Abstract
This study aimed to investigate the effects of nanohydroxyapatite–silica–glass ionomer cement (nanoHA–silica–GIC) on the differentiation of dental pulp stem cells (DPSCs) into odontogenic lineage. DPSCs were cultured in complete Minimum Essential Medium Eagle—Alpha Modification (α-MEM) with or without nanoHA–silica–GIC extract and conventional glass ionomer cement (cGIC) extract. Odontogenic differentiation of DPSCs was evaluated by real-time reverse transcription polymerase chain reaction (rRT–PCR) for odontogenic markers: dentin sialophosphoprotein (DSPP), dentin matrix protein 1 (DMP1), osteocalcin (OCN), osteopontin (OPN), alkaline phosphatase (ALP), collagen type I (COL1A1), and runt-related transcription factor 2 (RUNX2) on day 1, 7, 10, 14, and 21, which were normalized to the house keeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Untreated DPSCs were used as a control throughout the study. The expressions of DSPP and DMP1 were higher on days 7 and 10, that of OCN on day 10, those of OPN and ALP on day 14, and that of RUNX2 on day 1; COL1A1 exhibited a time-dependent increase from day 7 to day 14. Despite the above time-dependent variations, the expressions were comparable at a concentration of 6.25 mg/mL between the nanoHA–silica–GIC and cGIC groups. This offers empirical support that nanoHA–silica–GIC plays a role in the odontogenic differentiation of DPSCs.
1. Introduction
Dental pulp stem cells (DPSCs) were first isolated by Gronthos and his colleagues from human dental pulp [1]. DPSCs can be used directly for dental therapy as these cells have the ability to differentiate into odontoblasts. Besides that, they have also been used as an in vitro model to evaluate newly developed bioactive materials [2]. The use of glass ionomer cements (GICs) was reported by Wilson and Kent in 1970s [3]. GICs are widely used in dental application due to their many advantages such as biocompatibility, long-term release of fluoride which acts as an anticariogenic agent, elasticity similar to dentin, and ability to bond to the tooth structure directly [4,5]. Despite their advantages, they have certain limitations such as susceptibility to dehydration and poor physical and mechanical properties [6], which have limited the extensive use of GICs as a filling material in dentistry. In order to overcome the poor mechanical properties of GICs, a number of modifications of conventional GICs (cGICs) have been done such as incorporation of fiber-reinforcement, hydroxyapatite, and zirconia into GICs [7,8]. GIC is composed of two main ingredients required for maintaining its desirable properties, namely, polymeric water-soluble acid and ion-leachable glass. For this purpose, aluminosilicate glass is used to prepare the GIC powder that provides a constant source of metal ions for the cement-forming reaction [9]. Glasses used in the GIC are complex and have three major components: silica (SiO2), alumina (Al2O3), and calcium fluoride (CaF2). In addition, they also contain sodium fluoride (NaF) and cryolite (Na3AlF6) or aluminium phosphate (AlPO4) [10,11]. However, alumina and silica are the two main components of GIC powder that form the “backbone and skeletal structure of the glass” [10]. The second component of GIC is the liquid containing polyacids known as polyalkenoics. Since the early formulations of GIC comprised about 40–50% of aqueous solution of acrylic acid [9,12] and had few disadvantages such as high viscosity and a short shelf life, acrylic acid was later co-polymerized with various homopolymers or copolymers of carboxylic acids such as acrylic acid, maleic acid, itaconic acid, and tricarboxylic acid. Thus, glass ionomers are complex materials with a predominantly silica gel-like matrix as a result of the reaction between an aqueous poly acrylic acid solution and a fluoro–alumino–silicate glass powder. The partially dissolved remnant glass cores act as fillers within the matrix which is composed of poly salt bridges and polymer chains [13]. Barry et al. [14] also suggested that a set matrix of GIC is a highly intricate network of aluminium and calcium polyacrylate gel which contains ample fluoride inside. Nicholson in 2010 also reported that three regions can be identified in the structure of GIC which include a core of glass particles surrounded by a layer of silica and lastly the matrix of the cement [15].
Hydroxyapatite has excellent biocompatibility and can promote osteoconduction and osteointegration. It is preferred as the biomaterial of choice in both dentistry and orthopedics [16,17]. Due to the development in nanotechnology, nano-hydroxyapatite (nanoHA) has found a place in dental applications [18]. Besides that, nanoHA has been used as an additive material with the aim of improving the already existing dental materials in restorative dentistry [19]. Nano-hydroxyapatite–silica (nanoHA–silica) has been synthesized by a one-pot sol-gel technique [20,21]. The nanoHA–silica alone was demonstrated to be non-genotoxic based on a comet assay [22]. The nanoHA–silica consists of a mixture of spherical silica particles (~50 nm) and rod-shaped HA particles ranging between 100–200 nm; moreover, the incorporation of nanoHA–silica into cGIC resulted in better shear bond strength and mechanical properties (compressive and flexural strengths and Vickers hardness) [20,21,23]. Moheet and colleagues, based on their several characterization studies on nanoHA–silica powder and nanohydroxyapatite-silica-glass ionomer cement (nanoHA–silica–GIC) composite, concluded that the nanopowder was successfully incorporated into the cGIC based on the elemental peaks and molecular interactions [23]. It was also suggested that the homogenous particle distribution might have contributed to the enhancement of mechanical properties of the modified cement resulting in the enhancement of GIC cement matrix [20]. Moreover, the high degree of cross linking between silica and GIC makes the nanoHA–silica–GIC much stronger in hardness compared to cGIC [21]. Studies have been previously carried out to investigate the odontogenic differentiation potential of human dental pulp cells (hDPCs) from deciduous teeth using pre-reacted glass–ionomer cement [24], hydrogel scaffolds from decellularized bone extracellular matrix and collagen type 1 [25], dental pulp stem cells (DPSCs) on tricalcium phosphate (TCP) scaffolds [26], and three bioactive materials, namely, nanoHA, mineral trioxide aggregate, and calcium-enriched mixture cements [27]. Kwon and colleagues investigated the effects of triethylene glycol dimethacrylate (TEGDMA) and 2-hydroxyethyl methacrylate (HEMA) on the odontogenic differentiation of human dental pulp cells (HDPCs) [28]. However, Bakapoulou et al. reported that HEMA exhibited a cytotoxic effect on dental pulp cells that can disturb the odontogenic differentiation potential of HEMA which could lead to compromising pulp-tissue homeostasis and repair [29]. Thus, there has always been a quest to explore the odontogenic differentiation potential of materials in dentistry. However, there is still a dearth of information on their odontogenic potential in dental stem cells. Bearing in mind the above properties, the present study was principally aimed at evaluating the effect of nanoHA–silica–GIC on the differentiation of DPSCs into odontogenic lineage.
