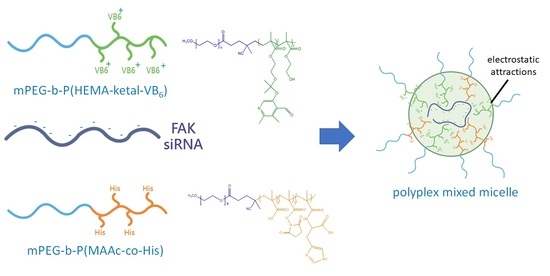
Development of a Rapid-Onset, Acid-Labile Linkage Polyplex-Mixed Micellar System for Anticancer Therapy
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
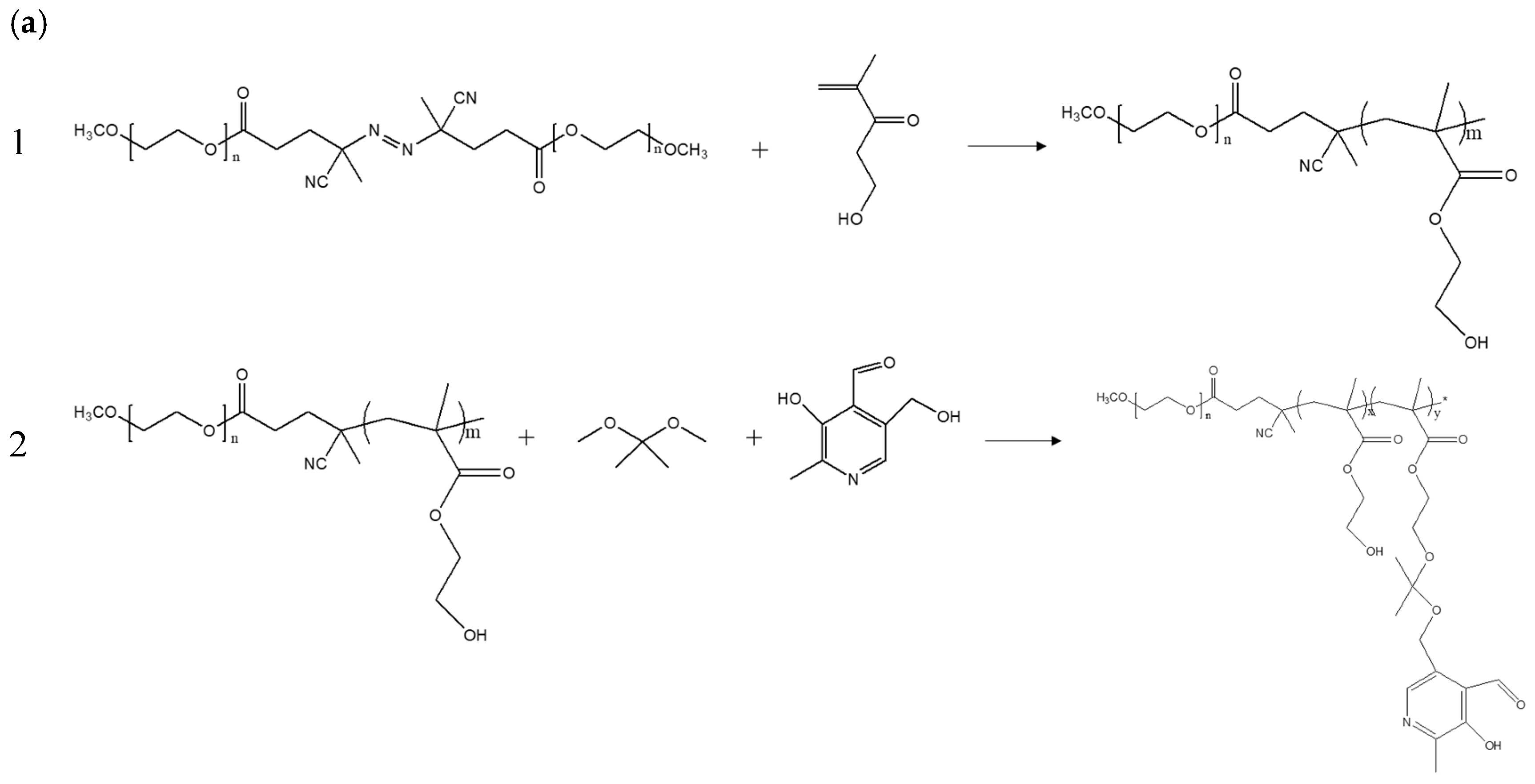
2.2. Preparation and Characterization of mPEG-b-P(HEMA-Ketal-VB6)
2.3. pH-Responsiveness of the Copolymer mPEG-b-P(HEMA-Ketal-VB6)
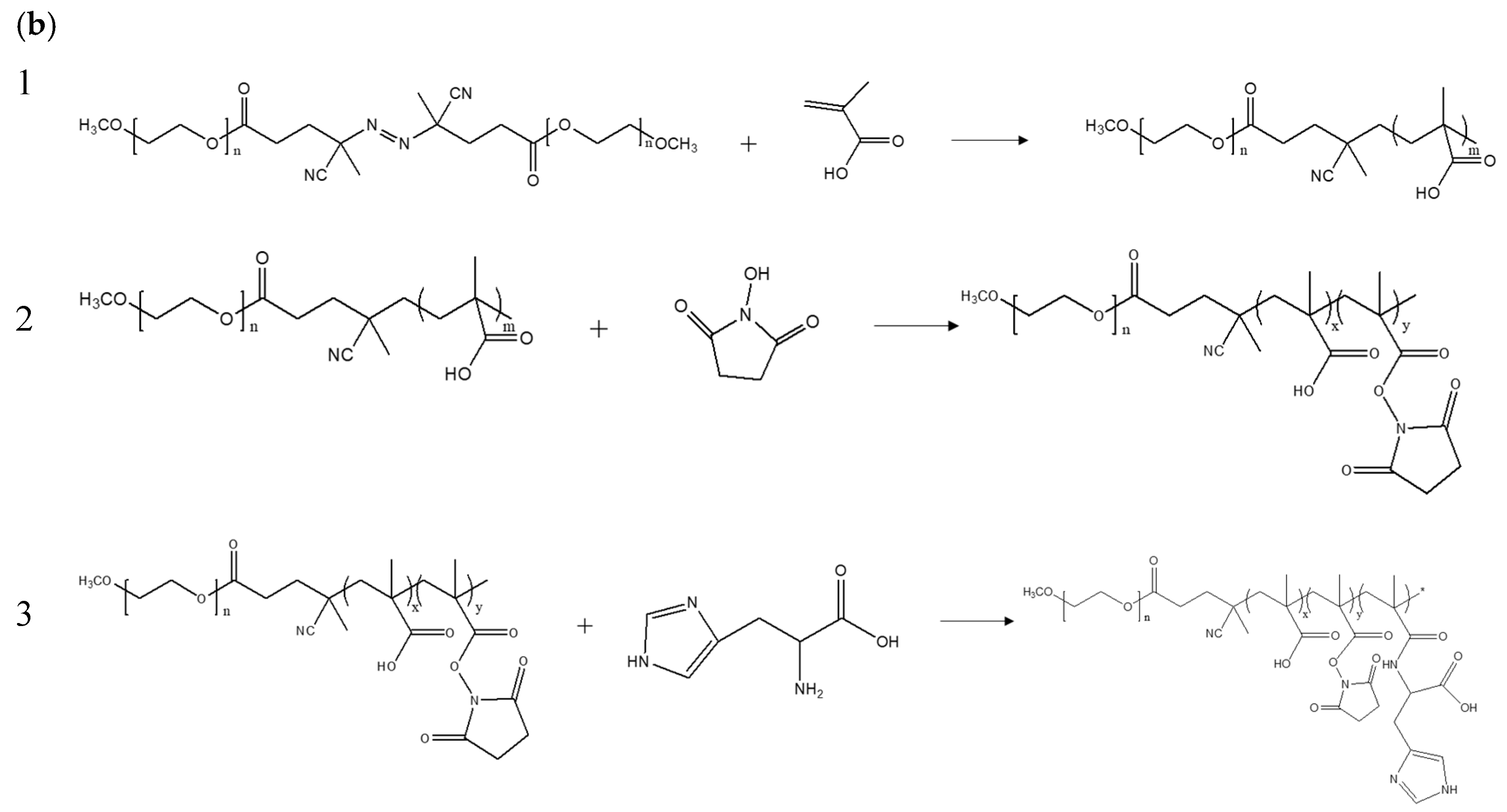
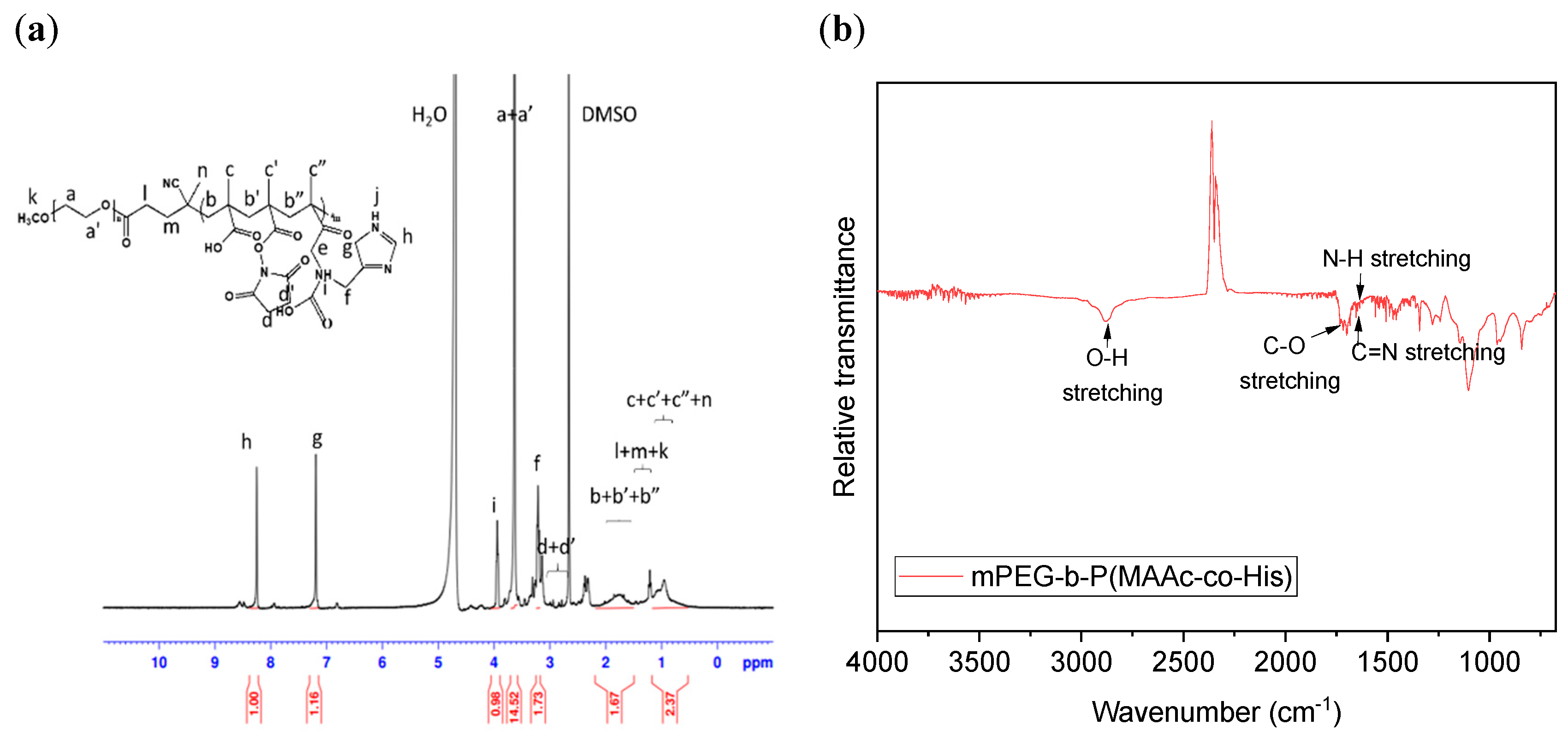
2.4. Preparation and Characterization of mPEG-b-P(MAAc-co-His)
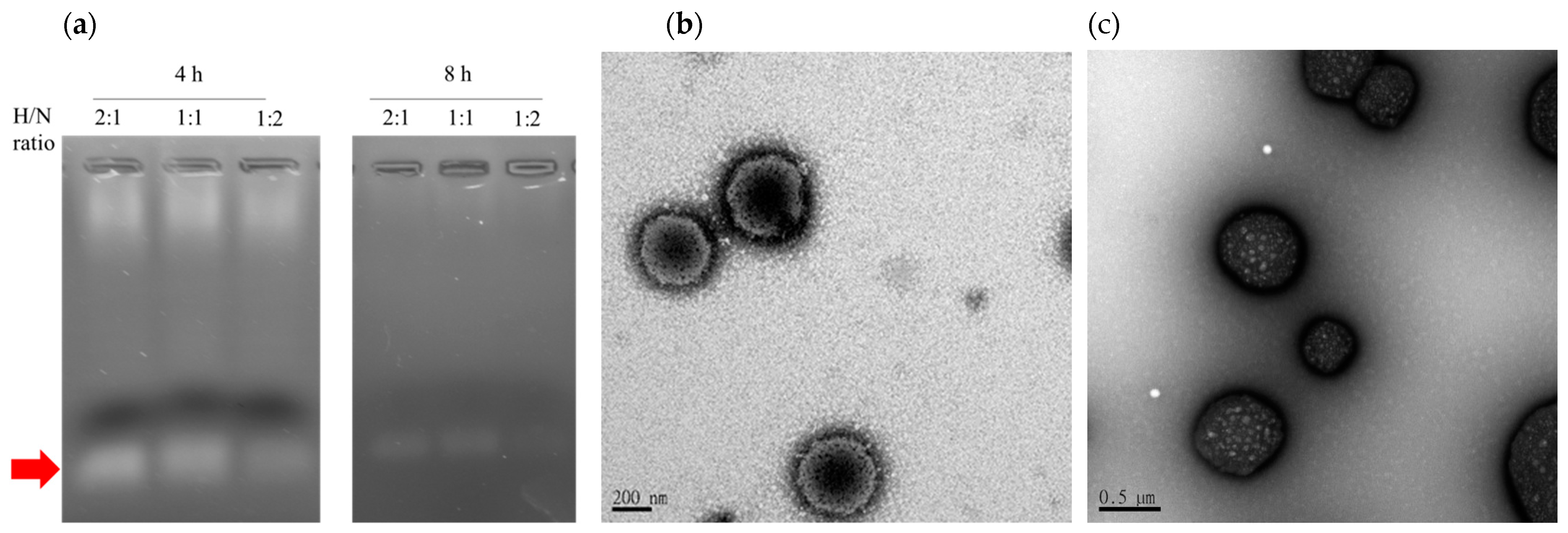
2.5. Polyplex Preparation and Characterization
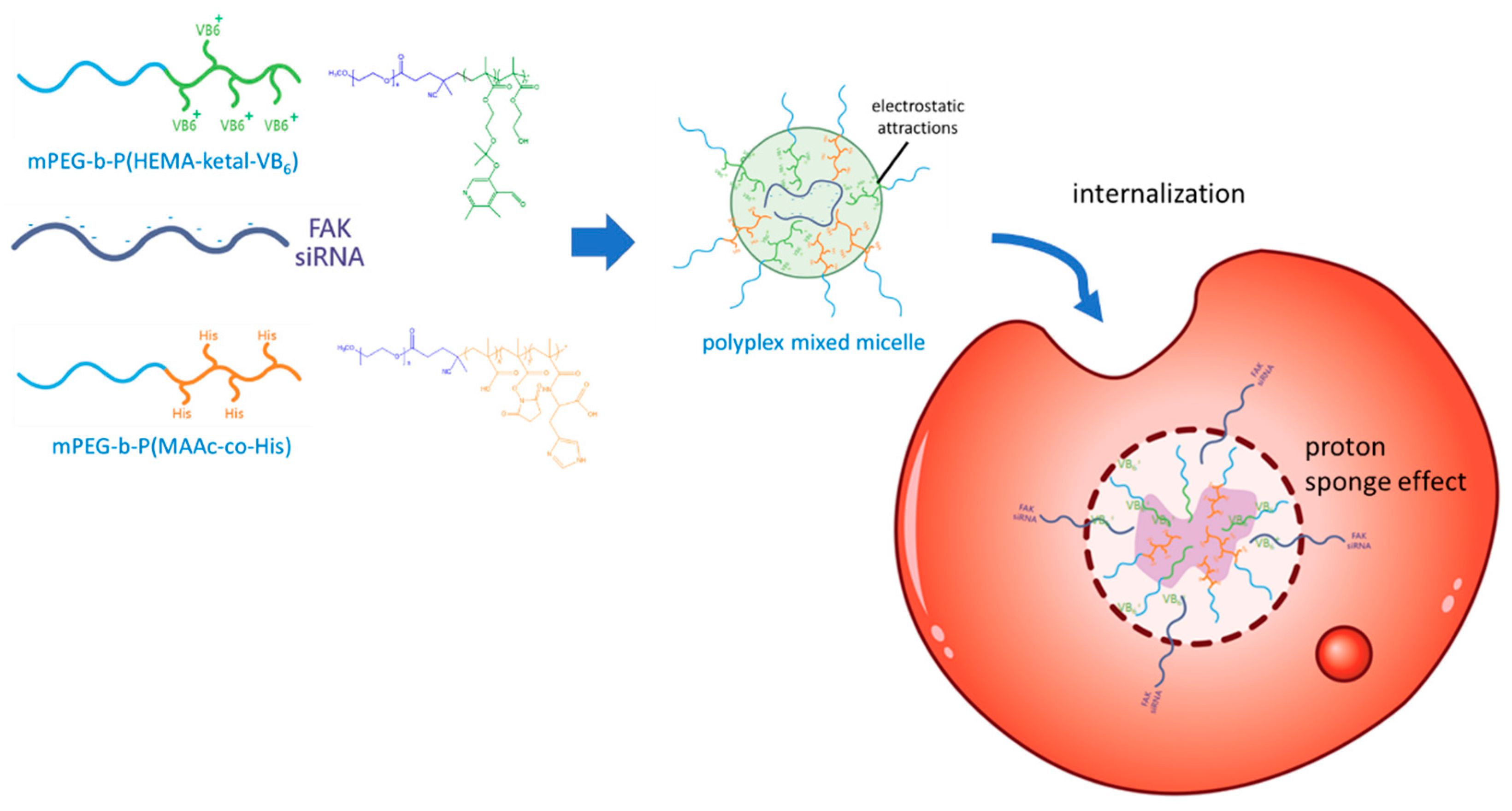
2.6. Polyplex-Mixed Micelle Preparation and Characterization
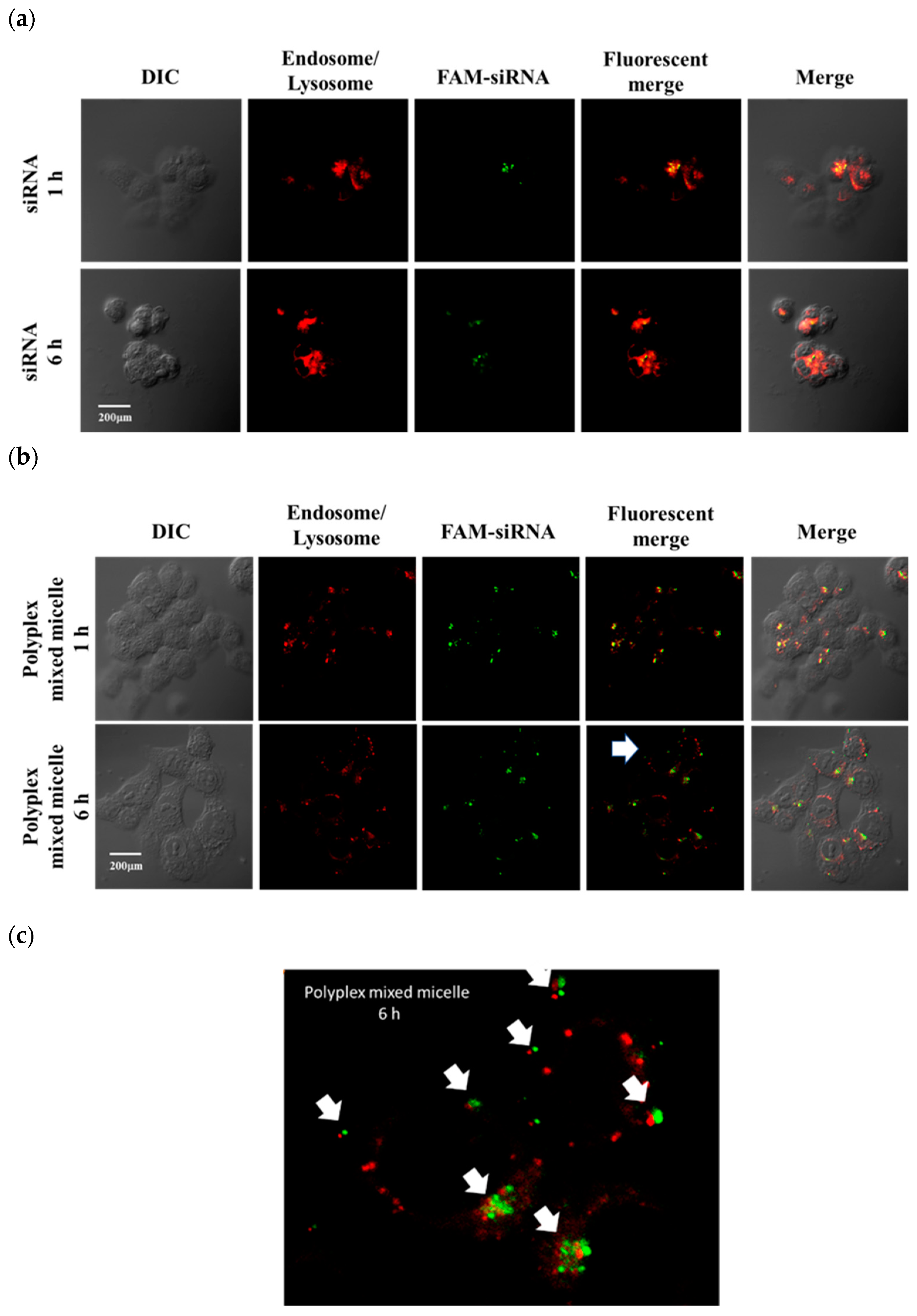
2.7. Internalization
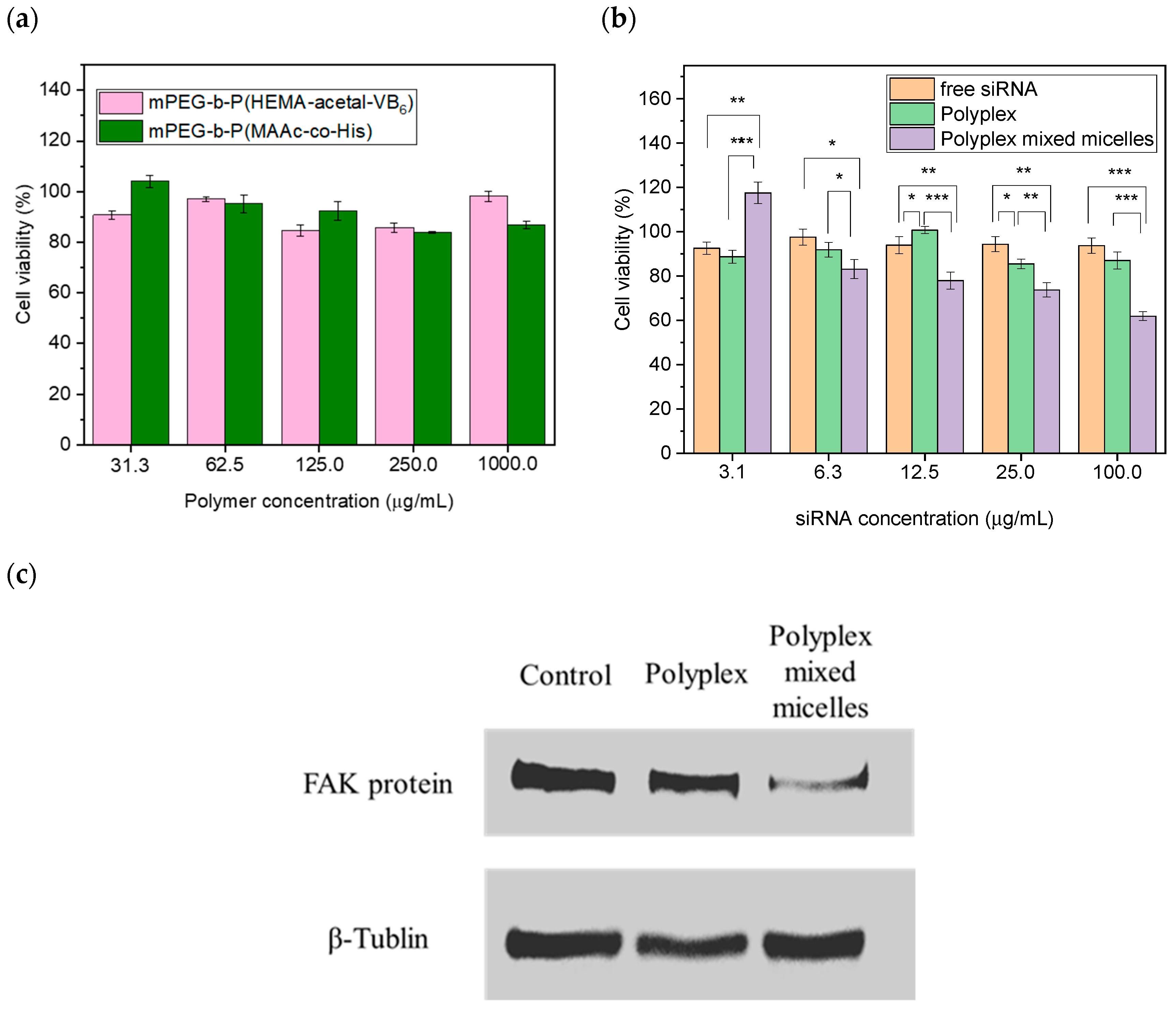
2.8. Cytotoxicity Evaluation
2.9. Western Blotting Analysis
3. Results and Discussions
3.1. Synthesis and Characterization of Copolymers mPEG-b-P(HEMA-ketal-VB6)
3.2. Synthesis and Characterization of Copolymers mPEG-b-P(MAAc-co-His)
3.3. Optimization and Characterization of the Polyplex-Mixed Micelles
3.4. Internalization, Intracellular Releasing Behaviors and Endosome Escape
3.5. In Vitro Cytotoxic Evaluation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Elbashir, S.M.; Harborth, J.; Lendeckel, W.; Yalcin, A.; Weber, K.; Tuschl, T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001, 411, 494–498. [Google Scholar] [CrossRef]
