Gene Cloning, Characterization, and Molecular Simulations of a Novel Recombinant Chitinase from Chitinibacter Tainanensis CT01 Appropriate for Chitin Enzymatic Hydrolysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmid Construction
2.2. Preparation of Colloidal Chitin
2.3. Culture Conditions and Protein Purification
2.4. Effects of pH and Temperature on Chitinase Activity
2.5. Chitinase Assay Using Thin-Layer Chromatography
2.6. Effects of Metal Ions and Coumpound on Enzyme Activity
2.7. Molecular Modeling of Chitinase 1198
2.8. SIE Free Energy Calculations
3. Results
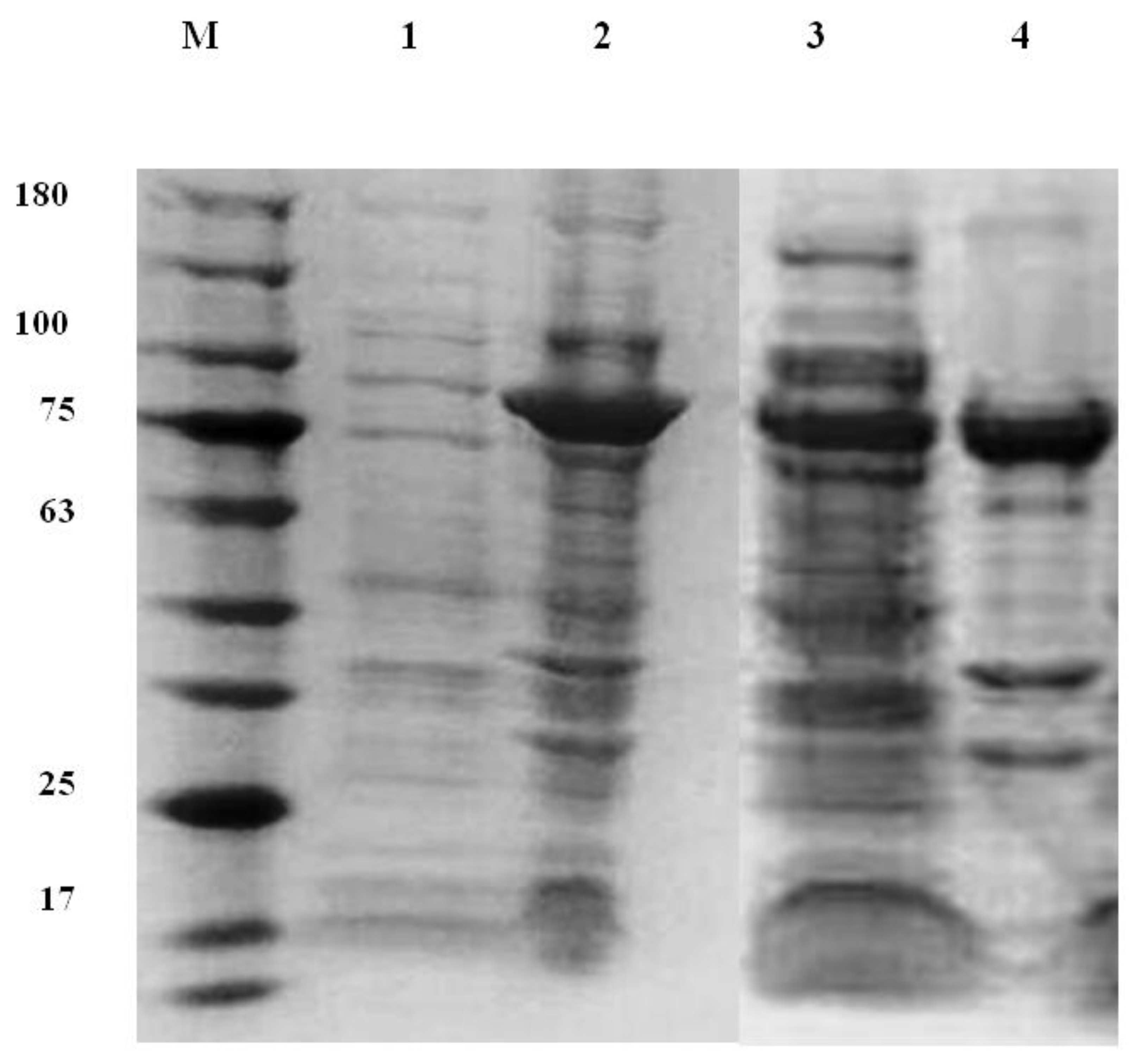
3.1. Overexpression and Purification of Chitinase 1198
3.2. Effects of pH and Temperature on Chitinase 1198 Activity
3.3. Effect of Metal Ions on Chitinase 1198 Activity
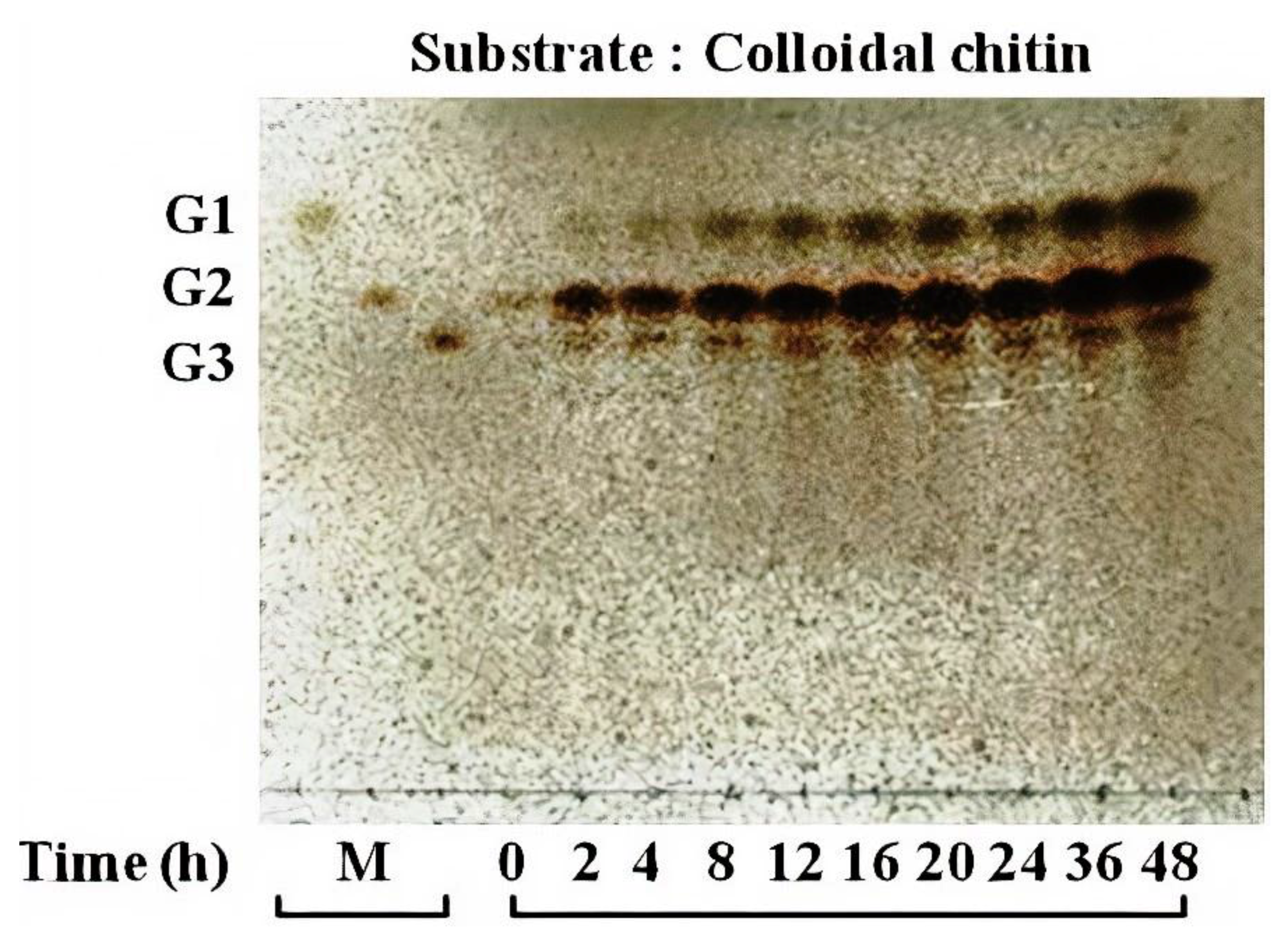
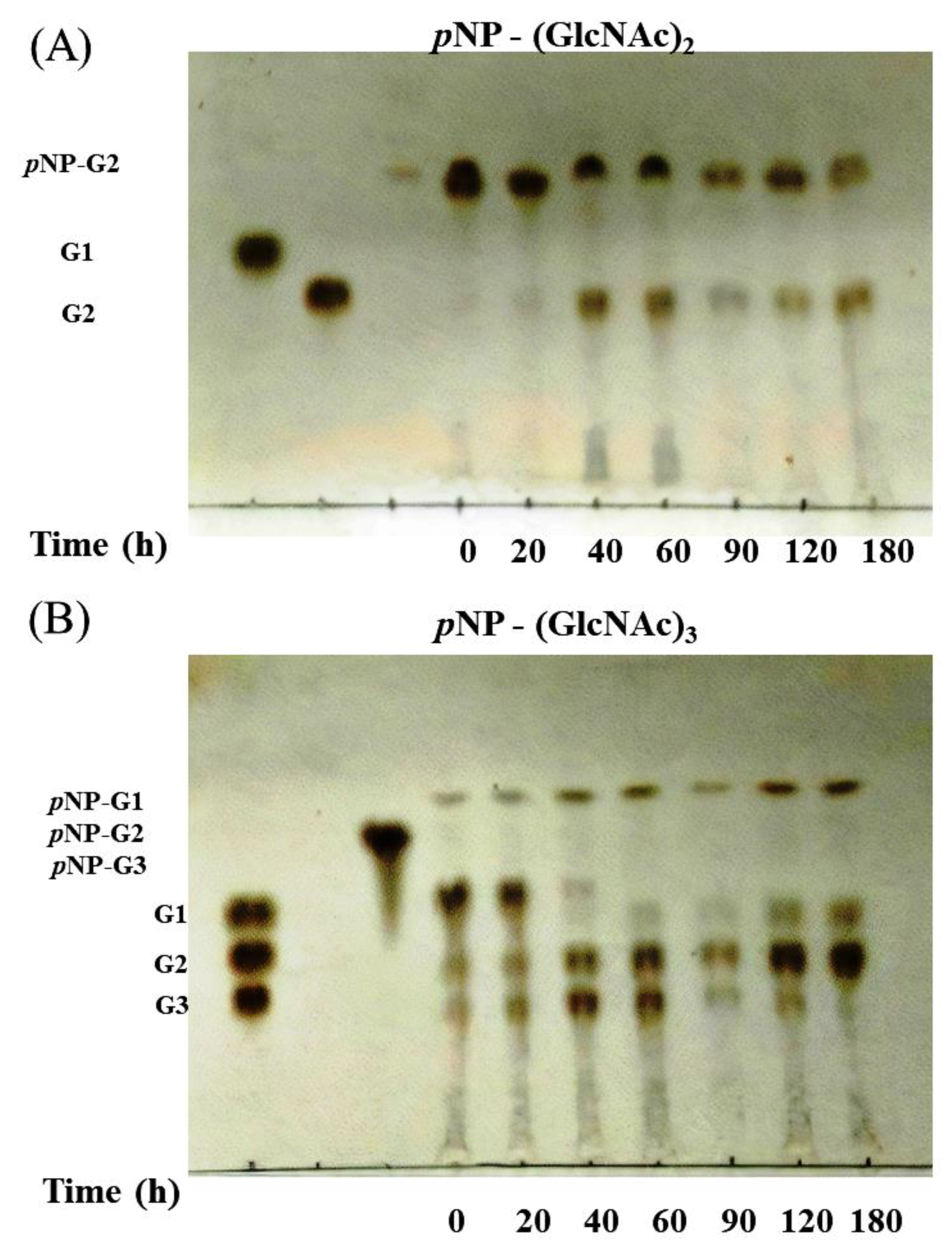
3.4. Activity of Chitinase 1198 on Colloidal Chitin and pNP-(GlcNAc)1–3 Substrates
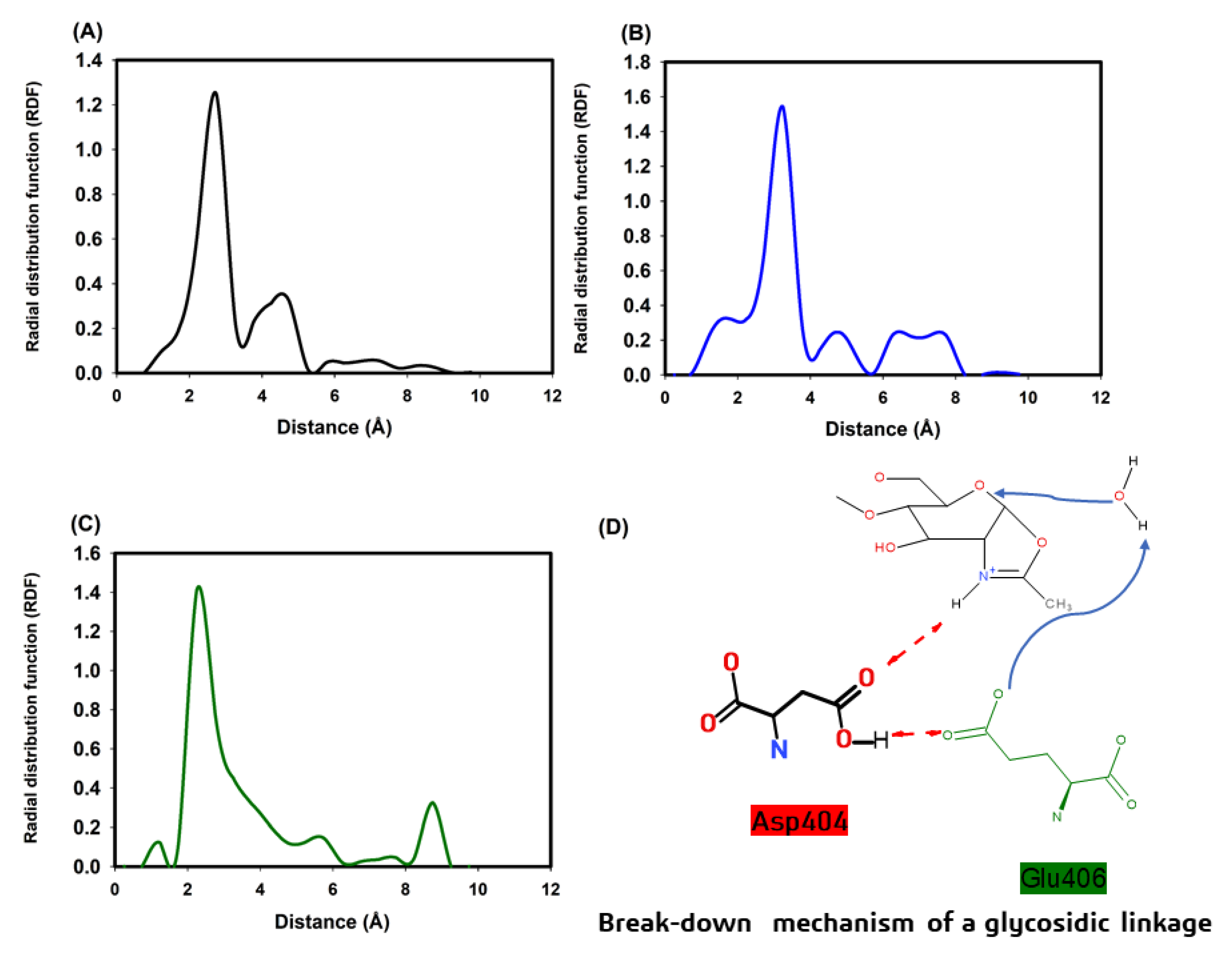
3.5. Molecular Modeling of Chitinase 1198 and These Enzymes with Chitin Oligomers (GlcNAc Repeat Unit: 12 and 3)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kidibule, P.E.; Santos-Moriano, P.; Jiménez-Ortega, E.; Ramírez-Escudero, M.; Limón, M.C.; Remacha, M.; Plou, F.J.; Sanz-Aparicio, J.; Fernández-Lobato, M. Use of chitin and chitosan to produce new chitooligosaccharides by chitinase Chit42: Enzymatic activity and structural basis of protein specificity. Microb. Cell Factories 2018, 17, 47. [Google Scholar] [CrossRef] [PubMed]