2. Materials and Methods
2.1. Cell Culture
DPSCs (AllCells, Alameda, CA, USA; Cat no. DP003F) were grown in Minimum Essential Medium Eagle—Alpha Modification (α-MEM, Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY, USA) and 1% of penicillin/streptomycin (100 U/mL penicillin and 100 g/mL streptomycin, Gibco, Grand Island, NY, USA) and were incubated at 37 °C in a 5% CO2 incubator until 70–80% confluence. The DPSCs were revived from cryopreservation and sub-cultured twice before seeding for treatment. A negative control group (DPSCs without treatment) was also included in this study. Both groups of DPSCs, one with nanoHA–silica–GIC and the other with cGIC, were incubated and harvested at different incubation times (day 1, 7, 10, 14, and 21). Passage 7 was used for the current study.
2.2. Material Preparation
NanoHA–silica–GIC and commercial cGIC Fuji IX GP (GC Corporation, Tokyo, Japan) were used in the current study. NanoHA–silica–GIC was prepared by the addition of nanoHA–silica to cGIC. cGIC was prepared according to the instructions provided by the manufacturer. Synthesis of nanoHA–silica powder was carried out as previously described [20]: 100 mg of nanoHA–silica powder was weighed and added to 1900 mg of cGIC powder to obtain a 5% nanoHA–silica–GIC powder mixture. This 5% nanoHA–silica–GIC powder mixture was ground manually using a pestle and mortar. The Fuji XI liquid was added to the powder mixture based on the manufacturer’s recommended ratio of 1:1 (powder: liquid), which was 0.36 g of powder to 0.10 g of liquid, and mixed using an agate spatula. The cement was then placed into a 10 mm × 2 mm mould and left for setting.
In the meantime, cGIC was prepared by spatulation of the powder into the Fuji XI liquid at a ratio of 1:1 (0.36 g of powder to 0.10 g of liquid) and mixed. This cement was also introduced into a 10 mm × 2 mm mould and left for setting. After 24 h of incubation, the cements were removed from the molds, weighed, and sterilized under ultraviolet radiation for 30 min. Then, the cements were introduced individually into a centrifuge tube with a suitable amount of complete growth medium (standardized at 200 mg/mL). The medium containing the materials was incubated at 37 °C with 5% CO2 for 72 h. After incubation, the material extracts were filtered using a 0.22 µm syringe filter into a centrifuge tube [30]. In this study, the material extract/indirect method was chosen over the direct method as in the latter, the cells are susceptible to trauma from abrasion or crushing. In addition, material extracts offer the advantage of measuring the dose–response relationship. A material extract can also be sterilized simply by filtration and offers the ability to evaluate its effect on cell cultures regardless of the cell proximity or lack of the material extract [31]. Concentrations of 3.125 and 6.25 mg/mL were selected for the nanoHA–silica–GIC material extract, and concentrations of 6.25 and 12.5 mg/mL were selected for the cGIC material extract in this study. This was based on a previous study where the authors evaluated the cell viability of DPSCs by treating the material extracts of nanoHA–silica–GIC and cGIC at 200, 100, 50, 25, 12.5, 6.25, and 3.125 mg/mL [32] using 3-[4,5-dimethylthiazole-2-yl]-2,5-diphenyltetrazolium bromide (MTT) assay. A previous study reported the highest cell viability percentage at concentrations of 3.125 mg/mL (96.57%) and 6.25 mg/mL (92.21%) for nanoHA–Silica–GIC and 6.25 mg/mL (92.65%) and 12.5 mg/mL (89.93%) for cGIC [33]. These concentrations were achieved by serially diluting half the original material extracts of nanoHA–silica–GIC and cGIC. This was done by transferring 2.5 mL of extract serially into a series of six 15 mL tubes that contained 2.5 mL of complete growth media. This resulted in material extracts with concentrations of 200, 100, 50, 25, 12.5, 6.25, and 3.125 mg/mL.
2.3. RNA Extraction
After each time interval, DPSCs were trypsinized and transferred into a 15 mL centrifuge tube. The cells were centrifuged at 12,000 rpm for 5 min. The supernatant was discarded, and each cell pellet was re-suspended using 1 mL of phosphate-buffered saline (PBS) and transferred into a 1.5 mL microcentrifuge tube and centrifuged at 13,000 rpm for 5 min. The supernatant was discarded, and the cell pellet was used for ribonucleic acid (RNA) extraction using an InnuPREP RNA Mini Kit (Analytik Jena, Jena, Germany).
2.4. Real-Time Reverse Transcription Polymerase Chain Reaction
A quantitative analysis of the gene expression of dentin sialophosphoprotein (DSPP), dentin matrix protein 1 (DMP1), osteocalcin (OCN), osteopontin (OPN), alkaline phosphatase (ALP), collagen type I (COL1A1), and runt-related transcription factor 2 (RUNX2) relevant to odontogenic differentiation of cells was performed. The extracted RNA was amplified using SensiFAST SYBR Hi-Rox One-step (Bioline, London, UK) according to the manufacturer’s instruction. The primer sequences of odontogenic marker genes were based on previous studies: DSPP [34], DMP1 [34], OCN [35], OPN [35], ALP [35], COL1A1 [34], and RUNX2 [36]. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the housekeeping gene [34]. Real-time reverse transcription polymerase chain reaction (rRT–PCR) conditions were as follows: 45 °C for 10 min, 95 °C for 2 min, followed by 40 cycles of 95 °C for 5 s; 60 °C for 10 s, and 72 °C for 5 s. The experiment was carried out in triplicate. The results were analyzed using the 2ΔΔCT method [37]. In this formula, CT values at each time point were normalized to the house keeping gene, GAPDH, in the same sample. Then, the CT values were further normalized to CT values of control samples at the corresponding time points. Briefly, the CT values of the gene of interest (GOI) in both the experimental sample(s) and calibrator(c) (control sample) were adjusted in relation to a normalizer (norm) gene’s (endogenous control/GAPDH) CT for the same two samples. The resulting 2−ΔΔCT value was incorporated to determine the fold change in expression using the equations below.
