Enzymatic Protein Biopolymers as a Tool to Synthetize Eukaryotic Messenger Ribonucleic Acid (mRNA) with Uses in Vaccination, Immunotherapy and Nanotechnology
Abstract
1. Introduction
2. Proteins and Core DNA Promoter Elements Involved in the Transcription Process
3. mRNA as a Tool for RNA-Based Vaccines
4. Other Uses of mRNA as Nanomedicines
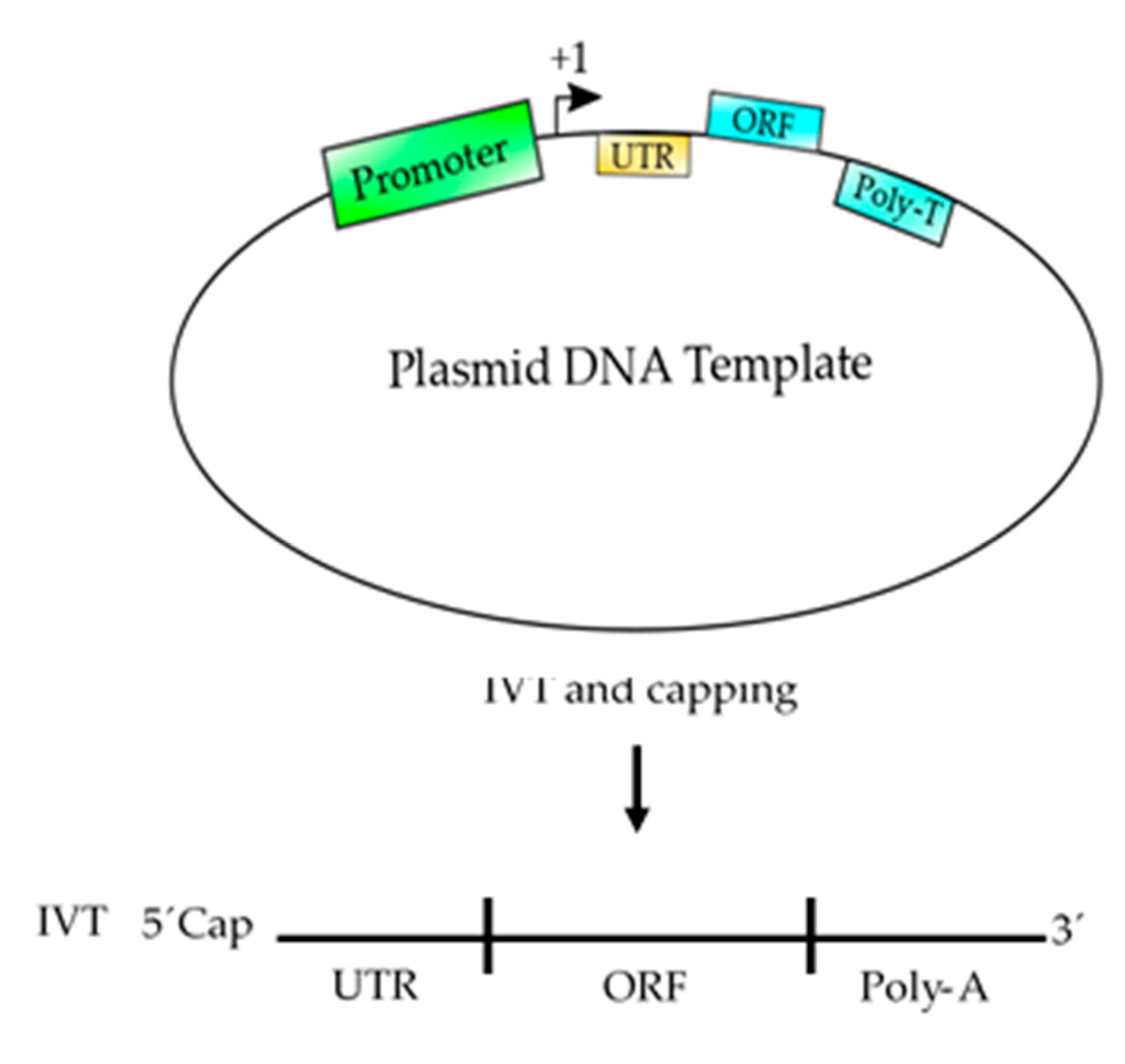
5. Basic Elements to Design an mRNA-Based Vaccine
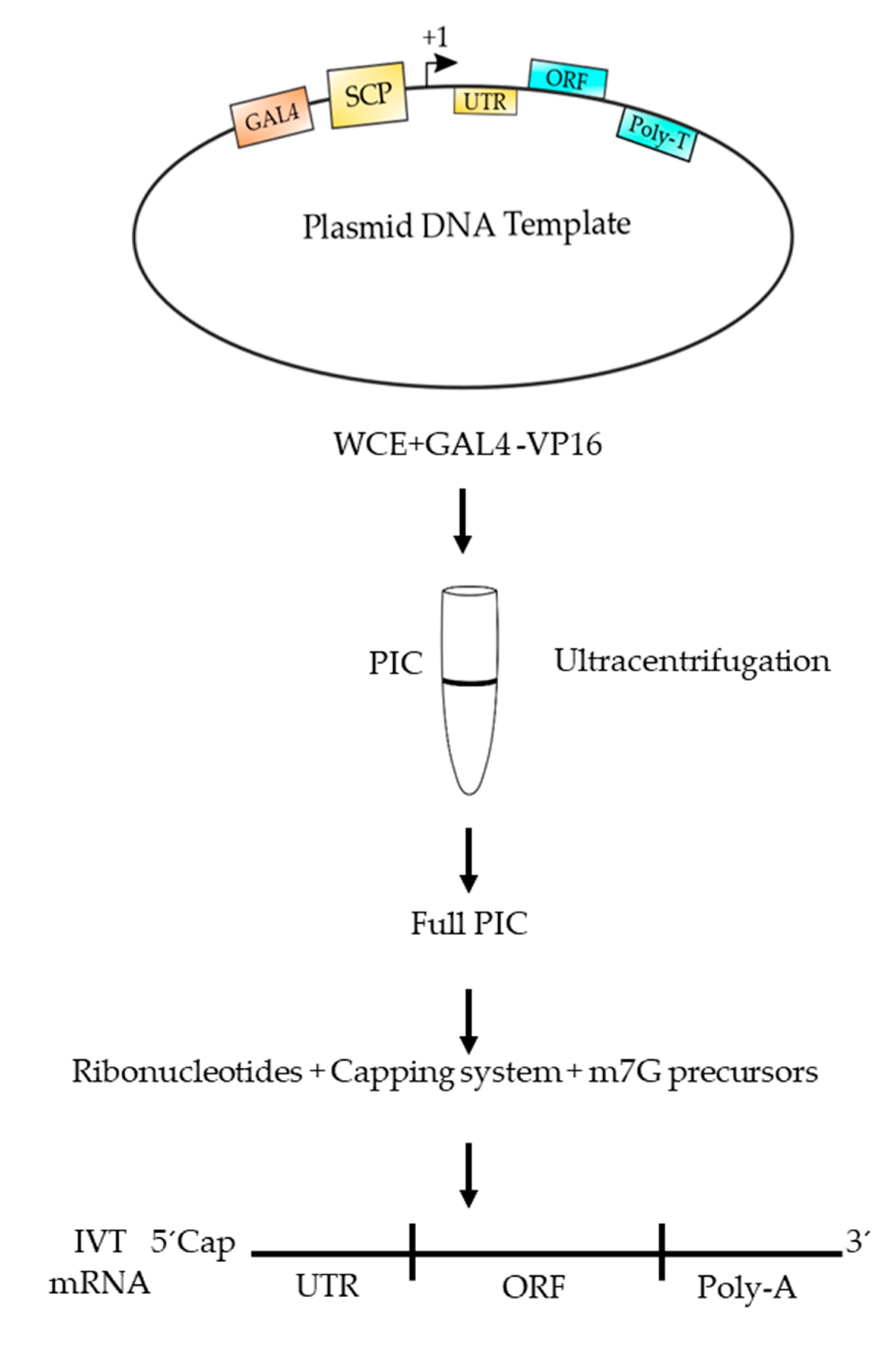
6. Advantages of Using Eukaryotic RNAPII to Synthetize mRNA
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hahn, S. Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Boil. 2004, 11, 394–403. [Google Scholar] [CrossRef]
- Cramer, P. Multisubunit RNA polymerases. Curr. Opin. Struct. Boil. 2002, 12, 89–97. [Google Scholar] [CrossRef]
- Bushnell, D.A.; Cramer, P.; Kornberg, R.D. Structural basis of transcription: Alpha-amanitin-RNA polymerase II cocrystal at 2.8 A resolution. Proc. Natl. Acad. Sci. USA 2002, 99, 1218–1222. [Google Scholar] [CrossRef] [PubMed]
- Phatnani, H.P.; Greenleaf, A.L. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006, 20, 2922–2936. [Google Scholar] [CrossRef]
- Meinhart, A.; Kamenski, T.; Hoeppner, S.; Baumli, S.; Cramer, P. A structural perspective of CTD function. Genes Dev. 2005, 19, 1401–1415. [Google Scholar] [CrossRef] [PubMed]
- Juven-Gershon, T.; Hsu, J.-Y.; Kadonaga, J.T. Perspectives on the RNA polymerase II core promoter. Biochem. Soc. Trans. 2006, 34, 1047–1050. [Google Scholar] [CrossRef]