- Aagaard, L.; Rossi, J.J. RNAi therapeutics: Principles, prospects and challenges. Adv. Drug Deliv. Rev. 2007, 59, 75–86. [Google Scholar] [CrossRef]
- Reddy, L.S.; Sarojamma, V.; Ramakrishna, V. Future of RNAi in medicine: A review. World J. Med. Sci. 2007, 2, 01–14. [Google Scholar]
- Murmann, A.E.; Yu, J.; Opal, P.; Peter, M.E. Trinucleotide repeat expansion diseases, RNAi, and Cancer. Trends Cancer 2018, 4, 684–700. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Liang, G.; Cui, K.; Liang, Y.; Wang, Q.; Lv, S.; Cheng, X.; Zhang, L. Insight into the Prospects for RNAi Therapy of Cancer. Front. Pharmacol. 2021, 12, 308. [Google Scholar] [CrossRef]
- Setten, R.L.; Rossi, J.J.; Han, S.-P. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 2019, 18, 421–446. [Google Scholar] [CrossRef]
- Ando, H.; Ishida, T. An RNAi therapeutic, DFP-10825, for intraperitoneal and intrapleural malignant cancers. Adv. Drug Deliv. Rev. 2020. [Google Scholar] [CrossRef] [PubMed]
- Haussecker, D.; Kay, M.A. Drugging RNAi. Science 2015, 347, 1069–1070. [Google Scholar] [CrossRef]
- Gary, D.J.; Puri, N.; Won, Y.-Y. Polymer-based siRNA delivery: Perspectives on the fundamental and phenomenological distinctions from polymer-based DNA delivery. J. Control. Release 2007, 121, 64–73. [Google Scholar] [CrossRef]
- Braasch, D.A.; Jensen, S.; Liu, Y.; Kaur, K.; Arar, K.; White, M.A.; Corey, D.R. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry 2003, 42, 7967–7975. [Google Scholar] [CrossRef] [PubMed]