- Kaczmarek, M.B.; Struszczyk-Swita, K.; Li, X.; Szczęsna-Antczak, M.; Daroch, M. Enzymatic Modifications of Chitin, Chitosan, and Chitooligosaccharides. Front. Bioeng. Biotechnol. 2019, 7, 243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawase, T.; Saito, A.; Sato, T.; Kanai, R.; Fujii, T.; Nikaidou, N.; Miyashita, K.; Watanabe, T. Distribution and Phylogenetic Analysis of Family 19 Chitinases in Actinobacteria. Appl. Environ. Microbiol. 2004, 70, 1135–1144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamid, R.; Khan, M.A.; Ahmad, M.; Ahmad, M.M.; Abdin, M.Z.; Musarrat, J.; Javed, S. Chitinases: An update. J. Pharm. Bioallied Sci. 2013, 5, 21–29. [Google Scholar] [CrossRef]
- Rottloff, S.; Stieber, R.; Maischak, H.; Turini, F.G.; Heubl, G.; Mithöfer, A. Functional characterization of a class III acid endochitinase from the traps of the carnivorous pitcher plant genus, Nepenthes. J. Exp. Bot. 2011, 62, 4639–4647. [Google Scholar] [CrossRef]
- Li, S.-C.; Li, Y.-T. Studies on the Glycosidases of Jack Bean Meal: III. CRYSTALLIZATION AND PROPERTIES OF β-N-ACETYLHEXOSAMINIDASE. J. Biol. Chem. 1970, 245, 5153–5160. [Google Scholar]
- Drexler, H.G.; Gaedicke, G.; Minowada, J. Isoenzyme studies in human leukemia-lymphoma cell lines — III. β-hexosaminidase (E.C. 3.2.1.30). Leuk. Res. 1985, 9, 549–559. [Google Scholar] [CrossRef]
- Bolar, J.P.; Norelli, J.L.; Harman, G.E.; Brown, S.K.; Aldwinckle, H.S. Synergistic activity of endochitinase and exochitinase from Trichoderma atroviride (T. harzianum) against the pathogenic fungus (Venturia inaequalis) in transgenic apple plants. Transgenic Res. 2001, 10, 533–543. [Google Scholar] [CrossRef]
