S. Fe3O4/graphene molecularly imprinted composite for selective separation of catecholamine neurotransmitters and their analysis in rat brain tissues. Talanta 2021, 224, 121843:1–121843:10. [Google Scholar] [CrossRef]
- Yuan, X.C.; Yuan, Y.X.; Gao, X.; Xiong, Z.L.; Zhao, L.S. Magnetic dummy-template molecularly imprinted polymers based on multi-walled carbon nanotubes for simultaneous selective extraction and analysis of phenoxy carboxylic acid herbicides in cereals. Food Chem. 2020, 333, 127540:1–127540:8. [Google Scholar] [CrossRef]
- Zhang, J.J.; Chen, Y.; Wu, W.Y.; Wang, Z.P.; Chu, Y.J.; Chen, X.H. Hollow porous dummy molecularly imprinted polymer as a sorbent of solid-phase extraction combined with accelerated solvent extraction for determination of eight bisphenols in plastic products. Microchem. J. 2019, 145, 1176–1184. [Google Scholar] [CrossRef]
- Zhao, M.; Hou, Z.H.; Lian, Z.R.; Qin, D.; Ge, C.Z. Direct extraction and detection of malachite green from marine sediments by magnetic nano-sized imprinted polymer coupled with spectrophotometric analysis. Mar. Pollut. Bull. 2020, 158, 111363:1–111363:8. [Google Scholar] [CrossRef]
- Feng, S.L.; Gao, F.; Chen, Z.W.; Grant, E.; Kitts, D.D.; Wang, S.; Lu, X.N. Determination of alpha-tocopherol in vegetable oils using a molecularly imprinted polymers-surface-enhanced raman spectroscopic biosensor. J. Agric. Food. Chem. 2013, 61, 10467–10475. [Google Scholar] [CrossRef]
- Gao, F.; Feng, S.L.; Chen, Z.W.; Li-Chan, E.C.Y.; Grant, E.; Lu, X.N. Detection and quantification of chloramphenicol in milk and honey using molecularly imprinted polymers: Canadian penny-based sers nano-biosensor. J. Food Sci. 2014, 79, N2542–N2549. [Google Scholar] [CrossRef]
- Gao, F.; Grant, E.; Lu, X.N. Determination of histamine in canned tuna by molecularly imprinted polymers-surface enhanced Raman spectroscopy. Anal. Chim. Acta 2015, 901, 68–75. [Google Scholar] [CrossRef]
- Gao, F.; Hu, Y.; Chen, D.; Li-Chan, E.C.Y.; Grant, E.; Lu, X. Determination of Sudan I in paprika powder by molecularly imprinted polymers-thin layer chromatography-surface enhanced Raman spectroscopic biosensor. Talanta 2015, 143, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.X.; Feng, S.L.; Gao, F.; Li-Chan, E.C.Y.; Grant, E.; Lu, X.N. Detection of melamine in milk using molecularly imprinted polymers-surface enhanced Raman spectroscopy. Food Chem. 2015, 176, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Lu, X. Rapid detection of melamine in tap water and milk using conjugated "one-step" molecularly imprinted polymers-surface enhanced raman spectroscopic sensor. J. Food Sci. 2016, 81, N1272-1280. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Xu, E.; Li, J.; Long, J.; Jiao, A.; Jin, Z. Highly sensitive determination of ethyl carbamate in alcoholic beverages by surface-enhanced Raman spectroscopy combined with a molecular imprinting polymer. Rsc Adv. 2016, 6, 109442–109452. [Google Scholar] [CrossRef]
- Feng, S.; Hu, Y.; Ma, L.; Lu, X. Development of molecularly imprinted polymers-surface-enhanced Raman spectroscopy/colorimetric dual sensor for determination of chlorpyrifos in apple juice. Sens. Actuators B 2017, 241, 750–757. [Google Scholar] [CrossRef]
- Cao, X.L.; Zhao, F.N.; Jiang, Z.J.; Hong, S.H.; Zhang, C.; She, Y.X.; Jin, F.; Jin, M.J.; Wang, J. Rapid analysis of bitertanol in agro-products using molecularly imprinted polymers-surface-enhanced raman spectroscopy. Food Anal. Methods 2018, 11, 1435–1443. [Google Scholar] [CrossRef]
- Decorbie, N.; Tijunelyte, I.; Gam-Derouich, S.; Solard, J.; Lamouri, A.; Decorse, P.; Felidj, N.; Gauchotte-Lindsay, C.; Rinnert, E.; Mangeney, C.; et al. Sensing polymer/paracetamol interaction with an independent component analysis-based SERS-MIP nanosensor. Plasmonics 2020, 15, 1533–1539. [Google Scholar] [CrossRef]
- Tan, J.; Wang, H.F.; Yan, X.P. Discrimination of saccharides with a fluorescent molecular imprinting sensor array based on phenylboronic acid functionalized mesoporous silica. Anal. Chem. 2009, 81, 5273–5280. [Google Scholar] [CrossRef] [PubMed]