- Dowdy, S.F. Overcoming cellular barriers for RNA therapeutics. Nat. Biotechnol. 2017, 35, 222–229. [Google Scholar] [CrossRef]
- Kim, B.; Park, J.H.; Sailor, M.J. Rekindling RNAi therapy: Materials design requirements for in vivo siRNA delivery. Adv. Mater. 2019, 31, 1903637. [Google Scholar] [CrossRef] [PubMed]
- Bholakant, R.; Qian, H.; Zhang, J.; Huang, X.; Huang, D.; Feijen, J.; Zhong, Y.; Chen, W. Recent Advances of Polycationic siRNA Vectors for Cancer Therapy. Biomacromolecules 2020, 21, 2966–2982. [Google Scholar] [CrossRef]
- Liu, Y.; Ji, X.; Tong, W.W.; Askhatova, D.; Yang, T.; Cheng, H.; Wang, Y.; Shi, J. Engineering multifunctional RNAi nanomedicine to concurrently target cancer hallmarks for combinatorial therapy. Angew. Chem. Int. Ed. 2018, 57, 1510–1513. [Google Scholar] [CrossRef] [PubMed]
- Merkel, O.M.; Kissel, T. Quo vadis polyplex? J. Control. Release 2014, 190, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Midoux, P.; Pichon, C.; Yaouanc, J.J.; Jaffrès, P.A. Chemical vectors for gene delivery: A current review on polymers, peptides and lipids containing histidine or imidazole as nucleic acids carriers. Br. J. Pharmacol. 2009, 157, 166–178. [Google Scholar] [CrossRef]
- Meng, Z.; Luan, L.; Kang, Z.; Feng, S.; Meng, Q.; Liu, K. Histidine-enriched multifunctional peptide vectors with enhanced cellular uptake and endosomal escape for gene delivery. J. Mater. Chem. B 2017, 5, 74–84. [Google Scholar] [CrossRef]
