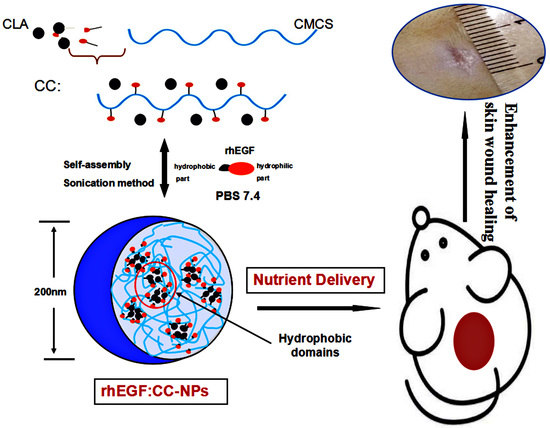
Enhancement of Skin Wound Healing by rhEGF-Loaded Carboxymethyl Chitosan Nanoparticles
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Cell Line
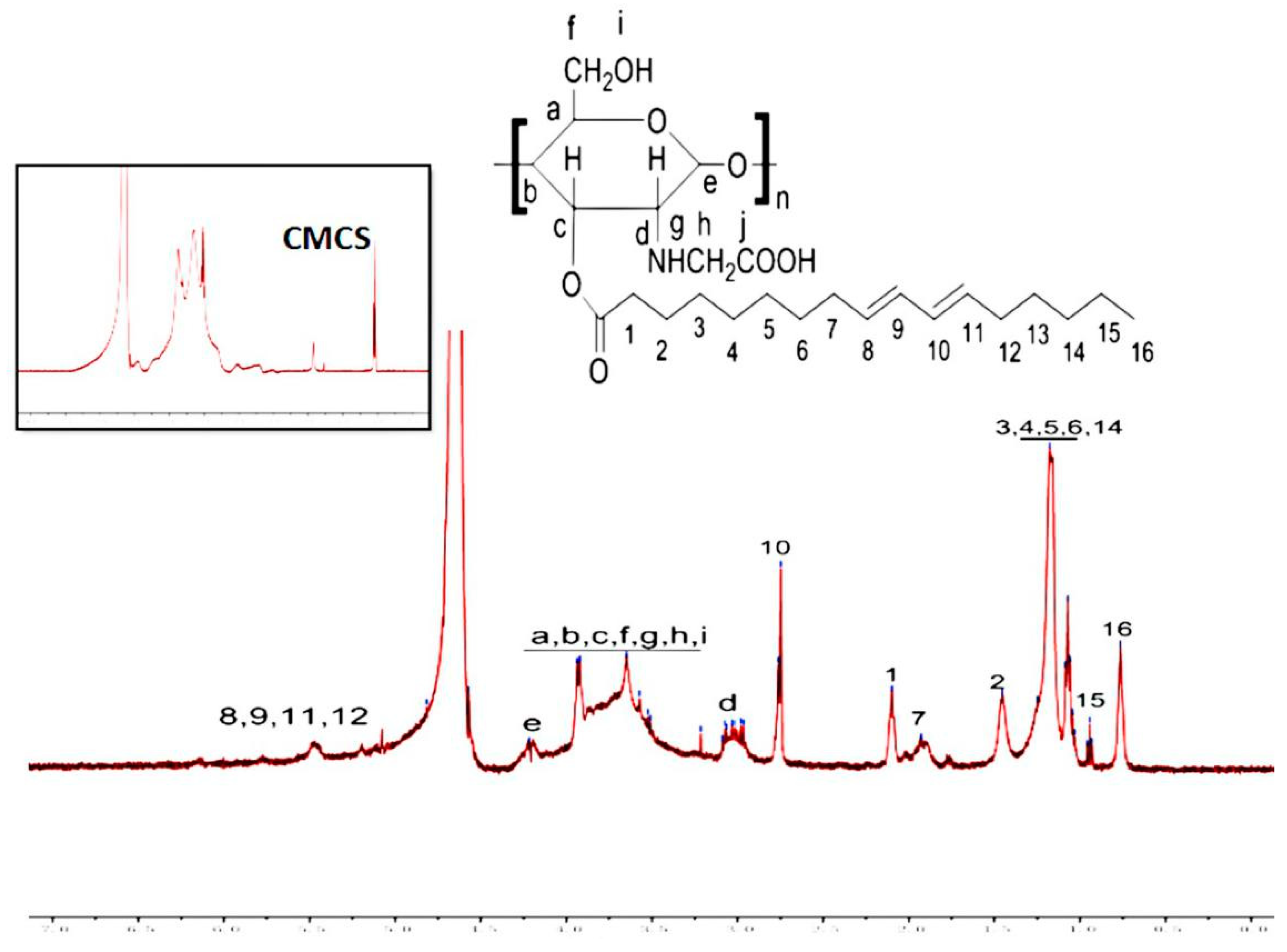
2.2. Synthesis of CLA–CMCS Conjugates
2.3. Preparation and Characterization of CC-NPs
2.4. In Vitro Studies
2.4.1. Drug Entrapment Efficiency and Release Studies
2.4.2. Cell Compatibility Assay
2.4.3. Migration Assay
2.5. In Vivo Studies
2.5.1. Animals Wound Induction