- Wangchareansak, T.; Thitithanyanont, A.; Chuakheaw, D.; Gleeson, M.P.; Lieberzeit, P.A.; Sangma, C. Influenza A virus molecularly imprinted polymers and their application in virus sub-type classification. J. Mater. Chem. B 2013, 1, 2190–2197. [Google Scholar] [CrossRef]
- Jha, S.K.; Liu, C.; Hayashi, K. Molecular imprinted polyacrylic acids based QCM sensor array for recognition of organic acids in body odor. Sens. Actuators B 2014, 204, 74–87. [Google Scholar] [CrossRef]
- Tan, J.; Li, R.; Jiang, Z.T. Discrimination of fresh fruit juices by a fluorescent sensor, array for carboxylic acids based on molecularly imprinted titania. Food Chem. 2014, 165, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Bueno, L.; El-Sharif, H.F.; Salles, M.O.; Boehm, R.D.; Narayan, R.J.; Paixão, T.R.L.C.; Reddy, S.M. MIP-based electrochemical protein profiling. Sens. Actuators B 2014, 204, 88–95. [Google Scholar] [CrossRef]
- Jha, S.K.; Hayashi, K. A quick responding quartz crystal microbalance sensor array based on molecular imprinted polyacrylic acids coating for selective identification of aldehydes in body odor. Talanta 2015, 134, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Jha, S.K.; Hayashi, K. Polyacrylic acid polymer and aldehydes template molecule based MIPs coated QCM sensors for detection of pattern aldehydes in body odor. Sens. Actuators B 2015, 206, 471–487. [Google Scholar] [CrossRef]
- Lu, W.; Dong, X.; Qiu, L.; Yan, Z.; Meng, Z.; Xue, M.; He, X.; Liu, X. Colorimetric sensor arrays based on pattern recognition for the detection of nitroaromatic molecules. J. Hazard. Mater. 2017, 326, 130–137. [Google Scholar] [CrossRef]
- Wang, S.; Wen, Y.; Wang, Y.; Ma, Y.; Liu, Z. Pattern recognition of cells via multiplexed imaging with monosaccharide-imprinted quantum dots. Anal. Chem. 2017, 89, 5646–5652. [Google Scholar] [CrossRef]
- El-Sharif, H.F.; Stevenson, D.; Reddy, S.M. MIP-based protein profiling: A method for interspecies discrimination. Sens. Actuators B 2017, 241, 33–39. [Google Scholar] [CrossRef]
- Chunta, S.; Suedee, R.; Singsanan, S.; Lieberzeit, P.A. Sensing array based on molecularly imprinted polymers for simultaneous assessment of lipoproteins. Sens. Actuators B 2019, 298, 126828:1–126828:8. [Google Scholar] [CrossRef]
- Herrera-Chacon, A.; Dinc-Zor, S.; Del Valle, M. Integrating molecularly imprinted polymer beads in graphite-epoxy electrodes for the voltammetric biosensing of histamine in wines. Talanta 2020, 208, 120348:1–120348:6. [Google Scholar] [CrossRef]
- Hardoyono, F.; Windhani, K.; Sambodo, H.; Pudjianto, H. Identification of bioactive compounds in ginger based on molecularly imprinted polymer quartz crystal microbalance gas sensor. Iop Conf. Ser. Mater. Sci. Eng. 2019, 546, 032012:1–032012:11. [Google Scholar] [CrossRef]
- Lin, Z.Z.; Li, L.; Fu, G.Y.; Lai, Z.Z.; Peng, A.H.; Huang, Z.Y. Molecularly imprinted polymer-based photonic crystal sensor array for the discrimination of sulfonamides. Anal. Chim. Acta 2020, 1101, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Rossignol, J.; Dujourdy, L.; Stuerga, D.; Cayot, P.; Gougeon, R.D.; Bou-Maroun, E. A first tentative for simultaneous detection of fungicides in model and real wines by microwave sensor coupled to molecularly imprinted sol-gel polymers. Sensors 2020, 20, 6224. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.W.; Jiao, T.T.; Xu, C.L.; Li, W.; Xiong, Y. Preparation of molecularly imprinted polymers for sensing of 2,4-dichlorophenoxyacetic acid residues in environmental water and mixed juice. J. Mater. Sci. 2020, 55, 6848–6860. [Google Scholar] [CrossRef]
- Mirmohseni, A.; Rastgouy-Houjaghan, M. Application of nanobalance technique and principal component analysis for detection of the soil fumigant telone residues in the air. J. Environ. Sci. Health. Part B 2012, 47, 677–686. [Google Scholar] [CrossRef]
- Liu, C.; Wyszynski, B.; Yatabe, R.; Hayashi, K.; Toko, K. Molecularly imprinted sol-gel-based QCM sensor arrays for the detection and recognition of volatile aldehydes. Sensors 2017, 17, 382. [Google Scholar] [CrossRef]