- Hunter, A.C. Molecular hurdles in polyfectin design and mechanistic background to polycation induced cytotoxicity. Adv. Drug Deliv. Rev. 2006, 58, 1523–1531. [Google Scholar] [CrossRef] [PubMed]
- Hunter, A.C.; Moghimi, S.M. Cationic carriers of genetic material and cell death: A mitochondrial tale. Biochim. Biophys. Acta (BBA) Bioenerg. 2010, 1797, 1203–1209. [Google Scholar] [CrossRef]
- Oishi, M.; Nagasaki, Y.; Itaka, K.; Nishiyama, N.; Kataoka, K. Lactosylated poly (ethylene glycol)-siRNA conjugate through acid-labile β-thiopropionate linkage to construct pH-sensitive polyion complex micelles achieving enhanced gene silencing in hepatoma cells. J. Am. Chem. Soc. 2005, 127, 1624–1625. [Google Scholar] [CrossRef]
- Lee, Y.; Mo, H.; Koo, H.; Park, J.-Y.; Cho, M.Y.; Jin, G.-w.; Park, J.-S. Visualization of the degradation of a disulfide polymer, linear poly (ethylenimine sulfide), for gene delivery. Bioconjugate Chem. 2007, 18, 13–18. [Google Scholar] [CrossRef]
- Forrest, M.L.; Koerber, J.T.; Pack, D.W. A degradable polyethylenimine derivative with low toxicity for highly efficient gene delivery. Bioconjugate Chem. 2003, 14, 934–940. [Google Scholar] [CrossRef]
- Wang, C.; Wang, X.; Du, L.; Dong, Y.; Hu, B.; Zhou, J.; Shi, Y.; Bai, S.; Huang, Y.; Cao, H. Harnessing pH-Sensitive Polycation Vehicles for the Efficient siRNA Delivery. ACS Appl. Mater. Interfaces 2021, 13, 2218–2229. [Google Scholar] [CrossRef]