- Sahai, A.S.; Manocha, M.S. Chitinases of fungi and plants: Their involvement in morphogenesis and host-parasite interaction. Fems Microbiol. Rev. 1993, 11, 317–338. [Google Scholar] [CrossRef]
- Khan, F.I.; Bisetty, K.; Gu, K.-R.; Singh, S.; Permaul, K.; Hassan, M.I.; Wei, D.-Q. Molecular dynamics simulation of chitinase I from Thermomyces lanuginosus SSBP to ensure optimal activity. Mol. Simul. 2017, 43, 480–490. [Google Scholar] [CrossRef]
- Bhattacharjee, B.; Pathaw, N.; Chrungoo, N.K.; Bhattacharjee, A. Molecular modelling, dynamics simulation and characterization of antifungal chitinase from Sechium edule. Gene 2017, 606, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Madhuprakash, J.; Tanneeru, K.; Karlapudi, B.; Guruprasad, L.; Podile, A.R. Mutagenesis and molecular dynamics simulations revealed the chitooligosaccharide entry and exit points for chitinase D from Serratia proteamaculans. Biochim. Biophys. Acta (Bba)—Gen. Subj. 2014, 1840, 2685–2694. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Okazaki, K.-I.; Furuta, T.; Sakurai, M.; Iino, R. Processive chitinase is Brownian monorail operated by fast catalysis after peeling rail from crystalline chitin. Nat. Commun. 2018, 9, 3814. [Google Scholar] [CrossRef] [Green Version]
- Arora, M.; Yennamalli, R.M.; Sen, T.Z. Application of Molecular Simulations toward Understanding Cellulase Mechanisms. Bioenergy Res. 2018, 11, 850–867. [Google Scholar] [CrossRef] [Green Version]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asif, T.; Javed, U.; Zafar, S.B.; Ansari, A.; Ul Qader, S.A.; Aman, A. Bioconversion of Colloidal Chitin Using Novel Chitinase from Glutamicibacter uratoxydans Exhibiting Anti-fungal Potential by Hydrolyzing Chitin Within Fungal Cell Wall. Waste Biomass Valoriz. 2019. [Google Scholar] [CrossRef]