- Rojas, D.A.; Moreira-Ramos, S.; Zock-Emmenthal, S.; Urbina, F.; Contreras-Levicoy, J.; Käufer, N.F.; Maldonado, E. Rrn7 Protein, an RNA Polymerase I Transcription Factor, Is Required for RNA Polymerase II-dependent Transcription Directed by Core Promoters with a HomolD Box Sequence*. J. Boil. Chem. 2011, 286, 26480–26486. [Google Scholar] [CrossRef]
- Witt, I.; Kivinen, K.; Käufer, N.F. Core Promoters in S. pombe: TATA and HomolD Boxes. In The Molecular Biology of Schizosaccharomyces Pombe; Springer: Berlin/Heidelberg, Germany, 2004; pp. 343–351. [Google Scholar]
- Greber, B.J.; Nogales, E. The Structures of Eukaryotic Transcription Pre-initiation Complexes and Their Functional Implications. Sub-Cell. Biochem. 2019, 93, 143–192. [Google Scholar] [CrossRef]
- Montes, M.; Moreira-Ramos, S.; Rojas, D.A.; Urbina, F.; Käufer, N.F.; Maldonado, E. RNA polymerase II components and Rrn7 form a preinitiation complex on the HomolD box to promote ribosomal protein gene expression inSchizosaccharomyces pombe. FEBS J. 2017, 284, 615–633. [Google Scholar] [CrossRef]
- Kim, Y.J.; Björklund, S.; Li, Y.; Sayre, M.H.; Kornberg, R.D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 1994, 77, 599–608. [Google Scholar] [CrossRef]
- Maldonado, E.; Shiekhattar, R.; Sheldon, M.; Cho, H.; Drapkin, R.; Rickert, P.; Lees, E.; Anderson, C.W.; Linn, S.; Reinberg, D. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature 1996, 381, 86–89. [Google Scholar] [CrossRef] [PubMed]