- Huang, X.; Sevimli, S.I.; Bulmus, V. pH-labile sheddable block copolymers by RAFT polymerization: Synthesis and potential use as siRNA conjugates. Eur. Polym. J. 2013, 49, 2895–2905. [Google Scholar] [CrossRef][Green Version]
- Wang, P.; Yan, Y.; Sun, Y.; Zhang, R.; Huo, C.; Li, L.; Wang, K.; Dong, Y.; Xing, J. Bioreducible and acid-labile polydiethylenetriamines with sequential degradability for efficient transgelin-2 siRNA delivery. J. Mater. Chem. B 2019, 7, 6994–7005. [Google Scholar] [CrossRef]
- Wong, S.; Kemp, J.A.; Shim, M.S.; Kwon, Y.J. Solvent-driven, self-assembled acid-responsive poly (ketalized serine)/siRNA complexes for RNA interference. Biomater. Sci. 2020, 8, 6718–6729. [Google Scholar] [CrossRef]
- Gannimani, R.; Walvekar, P.; Naidu, V.R.; Aminabhavi, T.M.; Govender, T. Acetal containing polymers as pH-responsive nano-drug delivery systems. J. Control. Release 2020. [Google Scholar] [CrossRef]
- Kwon, Y.J. Before and after endosomal escape: Roles of stimuli-converting siRNA/polymer interactions in determining gene silencing efficiency. Acc. Chem. Res. 2012, 45, 1077–1088. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.S.; Shin, H.J.; Na, K.; Bae, Y.H. Poly (l-histidine)–PEG block copolymer micelles and pH-induced destabilization. J. Control. Release 2003, 90, 363–374. [Google Scholar] [CrossRef]
- Mitra, S.K.; Hanson, D.A.; Schlaepfer, D.D. Focal adhesion kinase: In command and control of cell motility. Nat. Rev. Mol. Cell Biol. 2005, 6, 56–68. [Google Scholar] [CrossRef] [PubMed]