- Murthy, B.N.; Bleakley, B. Simplified Method of Preparing Colloidal Chitin Used For Screening of Chitinase- Producing Microorganisms. Internet J. Microbiol. 2012, 10, e2bc3. [Google Scholar]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Cheba, B.A.; Zaghloul, T.I.; El-Mahdy, A.R.; El-Massry, M.H. Effect of pH and Temperature on Bacillus sp. R2 Chitinase Activity and Stability. Procedia Technol. 2016, 22, 471–477. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Shen, N.; Wu, J.; Jiang, M.; Shi, S.; Wang, J.; Wei, Y.; Yang, L. Cloning, expression and characterization of a chitinase from Paenibacillus chitinolyticus strain UMBR 0002. PeerJ 2020, 8, e8964. [Google Scholar] [CrossRef]
- Haldane, J.B.S. Graphical Methods in Enzyme Chemistry. Nature 1957, 179, 832. [Google Scholar] [CrossRef]
- Tanaka, K.; Nishide, J.; Okazaki, K.; Kato, H.; Niwa, O.; Nakagawa, T.; Matsuda, H.; Kawamukai, M.; Murakami, Y. Characterization of a Fission Yeast SUMO-1 Homologue, Pmt3p, Required for Multiple Nuclear Events, Including the Control of Telomere Length and Chromosome Segregation. Mol. Cell. Biol. 1999, 19, 8660–8672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Case, D.A.; Cheatham, T.E.; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef] [Green Version]