- Bringmann, A.; Held, S.A.E.; Heine, A.; Brossart, P. RNA Vaccines in Cancer Treatment. J. Biomed. Biotechnol. 2010, 2010, 1–12. [Google Scholar] [CrossRef] [PubMed]
- McNamara, M.A.; Nair, S.K.; Holl, E.K. RNA-Based Vaccines in Cancer Immunotherapy. J. Immunol. Res. 2015, 2015, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ross, J. Control of messenger RNA stability in higher eukaryotes. Trends Genet. 1996, 12, 171–175. [Google Scholar] [CrossRef]
- Kreiter, S.; Diken, M.; Selmi, A.; Diekmann, J.; Attig, S.; Hüsemann, Y.; Koslowski, M.; Huber, C.; Türeci, Ö.; Sahin, U. FLT3 Ligand Enhances the Cancer Therapeutic Potency of Naked RNA Vaccines. Cancer Res. 2011, 71, 6132–6142. [Google Scholar] [CrossRef]
- Qiu, P.; Ziegelhoffer, P.; Sun, J.; Yang, N.S. Gene gun delivery of mRNA in situ results in efficient transgene expression and genetic immunization. Gene Ther. 1996, 3, 262–268. [Google Scholar]
- Weide, B.; Pascolo, S.; Scheel, B.; Derhovanessian, E.; Pflugfelder, A.; Eigentler, T.; Pawelec, G.; Hoerr, I.; Rammensee, H.-G.; Garbe, C. Direct Injection of Protamine-protected mRNA: Results of a Phase 1/2 Vaccination Trial in Metastatic Melanoma Patients. J. Immunother. 2009, 32, 498–507. [Google Scholar] [CrossRef]
- Hafner, A.M.; Corthésy, B.; Merkle, H.P. Particulate formulations for the delivery of poly(I:C) as vaccine adjuvant. Adv. Drug Deliv. Rev. 2013, 65, 1386–1399. [Google Scholar] [CrossRef]