- McLean, G.W.; Carragher, N.O.; Avizienyte, E.; Evans, J.; Brunton, V.G.; Frame, M.C. The role of focal-adhesion kinase in cancer—a new therapeutic opportunity. Nat. Rev. Cancer 2005, 5, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.-T.; Cheng, Y.-T.; Lu, C.-Y.; Yen, Y.-W.; Yu, L.-Y.; Yu, K.-S.; Lyu, S.-Y.; Yang, C.-Y.; Lo, C.-L. Polymer–liposome complexes with a functional hydrogen-bond cross-linker for preventing protein adsorption and improving tumor accumulation. Chem. Mater. 2013, 25, 4364–4372. [Google Scholar] [CrossRef]
- Lee, K.-Y.; Chiang, Y.-T.; Hsu, N.-Y.; Yang, C.-Y.; Lo, C.-L.; Ku, C.-A. Vitamin E containing polymer micelles for reducing normal cell cytotoxicity and enhancing chemotherapy efficacy. Acta Biomater. 2015, 24, 286–296. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.; Neu, M.; Germershaus, O.; Merkel, O.; Sitterberg, J.; Bakowsky, U.; Kissel, T. Influence of polyethylene glycol chain length on the physicochemical and biological properties of poly (ethylene imine)-graft-poly (ethylene glycol) block copolymer/SiRNA polyplexes. Bioconjugate Chem. 2006, 17, 1209–1218. [Google Scholar] [CrossRef] [PubMed]
- Ozorio, L.P.; Pianzolli, R.; Mota, M.B.S.; Mota, C.J. Reactivity of glycerol/acetone ketal (solketal) and glycerol/formaldehyde acetals toward acid-catalyzed hydrolysis. J. Braz. Chem. Soc. 2012, 23, 931–937. [Google Scholar] [CrossRef]