- Pontius, J.; Richelle, J.; Wodak, S.J. Deviations from Standard Atomic Volumes as a Quality Measure for Protein Crystal Structures. J. Mol. Biol. 1996, 264, 121–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Hoell, I.A.; Vaaje-Kolstad, G.; Eijsink, V.G.H. Structure and function of enzymes acting on chitin and chitosan. Biotechnol. Genet. Eng. Rev. 2010, 27, 331–366. [Google Scholar] [CrossRef] [Green Version]
- Salomon-Ferrer, R.; Götz, A.W.; Poole, D.; Le Grand, S.; Walker, R.C. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comput. 2013, 9, 3878–3888. [Google Scholar] [CrossRef]
- Le Grand, S.; Götz, A.W.; Walker, R.C. SPFP: Speed without compromise—A mixed precision model for GPU accelerated molecular dynamics simulations. Comput. Phys. Commun. 2013, 184, 374–380. [Google Scholar] [CrossRef]
- Götz, A.W.; Williamson, M.J.; Xu, D.; Poole, D.; Le Grand, S.; Walker, R.C. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 1. Generalized Born. J. Chem. Theory Comput. 2012, 8, 1542–1555. [Google Scholar] [CrossRef]
- Ryckaert, J.-P.; Ciccotti, G.; Berendsen, H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef] [Green Version]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N [center-dot] log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef] [Green Version]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef] [PubMed]
- Naïm, M.; Bhat, S.; Rankin, K.N.; Dennis, S.; Chowdhury, S.F.; Siddiqi, I.; Drabik, P.; Sulea, T.; Bayly, C.I.; Jakalian, A.; et al. Solvated Interaction Energy (SIE) for Scoring Protein−Ligand Binding Affinities. 1. Exploring the Parameter Space. J. Chem. Inf. Model. 2007, 47, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Purisima, E.O. Fast summation boundary element method for calculating solvation free energies of macromolecules. J. Comput. Chem. 1998, 19, 1494–1504. [Google Scholar] [CrossRef]