- Okumura, K.; Nakase, M.; Inui, M.; Nakamura, S.; Watanabe, Y.; Tagawa, T. Bax mRNA therapy using cationic liposomes for human malignant melanoma. J. Gene Med. 2008, 10, 910–917. [Google Scholar] [CrossRef]
- Grudzien, E.; Stepinski, J.; Jankowska-Anyszka, M.; Stolarski, R.; Darzynkiewicz, E.; Rhoads, R.E. Novel cap analogs for in vitro synthesis of mRNAs with high translational efficiency. RNA 2004, 10, 1479–1487. [Google Scholar] [CrossRef]
- Maruggi, G.; Zhang, C.; Li, J.; Ulmer, J.B.; Yu, D. mRNA as a Transformative Technology for Vaccine Development to Control Infectious Diseases. Mol. Ther. 2019, 27, 757–772. [Google Scholar] [CrossRef] [PubMed]
- Richner, J.M.; Himansu, S.; Dowd, K.A.; Butler, S.L.; Salazar, V.; Fox, J.M.; Julander, J.G.; Tang, W.W.; Shresta, S.; Pierson, T.C.; et al. Modified mRNA Vaccines Protect against Zika Virus Infection. Cell 2017, 168, 1114–1125.e10. [Google Scholar] [CrossRef] [PubMed]
- Schnee, M.; Vogel, A.B.; Voss, D.; Petsch, B.; Baumhof, P.; Kramps, T.; Stitz, L. An mRNA Vaccine Encoding Rabies Virus Glycoprotein Induces Protection against Lethal Infection in Mice and Correlates of Protection in Adult and Newborn Pigs. PLoS Negl. Trop. Dis. 2016, 10, e0004746. [Google Scholar] [CrossRef] [PubMed]
- Kyte, J.A.; Mu, L.; Aamdal, S.; Kvalheim, G.; Dueland, S.; Hauser, M.; Gullestad, H.P.; Ryder, T.; Lislerud, K.; Hammerstad, H.; et al. Phase I/II trial of melanoma therapy with dendritic cells transfected with autologous tumor-mRNA. Cancer Gene Ther. 2006, 13, 905–918. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, Y.; Miao, L.; Liu, Q.; Musetti, S.; Li, J.; Huang, L. Combination Immunotherapy of MUC1 mRNA Nano-vaccine and CTLA-4 Blockade Effectively Inhibits Growth of Triple Negative Breast Cancer. Mol. Ther. 2017, 26, 45–55. [Google Scholar] [CrossRef]
- DeRosa, F.; Guild, B.; Karve, S.; Smith, L.; Love, K.; Dorkin, J.R.; Kauffman, K.J.; Zhang, J.; Yahalom, B.; Anderson, D.G.; et al. Therapeutic efficacy in a hemophilia B model using a biosynthetic mRNA liver depot system. Gene Ther. 2016, 23, 699–707. [Google Scholar] [CrossRef]
- Bangel-Ruland, N.; Tomczak, K.; Fernández, E.F.; Leier, G.; Leciejewski, B.; Rudolph, C.; Rosenecker, J.; Weber, W.-M. Cystic fibrosis transmembrane conductance regulator-mRNA delivery: A novel alternative for cystic fibrosis gene therapy. J. Gene Med. 2013, 15, 414–426. [Google Scholar] [CrossRef]
- Zhu, X.; Yin, L.; Theisen, M.; Zhuo, J.; Siddiqui, S.; Levy, B.; Presnyak, V.; Frassetto, A.; Milton, J.; Salerno, T.; et al. Systemic mRNA Therapy for the Treatment of Fabry Disease: Preclinical Studies in Wild-Type Mice, Fabry Mouse Model, and Wild-Type Non-human Primates. Am. J. Hum. Genet. 2019, 104, 625–637. [Google Scholar] [CrossRef]
- Wang, H.; Yang, H.; Shivalila, C.S.; Dawlaty, M.M.; Cheng, A.W.; Zhang, F.; Jaenisch, R. One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering. Cell 2013, 153, 910–918. [Google Scholar] [CrossRef]