- Wang, Z.H.; Chen, W.; Gu, S.; Wang, J.; Wang, Y.W. Discrimination of wood borers infested Platycladus orientalis trunks using quartz crystal microbalance gas sensor array. Sens. Actuators B 2020, 309, 127767:1–127767:9. [Google Scholar] [CrossRef]
- Das, D.; Chatterjee, T.N.; Roy, R.B.; Tudu, B.; Hazarika, A.K.; Sabhapondit, S.; Bandyopadhyay, R. Titanium oxide nanocubes embedded molecularly imprinted polymer-based electrode for selective detection of caffeine in green tea. IEEE Sens. J. 2020, 20, 6240–6247. [Google Scholar] [CrossRef]
- Chatterjee, T.N.; Das, D.; Roy, R.B.; Tudu, B.; Sabhapondit, S.; Tamuly, P.; Pramanik, P.; Bandyopadhyay, R. Molecular imprinted polymer based electrode for sensing catechin (plus C) in green tea. IEEE Sens. J. 2018, 18, 2236–2244. [Google Scholar] [CrossRef]
- Valero-Navarro, A.; Damiani, P.C.; Fernández-Sánchez, J.F.; Segura-Carretero, A.; Fernández-Gutiérrez, A. Chemometric-assisted MIP-optosensing system for the simultaneous determination of monoamine naphthalenes in drinking waters. Talanta 2009, 78, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Rosengren, A.M.; Karlsson, J.G.; Andersson, P.O.; Nicholls, I.A. Chemometric models of template molecularly imprinted polymer binding. Anal. Chem. 2005, 77, 5700–5705. [Google Scholar] [CrossRef]
- Rosengren, A.M.; Golker, K.; Karlsson, J.G.; Nicholls, I.A. Dielectric constants are not enough: Principal component analysis of the influence of solvent properties on molecularly imprinted polymer–ligand rebinding. Biosens. Bioelectron. 2009, 25, 553–557. [Google Scholar] [CrossRef]
- Bitar, M.; Roudaut, G.; Maalouly, J.; Brandes, S.; Gougeon, R.D.; Cayot, P.; Bou-Maroun, E. Water sorption isotherms of molecularly imprinted polymers. Relation between water binding and iprodione binding capacity. React. Funct. Polym. 2017, 114, 1–7. [Google Scholar] [CrossRef]
- Rossetti, C.; Ore, O.G.; Sellergren, B.; Halvorsen, T.G.; Reubsaet, L. Exploring the peptide retention mechanism in molecularly imprinted polymers. Anal. Bioanal. Chem. 2017, 409, 5631–5643. [Google Scholar] [CrossRef]
- Liu, D.L.; Chen, Z.B.; Du, X.Y.; Liu, Z. Study of structural parameters on the adsorption selectivity of a molecularly imprinted polymer. J. Macromol. Sci. Part A Pure Appl. Chem. 2017, 54, 622–628. [Google Scholar] [CrossRef]
- Baggiani, C.; Anfossi, L.; Giovannoli, C.; Tozzi, C. Multivariate analysis of the selectivity for a pentachlorophenol-imprinted polymer. J. Chromatogr. B 2004, 804, 31–41. [Google Scholar] [CrossRef]
- Nantasenamat, C.; Tantimongcolwat, T.; Naenna, T.; Isarankura-Na-Ayudhya, C.; Prachayasittikul, V. Prediction of selectivity index of pentachlorophenol-imprinted polymers. Excli J. 2006, 5, 150–163. [Google Scholar]
- Nantasenamat, C.; Isarankura-Na-Ayudhya, C.; Naenna, T.; Prachayasittikul, V. Quantitative structure-imprinting factor relationship of molecularly imprinted polymers. Biosens. Bioelectron. 2007, 22, 3309–3317. [Google Scholar] [CrossRef]
- Nantasenamat, C.; Naenna, T.; Ayudhya, C.I.N.; Prachayasittikul, V. Quantitative prediction of imprinting factor of molecularly imprinted polymers by artificial neural network. J. Comput. Aided Mol. Des. 2005, 19, 509–524. [Google Scholar] [CrossRef]
- Nantasenamat, C.; Isarankura-Na-Ayudhya, C.; Bülow, L.; Ye, L.; Prachayasittikul, V. In silico design for synthesis of molecularly imprinted microspheres specific towards bisphenol A by precipitation polymerization. Excli J. 2006, 5, 103–114. [Google Scholar]
- Kempe, H.; Kempe, M. QSRR analysis of beta-lactam antibiotics on a penicillin G targeted MIP stationary phase. Anal. Bioanal. Chem. 2010, 398, 3087–3096. [Google Scholar] [CrossRef]
- Kempe, H.; Parareda Pujolràs, A.; Kempe, M. Molecularly imprinted polymer nanocarriers for sustained release of erythromycin. Pharm. Res. 2014, 32, 375–388. [Google Scholar] [CrossRef] [PubMed]
- Leite, M.S.; Santos, M.A.; Costa, E.M.F.; Balieiro, A.; Lima, A.S.; Sanchez, O.L.; Soares, C.M.F. Modeling of milk lactose removal by column adsorption using artificial neural networks: Mlp and Rbf. Chem. Ind. Chem. Eng. Q. 2019, 25, 369–382. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).