- Purisima, E.O.; Nilar, S.H. A simple yet accurate boundary element method for continuum dielectric calculations. J. Comput. Chem. 1995, 16, 681–689. [Google Scholar] [CrossRef]
- Bhat, S.; Purisima, E.O. Molecular surface generation using a variable-radius solvent probe. Proteins Struct. Funct. Bioinform. 2006, 62, 244–261. [Google Scholar] [CrossRef]
- Perdih, A.; Bren, U.; Solmajer, T. Binding free energy calculations of N-sulphonyl-glutamic acid inhibitors of MurD ligase. J. Mol. Model. 2009, 15, 983–996. [Google Scholar] [CrossRef]
- Farag, A.M.; Abd-Elnabey, H.M.; Ibrahim, H.A.H.; El-Shenawy, M. Purification, characterization and antimicrobial activity of chitinase from marine-derived Aspergillus terreus. Egypt. J. Aquat. Res. 2016, 42, 185–192. [Google Scholar] [CrossRef] [Green Version]
- Inglis, P.W.; Peberdy, J.F. Production and purification of a chitinase from Ewingella americana, a recently described pathogen of the mushroom, Agaricus bisporus. FEMS Microbiol. Lett. 1997, 157, 189–194. [Google Scholar] [CrossRef]
- Kim, Y.H.; Park, S.K.; Hur, J.Y.; Kim, Y.C. Purification and Characterization of a Major Extracellular Chitinase from a Biocontrol Bacterium, Paenibacillus elgii HOA73. Plant Pathol. J. 2017, 33, 318–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, T.; Uchida, M.; Kobori, K.; Tanaka, H. Site-directed Mutagenesis of the Asp-197 and Asp-202 Residues in Chitinase A1 of Bacillus circulans WL-12. Biosci. Biotechnol. Biochem. 