- Koblas, T.; Leontovyc, I.; Loukotova, S.; Kosinova, L.; Saudek, F. Reprogramming of Pancreatic Exocrine Cells AR42J Into Insulin-producing Cells Using mRNAs for Pdx1, Ngn3, and MafA Transcription Factors. Mol. Ther. Nucleic Acids 2016, 5, e320. [Google Scholar] [CrossRef]
- Van Hoecke, L.; Roose, K. How mRNA therapeutics are entering the monoclonal antibody field. J. Transl. Med. 2019, 17, 54. [Google Scholar] [CrossRef] [PubMed]
- Yue, Z.; Maldonado, E.; Pillutla, R.; Cho, H.; Reinberg, D.; Shatkin, A.J. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc. Natl. Acad. Sci. USA 1997, 94, 12898–12903. [Google Scholar] [CrossRef] [PubMed]
- Pillutla, R.C.; Yue, Z.; Maldonado, E.; Shatkin, A.J. Recombinant Human mRNA Cap Methyltransferase Binds Capping Enzyme/RNA Polymerase IIo Complexes. J. Boil. Chem. 1998, 273, 21443–21446. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, R.; Murakami, K. In vitro reconstitution of yeast RNA polymerase II transcription initiation with high efficiency. Methods 2019, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Even, D.Y.; Kedmi, A.; Basch-Barzilay, S.; Ideses, D.; Tikotzki, R.; Shir-Shapira, H.; Shefi, O.; Juven-Gershon, T. Engineered Promoters for Potent Transient Overexpression. PLoS ONE 2016, 11, e0148918. [Google Scholar] [CrossRef][Green Version]

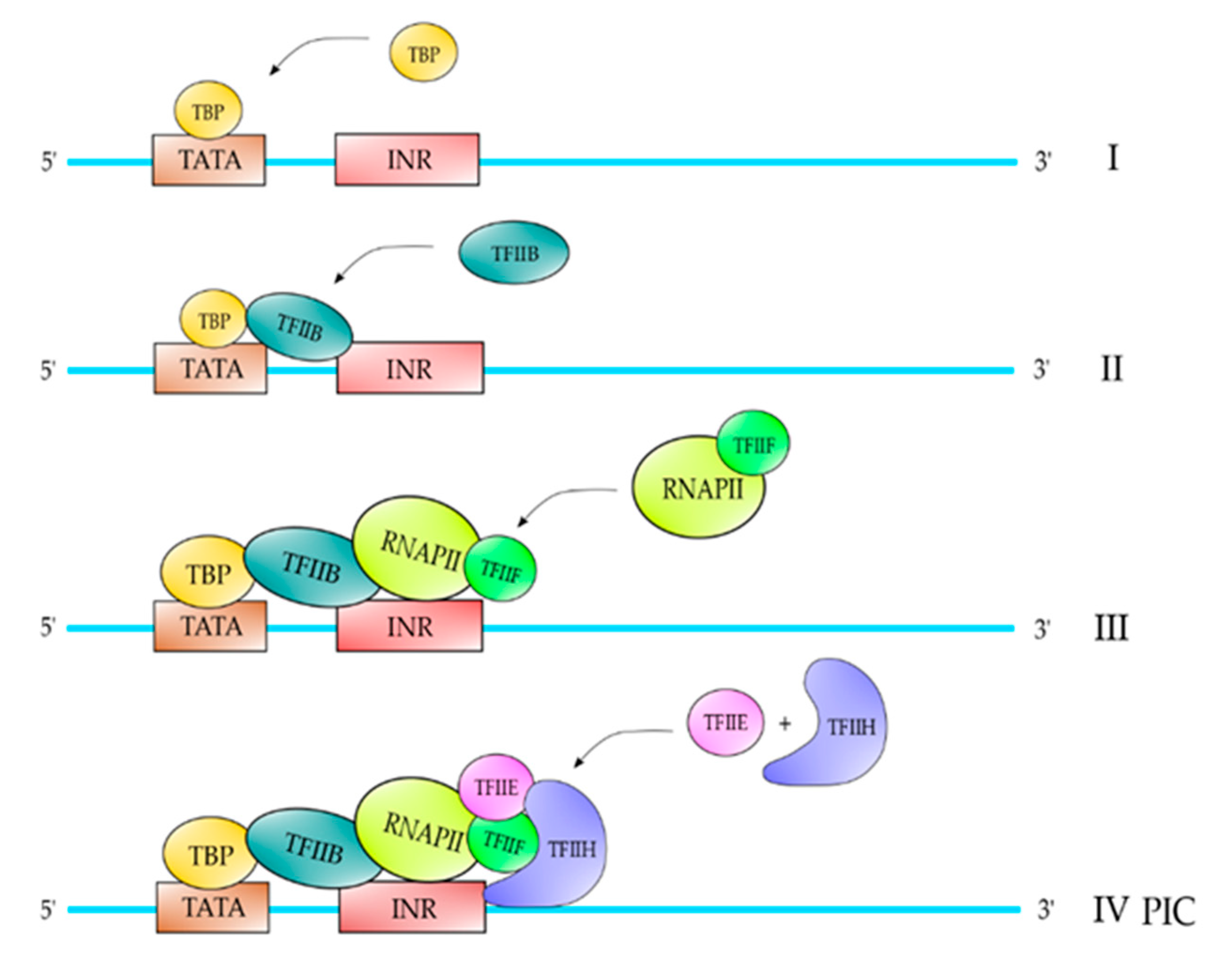
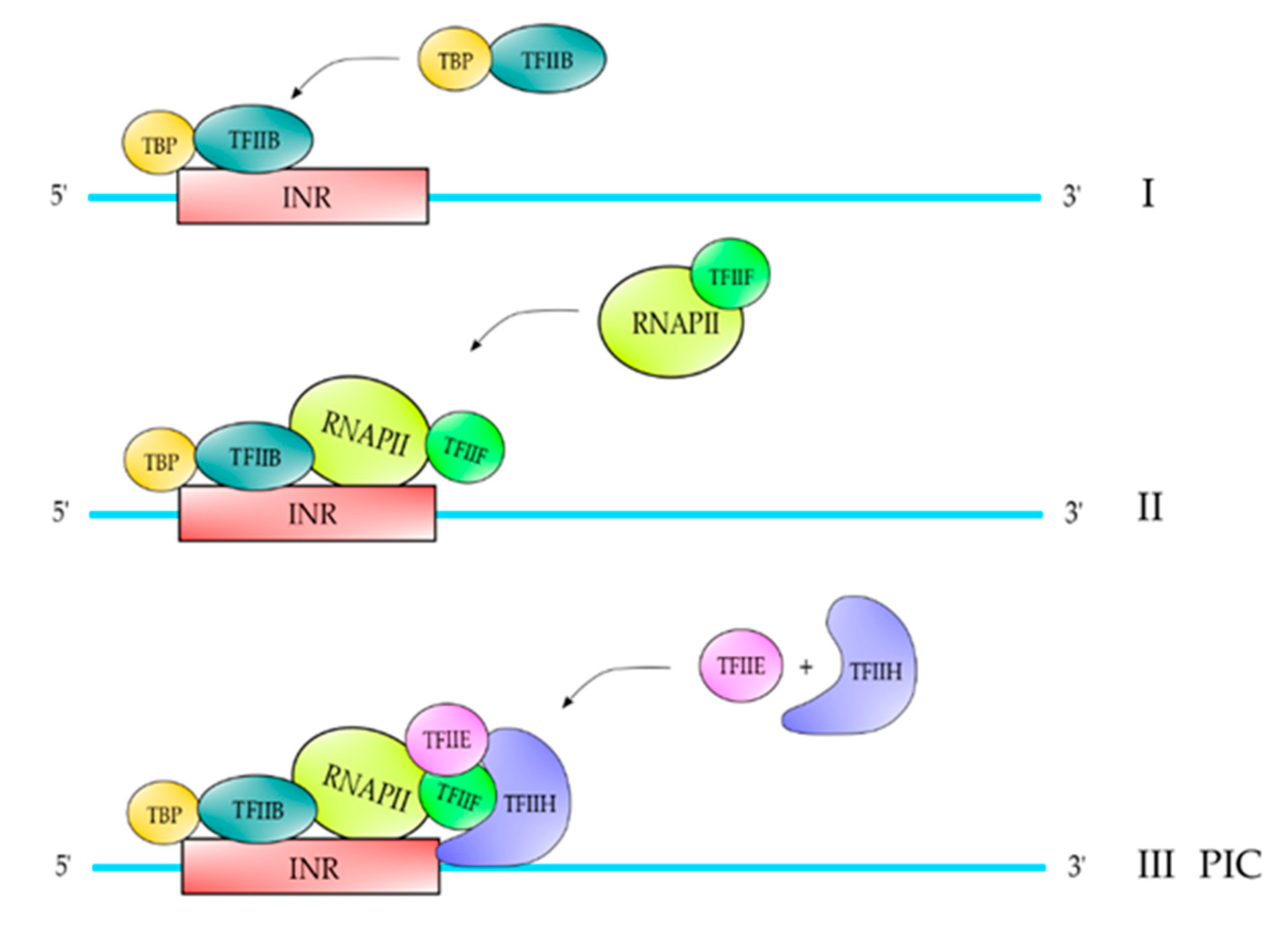


| TFII | Polypeptide Composition | Function |
|---|---|---|
| TBP | 1 | Recognize TATA elements and INR |
| TFIIB | 1 | Bridges TBP and RNAPII-TFIIF |
| TFIIF | 3 a | Helps to recruit RNAPII to the PIC |