The results are expressed as the mean ± standard error of mean (SEM).
2.5. Statistical Analysis
The data were analyzed using SPSS version 20 (SPSS Inc., Chicago, IL, USA). The Kolmogorov–-Smirnov test showed that the data were normally distributed, and therefore, parametric statistical tests were performed (analysis of variance, followed by Tukey’s test; Dunnett’s test for multiple comparisons). The significance level was set at p < 0.05.
3. Results
The gene expression analyses were carried out to determine the expression of odontogenic markers in DPSCs. The odontogenic markers selected for this study were DSPP, DMP1, OCN, OPN, ALP, COL1A1, and RUNX2. These genes were normalized with GAPDH. Untreated DPSCs were used as a control throughout the study. The results are expressed as mRNA relative expression. The mRNA expression of DSPP increased after day 1 and peaked on day 10 in all the groups (Figure 1A). However, the expression decreased after day 14 followed by day 21. Furthermore, the expression levels in the 6.25 mg/mL nanoHA–silica–GIC and cGIC were significantly higher than those in other treatment groups and control group on days 7 and 10. However, there was no significant difference between nanoHA–silica–GIC and cGIC groups on days 14 and 21. DMP1 mRNA expression showed an increase after day 1 and peaked on day 10 in all groups (Figure 1B). However, the expression decreased after day 14 followed by day 21. On day 10, the fold change of the expression in 6.25 mg/mL nanoHA–silica–GIC and cGIC was 50.18 and 49.97, which was higher than those in 3.125 mg/mL nanoHA–silica–GIC and 12.5 mg/mL cGIC and control groups, respectively, with no significant difference between them.
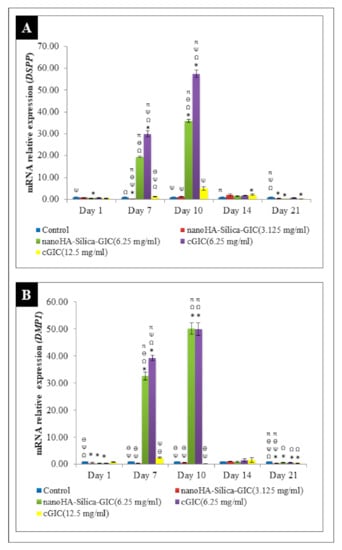
Figure 1.
Expression of odontogenic gene markers (A). Dentin sialophosphoprotein (DSPP) and (B). Dentin matrix protein 1 (DMP1) by real-time reverse transcription polymerase chain reaction (rRT–PCR) in dental pulp stem cells (DPSCs). The data are presented as the mean ± standard error of mean (SEM). * indicates a significant difference compared to the control. Ω indicates a significant difference compared to nanohydroxyapatite-silica-glass ionomer cement (nanoHA–silica–GIC) (3.125 mg/mL). Ψ indicates a significant difference compared to nanoHA–silica–GIC (6.25 mg/mL). Ө indicates a significant difference compared to conventional glass ionomer cement (cGIC) (6.25 mg/mL). π indicates a significant difference compared to cGIC (12.5 mg/mL).
OCN expression was not altered significantly among all groups on days 1 and 14 (Figure 2A). However, the fold change of OCN mRNA expression in the 6.25 mg/mL nanoHA–silica–GIC and cGIC groups was 1.09 and 2.49, which was higher than that in other treatment groups and control group, respectively, on day 10, which showed a significant difference between them. The expression of OPN fluctuated in all the groups as illustrated in Figure 2B. In addition, OPN expression in treatment groups was lower than that in the control group at all time points.
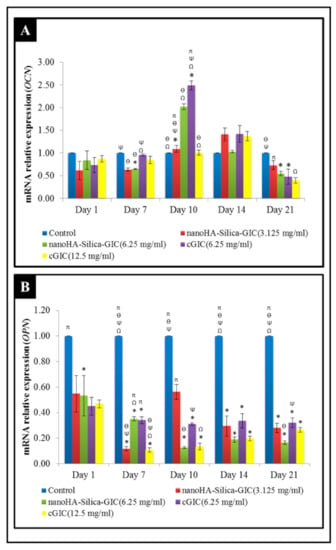
Figure 2.
Expression of odontogenic gene markers (A) Osteocalcin (OCN) and (B) Osteopontin (OPN) based on real-time reverse transcription polymerase chain reaction (rRT–PCR) in dental pulp stem cells (DPSCs). The data are presented as the mean ± standard error of mean (SEM). * indicates a significant difference compared to the control. Ω indicates a significant difference compared to nanohydroxyapatite-silica-glass ionomer cement (nanoHA–silica–GIC) (3.125 mg/mL). Ψ indicates a significant difference compared to nanoHA–silica–GIC (6.25 mg/mL). Ө indicates a significant difference compared to conventional glass ionomer cement (cGIC) (6.25 mg/mL). π indicates a significant difference compared to cGIC (12.5 mg/mL).
The mRNA expression of ALP increased on day 7 and peaked on day 14 in all the groups (Figure 3A). However, the expression of ALP declined after day 21. On day 14, 12.5 mg/mL cGIC had the highest fold change of 15.467 when compared with other groups. It was noted that there was significant difference between 12.5 mg/mL cGIC and other groups on day 14 (p < 0.05). For COL1A1 expression, cells treated with nanoHA–silica–GIC and cGIC exhibited a time-dependent increase from day 7 to day 14 (Figure 3B). Moreover, the highest up-regulation was seen at 6.25 mg/mL cGIC compared with other treatment groups and the control group. The mRNA expression of COL1A1 declined on day 21 where there was no significant difference between all groups (p > 0.05).
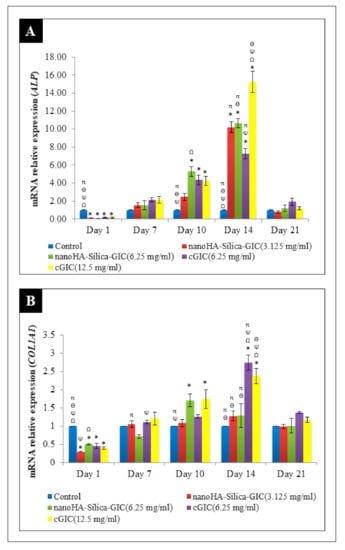
Figure 3.