1994, 58, 2283–2285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Q.; Fong, S.S. Cloning and characterization of a chitinase from Thermobifida fusca reveals Tfu_0580 as a thermostable and acidic endochitinase. Biotechnol. Rep. 2018, 19, e00274. [Google Scholar] [CrossRef]
- Chen, W.; Qu, M.; Zhou, Y.; Yang, Q. Structural analysis of group II chitinase (ChtII) catalysis completes the puzzle of chitin hydrolysis in insects. J. Biol. Chem. 2018, 293, 2652–2660. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Metal Ions | Relative Activity (%) |
|---|---|
| K+ (KCl) | 106.4 ± 0.1 |
| Na+ (NaCl) | 101.6 ± 1.9 |
| Ba2+ (BaCl2) | 106.3 ± 0.2 |
| Ca2+ (CaCl2) | 101.6 ± 1.2 |
| Co2+ (CoCl2) | 81.9 ± 0.8 |
| EDTA | 106.3 ± 3.2 |
| Cu2+ (CuSO4) | 36.1 ± 1.6 |
| Mn2+ (MnSO4) | 106.4 ± 0.1 |
| Mg2+ (MgSO4) | 104.7 ± 0.1 |
| Zn2+ (ZnSO4) | 65.3 ± 1.1 |
| Substrate | Specific Activity * (Units/mg) | Relative Activity (%) |
|---|---|---|
| pNP-GlcNAc | 0 | 0 |
| pNP-(GlcNAc)2 | 3.09 × 105 | 91.4 |
| pNP-(GlcNAc)3 | 3.38 × 105 | 100 |
| Chitinase1198 | kcat (s−1) | KM (μM) | kcat/KM (s−1μM−1) |
|---|---|---|---|
| Wild type (WT) | 1.91 ± 0.33 | 170.40 ± 27.65 | 11.43 × 10−3 |
| Truncated | 0.36 ± 0.04 | 58.30 ± 11.01 | 6.38 × 10−3 |
| Number of Peptide Inhibitors | Chitinase 1198 with the Chitin Oligomers (GlcNAc Repeat Unit: 12) (the Asp440-2nd GlcNAc Repeat Unit) | Chitinase 1198 with the Chitin Oligomers (GlcNAc Repeat Unit: 12) (the Asp440-3rd GlcNAc Repeat Unit) | Chitinase 1198 with the Chitin Oligomers (GlcNAc Repeat Unit: 3) (the Asp440-2nd GlcNAc Repeat Unit) |