| TFIIE | 2 | Stabilizes RNAPII-TFIIF to the PIC |
| TFIIH | 10 | Kinase and helicase activities |
| Core RNAPII | 12–14 | Synthesis of the mRNA |
| Mediator | 24 b | Target of regulatory factors Bridge the PIC with activator factors |
| Rrn7 | 1 | In S. pombe, this transcription factor recognizes the HomolD-box of RPG promoters |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urbina, F.; Morales-Pison, S.; Maldonado, E. Enzymatic Protein Biopolymers as a Tool to Synthetize Eukaryotic Messenger Ribonucleic Acid (mRNA) with Uses in Vaccination, Immunotherapy and Nanotechnology. Polymers 2020, 12, 1633. https://doi.org/10.3390/polym12081633
Urbina F, Morales-Pison S, Maldonado E. Enzymatic Protein Biopolymers as a Tool to Synthetize Eukaryotic Messenger Ribonucleic Acid (mRNA) with Uses in Vaccination, Immunotherapy and Nanotechnology. Polymers. 2020; 12(8):1633. https://doi.org/10.3390/polym12081633
Chicago/Turabian StyleUrbina, Fabiola, Sebastián Morales-Pison, and Edio Maldonado. 2020. "Enzymatic Protein Biopolymers as a Tool to Synthetize Eukaryotic Messenger Ribonucleic Acid (mRNA) with Uses in Vaccination, Immunotherapy and Nanotechnology" Polymers 12, no. 8: 1633. https://doi.org/10.3390/polym12081633
APA StyleUrbina, F., Morales-Pison, S., & Maldonado, E. (2020). Enzymatic Protein Biopolymers as a Tool to Synthetize Eukaryotic Messenger Ribonucleic Acid (mRNA) with Uses in Vaccination, Immunotherapy and Nanotechnology. Polymers, 12(8), 1633. https://doi.org/10.3390/polym12081633