- Hwang, H.S.; Hu, J.; Na, K.; Bae, Y.H. Role of polymeric endosomolytic agents in gene transfection: A comparative study of poly (L-lysine) grafted with monomeric L-histidine analogue and poly (L-histidine). Biomacromolecules 2014, 15, 3577–3586. [Google Scholar] [CrossRef]
- Spagnou, S.; Miller, A.D.; Keller, M. Lipidic carriers of siRNA: Differences in the formulation, cellular uptake, and delivery with plasmid DNA. Biochemistry 2004, 43, 13348–13356. [Google Scholar] [CrossRef]
- Grayson, A.C.R.; Doody, A.M.; Putnam, D. Biophysical and structural characterization of polyethylenimine-mediated siRNA delivery in vitro. Pharm. Res. 2006, 23, 1868–1876. [Google Scholar] [CrossRef]
- Shi, J.; Choi, J.L.; Chou, B.; Johnson, R.N.; Schellinger, J.G.; Pun, S.H. Effect of polyplex morphology on cellular uptake, intracellular trafficking, and transgene expression. ACS Nano 2013, 7, 10612–10620. [Google Scholar] [CrossRef]
- Von Gersdorff, K.; Sanders, N.N.; Vandenbroucke, R.; De Smedt, S.C.; Wagner, E.; Ogris, M. The internalization route resulting in successful gene expression depends on both cell line and polyethylenimine polyplex type. Mol. Ther. 2006, 14, 745–753. [Google Scholar] [CrossRef]
- Rejman, J.; Oberle, V.; Zuhorn, I.S.; Hoekstra, D. Size-dependent internalization of particles via the pathways of clathrin-and caveolae-mediated endocytosis. Biochem. J. 2004, 377, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhou, J.; Wu, Y.; Dong, Y.; Du, L.; Yang, T.; Wang, Y.; Guo, S.; Zhang, M.; Hussain, A. Core Role of Hydrophobic Core of Polymeric Nanomicelle in Endosomal Escape of siRNA. Nano Lett. 2021, 21, 3680–3689. [Google Scholar] [CrossRef] [PubMed]