Expression of odontogenic gene markers (A) Alkaline phosphatase (ALP) and (B) Collagen type I (COL1A1) based on real time reverse transcription polymerase chain reaction (rRT–PCR) in dental pulp stem cells (DPSCs). The data are presented as the mean ± standard error of mean (SEM). * indicates a significant difference compared to C (control). Ω indicates a significant difference compared to nanohydroxyapatite-silica-glass ionomer cement (nanoHA–silica–GIC) (3.125 mg/mL). Ψ indicates a significant difference compared to nanoHA–silica–GIC (6.25 mg/mL). Ө indicates a significant difference compared to conventional glass ionomer cement (cGIC) (6.25 mg/mL). π indicates a significant difference compared to cGIC (12.5 mg/mL).
The mRNA expression of RUNX2 increased on day 1 where the difference was not significant among all the groups (Figure 4). However, a low expression of RUNX2 was detected in all the groups after day 7.
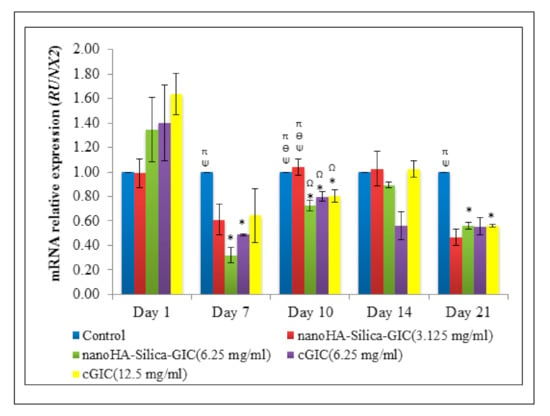
Figure 4.
Expression of odontogenic gene marker, Runt-related transcription factor 2 (RUNX2) based on real time reverse transcription polymerase chain reaction (rRT–PCR) in dental pulp stem cells (DPSCs). The data are presented as the mean ± standard error of mean (SEM). * indicates a significant difference compared to C (control). Ω indicates a significant difference compared to nanohydroxyapatite-silica-glass ionomer cement (nanoHA–silica–GIC) (3.125 mg/mL). Ψ indicates a significant difference compared to nanoHA–silica–GIC (6.25 mg/mL). Ө indicates a significant difference compared to conventional glass ionomer cement (cGIC) (6.25 mg/mL). π indicates a significant difference compared to cGIC (12.5 mg/mL).
4. Discussion
GICs have wide application in clinical dentistry including as liners and bases, fissure sealants, restorative materials, and also as bonding agents for orthodontic brackets [38]. The addition of HA to GICs improves the biocompatibility of GICs and also the mechanical characteristics. Additionally, it has the ability to enhance the bond strength to the tooth structure because of its similar composition and structure to enamel and dentin [39]. In addition, Gu and colleagues reported that GICs containing 4 wt% HA particles exhibited enhanced mechanical properties in comparison with commercial GICs which could be due to the continuous formation of aluminium salt bridges, which provided the final strength of the cements [7]. NanoHA has significant remineralizing effects on initial enamel lesions [18,19]. Moshaverina and colleagues focused on the addition of nanoHA and fluorapatite (FA) to cGICs and reported that the nanoHA/FA added cements exhibited higher mechanical strength and higher bond strength to dentin as compared with the control group [18]. It was also reported that both nanoHA and FA are involved in the acid–base reaction of the GIC and react with inorganic/organic components of the GIC network through their phosphate and calcium ions. During the reaction, after H+ ions attack the ceramic particles, there would be more Ca2+ ions available for cement formation, polysalt bridge formation, and cross-linking, therefore reinforcing the GIC matrix [18].
Despite this, research continues to enhance the mechanical properties of GICs with the aim of expanding their indications and clinical applications. A number of markers have been well identified to be directly and indirectly involved in odontogenic differentiation. These include DSPP, DMP1, OCN, ALP, OPN, COL1A1, and RUNX2 [34,35,36]. Therefore, these genes were investigated with GAPDH as the housekeeping gene in this study.
DSPP, a member of small integrin-binding ligand N-linked glycoproteins (SIBLINGs), is widely regarded as a specific marker of odontoblast. DSPP is expressed more in the dentin than in the bone and it regulates the progress of dentin formation [40]. In the current study, the gene expression level of DSPP was up-regulated significantly on day 7 and reached a peak on day 10 in treatment groups. This is in accordance with the fact that DSPP is a marker for early odontogenic differentiation as reported previously where the expression of DSPP was higher during primary dentinogenesis than secondary dentinogenesis in odontoblast formation [41]. This showed that DSPP functions during primary dentinogenesis and is involved in odontoblast differentiation [41]. On day 14 and 21, the expression of DSPP was down-regulated which may be due to the cells entering their terminal differentiation state. The expression of DSPP mRNA was significantly increased in the 6.25 mg/mL nanoHA–silica–GIC and cGIC groups compared with the control group and in 3.125 mg/mL nanoHA–silica–GIC and 12.5 mg/mL cGIC groups on days 7 and 10. These results indicated that nanoHA–silica–GIC and cGIC at certain concentrations may increase the expression of DSPP, especially during the early stages of differentiation, which contributed to the odontoblastic differentiation of DPSCs.
DMP1 plays a regulatory role in collagen matrix organization and dentin mineralization. It is expressed during early odontoblast differentiation [25,26]. Previous studies reported that DMP1 is expressed prior to the expression of DSPP and regulates DSPP gene transcription [42,43,44]. In the present study, DMP1 expression was up-regulated from day 7 to day 10 and reached a maximum on day 10, suggesting that DMP1 expression is necessary in the early stage of odontogenesis. However, after day 14 and day 21, the gene expression of DMP1 was down-regulated until day 21 indicating that the cells entered terminal differentiation. Moreover, DMP1 is shown to bind specifically with the DSPP promoter during early odontogenic differentiation. It was reported that DMP1 activates DSPP transcription which explains the synchronized expression of DSPP and DMP1 in the study [45]. Moreover, the expression levels in 6.25 mg/mL nanoHA–silica–GIC and cGIC on day 10 were 50.18 and 49.97 times those in 3.125 mg/mL nanoHA–silica–GIC and 12.5 mg/mL cGIC and control groups, respectively, with no significant difference. The result showed that nanoHA–silica–GIC and cGIC may promote early odontogenic differentiation.