|---|---|---|---|
| Inter vdW | −92.36 ± 1.20 | −90.26 ± 0.67 | −66.44 ± 0.82 |
| Inter Coulomb | −23.33 ± 0.56 | −36.72 ± 0.87 | −23.91 ± 0.53 |
| Reaction Field | 39.14 ± 0.53 | 58.26 ± 0.69 | 33.64 ± 0.38 |
| Cavity | −14.81 ± 0.19 | −15.98 ± 0.73 | −11.09 ± 0.06 |
| Constant | −2.89 | −2.89 | −2.89 |
| △GBinding (kcal/mol) | −12.46 ± 0.14 | −11.76 ± 0.09 | −9.99 ± 0.11 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.-T.; Wu, P.-L. Gene Cloning, Characterization, and Molecular Simulations of a Novel Recombinant Chitinase from Chitinibacter Tainanensis CT01 Appropriate for Chitin Enzymatic Hydrolysis. Polymers 2020, 12, 1648. https://doi.org/10.3390/polym12081648
Wang Y-T, Wu P-L. Gene Cloning, Characterization, and Molecular Simulations of a Novel Recombinant Chitinase from Chitinibacter Tainanensis CT01 Appropriate for Chitin Enzymatic Hydrolysis. Polymers. 2020; 12(8):1648. https://doi.org/10.3390/polym12081648
Chicago/Turabian StyleWang, Yeng-Tseng, and Po-Long Wu. 2020. "Gene Cloning, Characterization, and Molecular Simulations of a Novel Recombinant Chitinase from Chitinibacter Tainanensis CT01 Appropriate for Chitin Enzymatic Hydrolysis" Polymers 12, no. 8: 1648. https://doi.org/10.3390/polym12081648
APA StyleWang, Y.-T., & Wu, P.-L. (2020). Gene Cloning, Characterization, and Molecular Simulations of a Novel Recombinant Chitinase from Chitinibacter Tainanensis CT01 Appropriate for Chitin Enzymatic Hydrolysis. Polymers, 12(8), 1648. https://doi.org/10.3390/polym12081648