- Patil, M.L.; Zhang, M.; Minko, T. Multifunctional triblock nanocarrier (PAMAM-PEG-PLL) for the efficient intracellular siRNA delivery and gene silencing. ACS Nano 2011, 5, 1877–1887. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hung, S.-F.; Wen, Y.-H.; Yu, L.-Y.; Chiu, H.-C.; Chiang, Y.-T.; Lo, C.-L. Development of a Rapid-Onset, Acid-Labile Linkage Polyplex-Mixed Micellar System for Anticancer Therapy. Polymers 2021, 13, 1823. https://doi.org/10.3390/polym13111823
Hung S-F, Wen Y-H, Yu L-Y, Chiu H-C, Chiang Y-T, Lo C-L. Development of a Rapid-Onset, Acid-Labile Linkage Polyplex-Mixed Micellar System for Anticancer Therapy. Polymers. 2021; 13(11):1823. https://doi.org/10.3390/polym13111823
Chicago/Turabian StyleHung, Shiou-Fen, Yu-Han Wen, Lu-Yi Yu, Hsin-Cheng Chiu, Yi-Ting Chiang, and Chun-Liang Lo. 2021. "Development of a Rapid-Onset, Acid-Labile Linkage Polyplex-Mixed Micellar System for Anticancer Therapy" Polymers 13, no. 11: 1823. https://doi.org/10.3390/polym13111823
APA StyleHung, S.-F., Wen, Y.-H., Yu, L.-Y., Chiu, H.-C., Chiang, Y.-T., & Lo, C.-L. (2021). Development of a Rapid-Onset, Acid-Labile Linkage Polyplex-Mixed Micellar System for Anticancer Therapy. Polymers, 13(11), 1823. https://doi.org/10.3390/polym13111823