OCN, a gamma-carboxyglutamic acid containing protein, is always expressed during the late period of odontoblast and osteoblast differentiation [46]. The expression of OCN showed an up-regulation on day 10, where high expression was exhibited in the 6.25 mg/mL nanoHA–silica–GIC and cGIC groups. This result indicated that nanoHA–silica–GIC and cGIC at a particular concentration might promote odontoblast differentiation. OPN is a phosphoprotein expressed in differentiating osteoblasts. OPN is highly expressed during the last stage of bone formation, the mineralization period [35]. The results demonstrated that OPN expression in treatment groups was lower than that in the control group at all times which indicated that neither nanoHA–silica–GIC nor cGIC promoted osteogenic differentiation.
ALP plays a crucial role in mineral deposition and is an important marker during the early stage of differentiation [47,48]. In the present study, the expression of ALP gradually increased and showed the highest up-regulation on day 14 followed by a decline in all groups on day 21. Similarly, it was demonstrated based on RT–PCR that the expression of ALP progressively increased in DPSCs after 5 and 10 days of culture [49]. However, in their study, osteogenic medium was used instead of αMEM to culture the cells. In addition, our result also showed that 12.5 mg/mL cGIC had the highest fold change of 15.467 when compared with other groups on day 14. The findings demonstrated that cGIC at a higher concentration induces early DPSC differentiation compared to cGIC at a lower concentration.
COL1A1 is the predominant collagen in dentin and constitutes the fundamental framework that supports cellular proliferation, migration, and mineralization. It is expressed by osteoblastic and odontoblastic cells at all stages during development and throughout life [50,51]. In addition, it forms a template for the controlled deposition of calcium phosphate [52]. The current findings showed that the expression of COL1A1 was up-regulated from day 7 to day 14 indicating that odontoblast differentiation had taken place in DPSCs. The findings are consistent with the previous study that reported that COL1A1 is one of the first extracellular matrix components to be expressed [52].
RUNX2 regulates tooth and bone development in the early stages. Besides direct regulation of tooth and bone development, RUNX2 also regulates tooth and bone development through RUNX2-related signaling pathways such as Osterix (Osx) [53]. Based on the current result, RUNX2 was up-regulated on day 1 and down-regulated from day 7 onwards. The results are in agreement with the previous findings in an in vivo study which stated that RUNX2 expression was down-regulated in the dental pulp cells and odontoblasts at the later stages of tooth development [53]. Thus, it was reported that RUNX2 was not involved in the dental pulp cell and odontoblast differentiation at the late stage. Interestingly, few studies have also demonstrated that RUNX2 can inhibit terminal differentiation of odontoblasts [54,55].
Fujita et al. in 2016 studied the effects of pre-reacted glass-ionomer (PRG) cement on the odontogenic differentiation of human dental pulp cells derived from deciduous teeth (hDPC-Ds) using alkaline phosphatase (ALP) activity and immunocytochemistry [24]. These authors reported that the PRG cement extracts significantly enhanced the ALP activity, ALP staining in the extracellular matrix of hDPC-Ds, and also the release of F and Al ions which enhanced the differentiation of hDPC-Ds. Another group of researchers investigated the odontogenic differentiation of DPSCs cultured for three weeks on hydrogel scaffolds derived from bone extracellular matrix (bECM) and compared that with those seeded on collagen I (Col-I) [25]. They evaluated the gene expression of DSPP, DMP1, and matrix extracellular phosphoglycoprotein (MEPE) using quantitative reverse transcription–polymerase chain reaction (qRT–PCR) and mineral deposition using Von Kossa staining. The mRNA expression levels of DSPP, DMP1, and MEPE were significantly upregulated with DPSCs cultured on bECM hydrogels in comparison to those cultured on Col-I scaffolds; there was more mineral deposition observed on bECM hydrogel scaffolds based on Von Kossa staining, establishing the potential of bECM hydrogel scaffolds in odontogenic differentiation of DPSCs [25]. In another study [26], the mRNA levels of DMP1 and DSPP using real-time RT–PCR were analyzed to study the odontogenic differentiation of DPSCs on TCP scaffolds (three-dimensional culture) cultured for 21 days. They reported that both the genes were up-regulated and concluded that TCP possessed odontogenic-inducing potential. The results of the current research are in agreement with the previous studies [25,26] where an up-regulation was observed in the case of both DSPP and DMP1. Mohamed and colleagues studied the effect of three bioactive materials, namely, nanoHA, mineral trioxide aggregate, and calcium-enriched mixture cements, on the odontogenic differentiation of DPSCs isolated from human third molars [27]. They classified the cultured cells incubated for 14 days according to biomaterial supplementation either in odontogenic differentiation medium or in growth medium, studied the relative expressions of Enamlysin and DSPP by real-time RT–PCR, and reported that all the materials in their study promoted the odontogenic differentiation of DPSCs. Another study evaluated the effects of TEGDMA and HEMA on odontogenic differentiation of HDPCs [28]. They mimicked the clinical situations by treating the HDPCs with resin monomers for 24 h prior to analyzing the mRNA expression of genes related to pulp cell differentiation. They found that the mRNA expression of DSPP, OCN, and OPN was downregulated by resin monomers after a culture period of 12 days [28]. In line with that, Bakapoulou et al. also reported that deciduous teeth stem cells exposed to HEMA and TEGDMA reduced or completely inhibited the expression of markers BSP, DSPP, and OCN and hence the odontogenic differentiation potential leading to compromising pulp-tissue homeostasis and repair [29]. The attachment of cells to material surfaces has been shown to participate in cell proliferation, migration, and differentiation [56] for which scanning electron microscopy (SEM) has been suggested to improve visualisation through observation of cell morphology and material–cell interactions [57]. Hii et al., based on their SEM study, reported that nano-HA–silica–GIC and cGIC favored the attachment of dental pulp stem cells [33].
In conclusion, the expressions of both DSPP and DMP1 were higher on days 7 and 10 and also comparable at a concentration of 6.25 mg/mL between the nanoHA–silica–GIC and cGIC groups. In the case of OCN, the mRNA expression in the 6.25 mg/mL nanoHA–silica–GIC and cGIC groups was higher than that in other treatment groups and the control group, respectively, on day 10, which were also comparable. However, the expression of OPN in treatment groups was lower than that in the control group at all time points. Despite the mRNA expression of ALP peaking on day 14 in all the groups, the expression was comparable between nanoHA–silica–GIC and cGIC at 6.25 mg/mL on day 10. For COL1A1 expression, cells treated with nanoHA–silica–GIC and cGIC exhibited a time-dependent increase from day 7 to day 14 with the expression comparable between nanoHA–silica–GIC and cGIC at 6.25 mg/mL on day 10. The mRNA expression of RUNX2 increased on day 1 where the difference was not significant in all the groups, with the expression being comparable between nanoHA–silica–GIC and cGIC at 6.25 mg/mL. The promotion effect could be due to the bioactive properties of GICs. As mentioned earlier, GICs release sodium, fluoride, silicate, and phosphate ions into surrounding aqueous media [58]. It was reported that silicate promotes osteoblast proliferation and gene expression through bone mineralization, collagen synthesis, cross-linking of the connective tissue, and metabolism [59]. The results of this study are based on an in vitro model to evaluate the odontogenic differentiation potential of the test materials which may not typically simulate the clinical situation as the material is applied to vital tissues comprising different types of cells such as ameloblasts and odontoblasts, blood, and interstitial fluids. Moreover, the response of related cell populations to the material may be affected by the placement of test materials in the oral cavity. This finding offers empirical evidence indicating that nanoHA–silica–GIC plays a role in the odontogenic differentiation of DPSCs and hence can be used as a potential restorative material in clinical dentistry.
Author Contributions
Conceptualization, K.T.P. and N.L.; methodology, K.T.P., I.A.R. and N.R.N.A.G.; visualization, H.S.C. and K.T.P.; data curation, H.S.C. and K.T.P.; validation, K.T.P.; writing—original draft preparation, H.S.C.; writing—review and editing, K.T.P., N.L., I.A.R., N.R.N.A.G. and H.S.C.; supervision, K.T.P., N.L., I.A.R. and N.R.N.A.G.; project administration, N.L. and K.T.P.; funding acquisition, N.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Fundamental Research Grant Scheme, Ministry of Higher Education Malaysia (203/PPSG/6171173).
Acknowledgments
The authors acknowledge the staff of Craniofacial Science Laboratory, School of Dental Sciences, Universiti Sains Malaysia, Malaysia, for their technical support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gronthos, S.; Mankani, M.; Brahim, J.; Robey, P.G.; Shi, S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc. Natl. Acad. Sci. USA 2000, 97, 13625–13630. [Google Scholar] [CrossRef] [PubMed]
- Mauth, C.; Huwig, A.; Graf-Hausner, U.; Roulet, J.F. Restorative applications for dental pulp therapy. Topics Tissue Eng. 2007, 3, 1–32. [Google Scholar]
- Wilson, A. Alumino-silicate polyacrylic acid and related cements. Polym. Int. 1974, 6, 165–179. [Google Scholar] [CrossRef]
- Kent, B.E.; Lewis, B.G.; Wilson, A.D. Glass ionomer cement formulations: I. The preparation of novel fluoroaluminosilicate glasses high in fluorine. J. Dent. Res. 1979, 58, 1607–1619. [Google Scholar] [CrossRef]
- Smith, D. Polyacrylic acid-based cements: Adhesion to enamel and dentin. Oper. Dent. 1992, 5, 177–183. [Google Scholar]
- Pelka, M.; Ebert, J.; Schneider, H.; Kramer, N.; Petschelt, A. Comparison of two-and three-body wear of glass-ionomers and composites. Eur. J. Oral. Sci. 1996, 104, 132–137. [Google Scholar] [CrossRef]
- Gu, Y.; Yap, A.; Cheang, P.; Khor, K. Effects of incorporation of HA/ZrO2 into glass ionomer cement (GIC). Biomaterials 2005, 26, 713–720. [Google Scholar] [CrossRef]
- Lohbauer, U.; Walker, J.; Nikolaenko, S.; Werner, J.; Clare, A.; Petschelt, A.; Greil, P. Reactive fibre reinforced glass ionomer cements. Biomaterials 2003, 24, 2901–2907. [Google Scholar] [CrossRef]
- Nicholson, J.W. Chemistry of glass-ionomer cements: A review. Biomaterials 1998, 19, 485–494. [Google Scholar] [CrossRef]
- Culbertson, B.M. Glass-ionomer dental restoratives. Prog. Polym. Sci. 2001, 26, 577–604. [Google Scholar] [CrossRef]
- Lohbauer, U. Dental glass ionomer cements as permanent filling materials?—Properties, limitations and future trends. Materials 2010, 3, 76. [Google Scholar] [CrossRef]
- Moshaverinia, A.; Roohpour, N.; Chee, W.W.; Schricker, S.R. A review of powder modifications in conventional glass-ionomer dental cements. J. Mater. Chem. 2011, 21, 1319–1328. [Google Scholar] [CrossRef]
- Wasson, E.; Nicholson, J. New aspects of the setting of glass-ionomer cements. J. Dent. Res. 1993, 72, 481–483. [Google Scholar] [CrossRef] [PubMed]
- Barry, T.I.; Clinton, D.J.; Wilson, A.D. Structure of a glass-ionomer cement and its relationship to the setting process. J. Dent. Res. 1979, 58, 1072–1079. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.W. Glass ionomer dental cements: Update. Mater. Technol. 2010, 25, 8–13. [Google Scholar] [CrossRef]
- Mendelson, B.C.; Jacobson, S.R.; Lavoipierre, A.M.; Huggins, R.J. The fate of porous hydroxyapatite granules used in facial skeletal augmentation. Aesthetic. Plast. Surg. 2010, 34, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Sakkers, R.J.; Dalmeyer, R.A.; Brand, R.; Rozing, P.M.; Van Blitterswijk, C.A. Assessment of bioactivity for orthopedic coatings in a gap-healing model. J. Biomed. Mater. Res. 1997, 36, 265–273. [Google Scholar] [CrossRef]
- Moshaverinia, A.; Ansari, S.; Moshaverinia, M.; Roohpour, N.; Darr, J.A.; Rehman, I. Effects of incorporation of hydroxyapatite and fluoroapatite nanobioceramics into conventional glass ionomer cements (GIC). Acta Biomater. 2008, 4, 432–440. [Google Scholar] [CrossRef]
- Pepla, E.; Besharat, L.K.; Palaia, G.; Tenore, G.; Migliau, G. Nano-hydroxyapatite and its applications in preventive, restorative and regenerative dentistry: A review of literature. Ann. Stomatol. 2014, 5, 108–114. [Google Scholar] [CrossRef]
- Ab Rahman, I.; Sam’an, M.M.; Luddin, N.; Shiekh, R.A. One-pot synthesis of hydroxyapatite-silica nanopowder composite for hardness enhancement of glass ionomer cement (GIC). Bull. Mater. Sci. 2014, 37, 213–219. [Google Scholar] [CrossRef]
- Ahmad Shiekh, R.; Ab Rahman, I.; Malik Masudi, S.; Luddin, N. Modification of glass ionomer cement by incorporating hydroxyapatite-silica nano-powder composite: Sol-gel synthesis and characterization. Ceram. Int. 2014, 40, 3165–3170. [Google Scholar] [CrossRef]
- Musa, M.; Kannan, T.P.; Ab Rahman, I. Assessment of DNA damage caused by locally produced hydroxyapatite-silica nanocomposite using Comet assay on human lung fibroblast cell line. Mol. Cell. Toxicol. 2012, 8, 53–60. [Google Scholar] [CrossRef]
- Moheet, I.A.; Luddin, N.; Ab Rahman, I.; Kannan, T.P.; Ghani, N.R.N.A. Evaluation of mechanical properties and bond strength of nano-hydroxyapatite-silica added glass ionomer cement. Ceram. Int. 2018, 44, 9899–9906. [Google Scholar] [CrossRef]
- Fujita, M.; Mikuni-Takagaki, T.; Komori, R.; Okubo, K.; Yasuda, M.; Kimoto, S. Effects of pre-reacted glass-ionomer cement on the viability and odontogenic differentiation of human dental pulp cells derived from deciduous teeth. Pediatr. Dent. J. 2016, 26, 74–82. [Google Scholar] [CrossRef]
- Paduano, F.; Marrelli, M.; White, L.J.; Shakesheff, K.M.; Tatullo, M. Odontogenic differentiation of human dental pulp stem cells on hydrogel scaffolds derived from decellularized bone extracellular matrix and collagen type I. PLoS ONE. 2016, 11, e0148225. [Google Scholar] [CrossRef]
- Eslaminejad, M.B.; Bordbar, S.; Nazarian, H. Odontogenic differentiation of dental pulp-derived stem cells on tricalcium phosphate scaffolds. J. Dent. Sci. 2013, 8, 306–313. [Google Scholar] [CrossRef]
- Mohamed, D.A.; Fayyad, D.M. The effect of different bioactive materials on the odontogenic differentiation potential of dental pulp stem cells using two different culture mediums. Tanta Dent. J. 2017, 14, 120–128. [Google Scholar] [CrossRef]
- Kwon, J.H.; Park, H.C.; Zhu, T.; Yang, H.C. Inhibition of odontogenic differentiation of human dental pulp cells by dental resin monomers. Biomater. Res. 2015, 19, 8. [Google Scholar] [CrossRef]
- Bakopoulou, A.; Leyhausen, G.; Volk, J.; Tsiftsoglou, A.; Garefis, P.; Koidis, P.; Geurtsen, W. Effects of HEMA and TEDGMA on the in vitro odontogenic differentiation potential of human pulp stem/progenitor cells derived from deciduous teeth. Dent. Mater. 2011, 27, 608–617. [Google Scholar] [CrossRef]
- Ahmed, H.M.A.; Omar, N.S.; Luddin, N.; Saini, R.; Saini, D. Cytotoxicity evaluation of a new fast set highly viscous conventional glass ionomer cement with L929 fibroblast cell line. J. Conserv. Dent. 2011, 14, 406–408. [Google Scholar] [CrossRef]
- Subhi, H.; Reza, F.; Husein, A.; Nurul, A. Cytotoxicity of gypsum-based biomaterial for direct pulp capping using stem cells from human exfoliated deciduous teeth. J. Conserv. Dent. 2018, 21, 21–25. [Google Scholar] [PubMed]
- International Organization for Standardization. ISO 10993-12. Biological Evaluation of Medical Devices. Part 12: Sample Preparation and Reference Materials; International Organization for Standardization: Geneva, Switzerland, 2012; Volume 4, pp. 1–20. [Google Scholar]
- Hii, S.C.; Luddin, N.; Kannan, T.P.; Ab Rahman, I.; Nik Abdul Ghani, N.R. The biological evaluation of conventional and nano-hydroxyapatite-silica glass ionomer cement on dental pulp stem cells: A comparative study. Contemp. Clin. Dent. 2019, 10, 324–332. [Google Scholar] [PubMed]
- Drissi, H.; Luc, Q.; Shakoori, R.; Chuva De Sousa Lopes, S.; Choi, J.Y.; Terry, A.; Hu, M.; Jones, S.; Neil, J.C.; Lian, J.B.; et al. Transcriptional autoregulation of the bone related CBFA1/RUNX2 gene. J. Cell Physiol. 2000, 184, 341–350. [Google Scholar] [CrossRef]
- Khanna-Jain, R.; Vanhatupa, S.; Vuorinen, A.; Sandor, G.; Suuronen, R.; Mannerstrom, B.; Miettinen, S. Growth and differentiation of human dental pulp stem cells maintained in fetal bovine serum, human serum and serum-free/xeno-free culture media. J. Stem Cell Res. Ther. 2012, 2, 2. [Google Scholar] [CrossRef]
- Guo, L.; Li, J.; Qiao, X.; Yu, M.; Tang, W.; Wang, H.; Guo, W.; Tian, W. Comparison of odontogenic differentiation of human dental follicle cells and human dental papilla cells. PLoS ONE 2013, 8, e62332. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Sidhu, S.K.; Nicholson, J.W. A review of glass-ionomer cements for clinical dentistry. J. Funct. Biomater. 2016, 7, 16. [Google Scholar] [CrossRef]
- Lucas, M.E.; Arita, K.; Nishino, M. Toughness, bonding and fluoride-release properties of hydroxyapatite-added glass ionomer cement. Biomaterials 2003, 24, 3787–3794. [Google Scholar] [CrossRef]
- Suzuki, S.; Sreenath, T.; Haruyama, N.; Honeycutt, C.; Terse, A.; Cho, A.; Kohler, T.; Müller, R.; Goldberg, M.; Kulkarni, A.B. Dentin sialoprotein and dentin phosphoprotein have distinct roles in dentin mineralization. Matrix Biol. 2009, 28, 221–229. [Google Scholar] [CrossRef]
- Simon, S.; Smith, A.; Lumley, P.; Berdal, A.; Smith, G.; Finney, S.; Cooper, P.R. Molecular characterization of young and mature odontoblasts. Bone 2009, 45, 693–703. [Google Scholar] [CrossRef]
- Massa, L.F.; Ramachandran, A.; George, A.; Arana-Chavez, V.E. Developmental appearance of dentin matrix protein 1 during the early dentinogenesis in rat molars as identified by high-resolution immunocytochemistry. Histochem. Cell Biol. 2005, 124, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, K.; Gajjeraman, S.; Ramachandran, A.; Hao, J.; George, A. Dentin matrix protein 1 regulates dentin sialophosphoprotein gene transcription during early odontoblast differentiation. J. Biol. Chem. 2006, 281, 19064–19071. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Y.; Chen, L.; Liu, L.; Lin, Y.F.; Li, S.W.; Tian, W.D. Odontogenic potential of bone marrow mesenchymal stem cells. J. Oral Maxillofac. Surg. 2007, 65, 494–500. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Ye, L.; Yu, S.; Zhang, S.; Xie, Y.; McKee, M.D.; Li, Y.C.; Kong, J.; David Eick, J.; Dallas, S.L.; et al. Rescue of odontogenesis in Dmp1-deficient mice by targeted re-expression of DMP1 reveals roles for DMP1 in early odontogenesis and dentin apposition in vivo. Dev. Biol. 2007, 303, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Cutarelli, A.; Marini, M.; Tancredi, V.; D’arcangelo, G.; Murdocca, M.; Frank, C.; Tarantino, M. Adenosine Triphosphate stimulates differentiation and mineralization in human osteoblast-like Saos-2 cells. Dev. Growth Differ. 2016, 58, 400–408. [Google Scholar] [CrossRef]
- Park, B.W.; Hah, Y.S.; Choi, M.J.; Ryu, Y.M.; Lee, S.G.; Kim, D.R.; Kim, J.R.; Byun, J.H. In vitro osteogenic differentiation of cultured human dental papilla-derived cells. J. Oral Maxillofac. Surg. 2009, 67, 507–514. [Google Scholar] [CrossRef]
- Hong, D.; Chen, H.X.; Yu, H.Q.; Liang, Y.; Wang, C.; Lian, Q.Q.; Deng, H.T.; Ge, R.S. Morphological and proteomic analysis of early stage of osteoblast differentiation in osteoblastic progenitor cells. Exp. Cell Res. 2010, 316, 2291–2300. [Google Scholar] [CrossRef]
- Mori, G.; Brunetti, G.; Oranger, A.; Carbone, C.; Ballini, A.; Lo Muzio, L.; Colucci, S.; Mori, C.; Grassi, F.R.; Grano, M. Dental pulp stem cells: Osteogenic differentiation and gene expression. Ann. N. Y. Acad. Sci. 2011, 1237, 47–52. [Google Scholar] [CrossRef]
- Yang, X.; van den Dolder, J.; Walboomers, X.F.; Zhang, W.; Bian, Z.; Fan, M.; Jansen, J.A. The odontogenic potential of STRO-1 sorted rat dental pulp stem cells in vitro. J. Tissue Eng. Regen. Med. 2007, 1, 66–73. [Google Scholar] [CrossRef]
- Sandberg, M.; Autio-Harmainen, H.; Vuorio, E. Localization of the expression of types I, III, and IV collagen, TGF-β1 and c-fos genes in developing human calvarial bones. Dev. Biol. 1988, 130, 324–334. [Google Scholar] [CrossRef]
- Arana-Chavez, V.E.; Massa, L.F. Odontoblasts: The cells forming and maintaining dentine. Int. J. Biochem. Cell Biol. 2004, 36, 1367–1373. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Gluhak-Heinrich, J.; Wang, Y.; Wu, Y.; Chuang, H.; Chen, L.; Yuan, G.H.; Dong, J.; Gay, I.; MacDougall, M. Runx2, Osx, and Dspp in tooth development. J. Dent. Res. 2009, 88, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, T.; Kanatani, N.; Rokutanda, S.; Yoshida, C.; Toyosawa, S.; Nakamura, R.; Takada, S.; Komori, T. Inhibition of the terminal differentiation of odontoblasts and their transdifferentiation into osteoblasts in Runx2 transgenic mice. Arch. Histol. Cytol. 2008, 71, 131–146. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Haruyama, N.; Nishimura, F.; Kulkarni, A.B. Dentin sialophosphoprotein and dentin matrix protein-1: Two highly phosphorylated proteins in mineralized tissues. Arch. Oral Biol. 2012, 57, 1165–1175. [Google Scholar] [CrossRef]
- Shie, M.Y.H.C.C.; Ding, S.J. Effects of altering the Si/Ca molar ratio of a calcium silicate cement on in vitro cell attachment. Int. Endod. J. 2012, 45, 337–345. [Google Scholar] [CrossRef]
- Ahmed, H.M.; Luddin, N.; Kannan, T.P.; Mokhtar, K.I.; Ahmad, A. Cell attachment properties of portland cement-based endodontic materials: Biological and methodological considerations. J. Endod. 2014, 40, 1517–1523. [Google Scholar] [CrossRef]
- Nicholson, J.; Czarnecka, B.; Limanowska-Shaw, H. The long-term interaction of dental cements with lactic acid solutions. J. Mater. Sci. Mater. Med. 1999, 10, 449–452. [Google Scholar] [CrossRef]
- Huang, Y.; Jin, X.; Zhang, X.; Sun, H.; Tu, J.; Tang, T.; Chang, J.; Dai, K. In vitro and in vivo evaluation of akermanite bioceramics for bone regeneration. Biomaterials 2009, 30, 5041–5048. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).