2.5.2. Serial Wound Analysis
2.5.3. Histological Analysis
2.6. Statistical Analysis
3. Results and Discussion
3.1. Characterization of CC Sample
3.2. Formation and Characteristics of CC-NPs
3.2.1. Characterization of CC-NPs
3.2.2. TEM, Stability, and Occlusion Test of CC-NPs
3.3. In Vitro Studies
3.3.1. rhEGF Loading Efficiency and Release Study
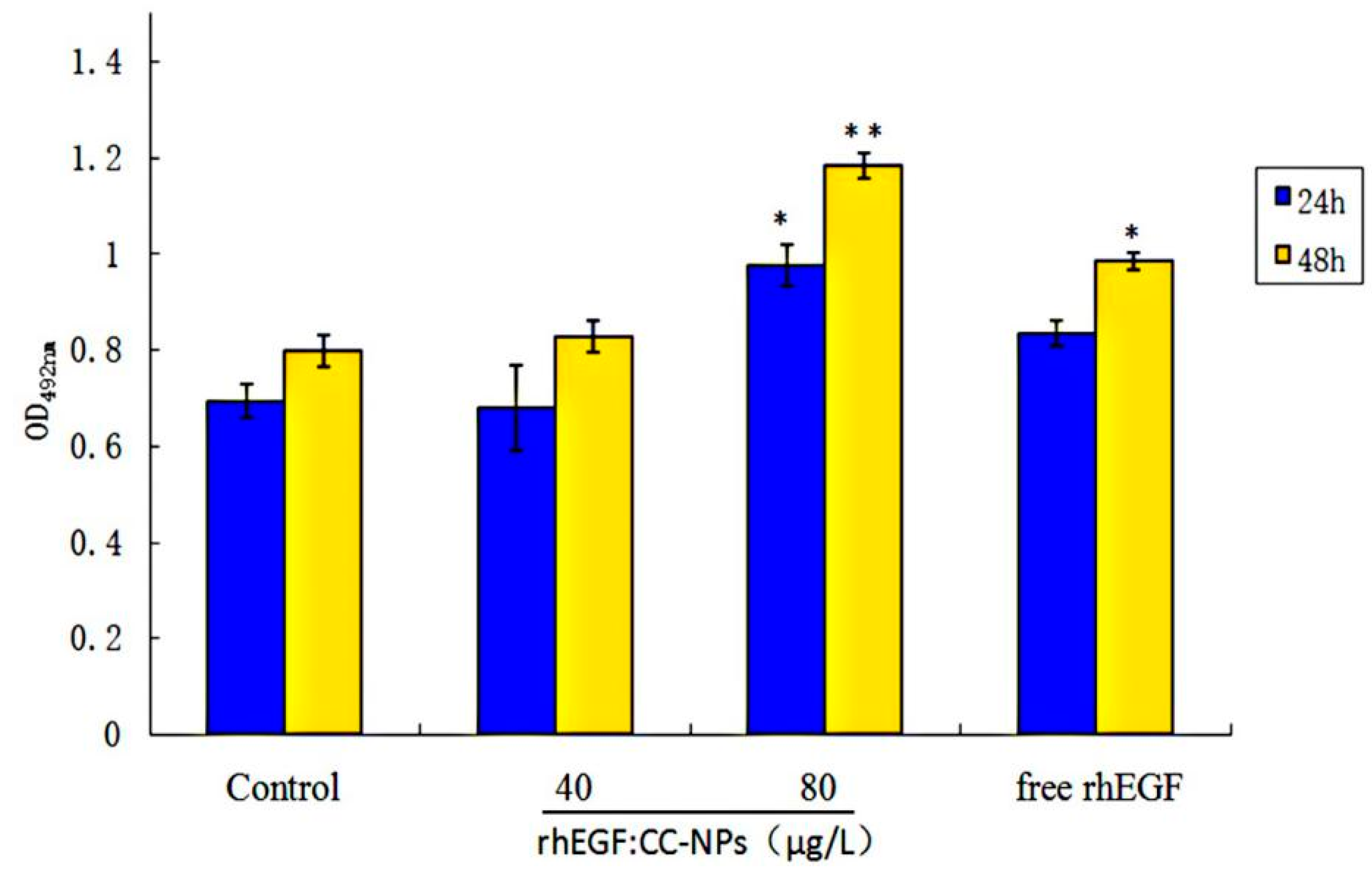
3.3.2. Cell Compatibility of CC-NPs
3.3.3. Enhancement of Cell Proliferation and Migration by rhEGF:CC-NPs Treatment
3.4. In Vivo Studies
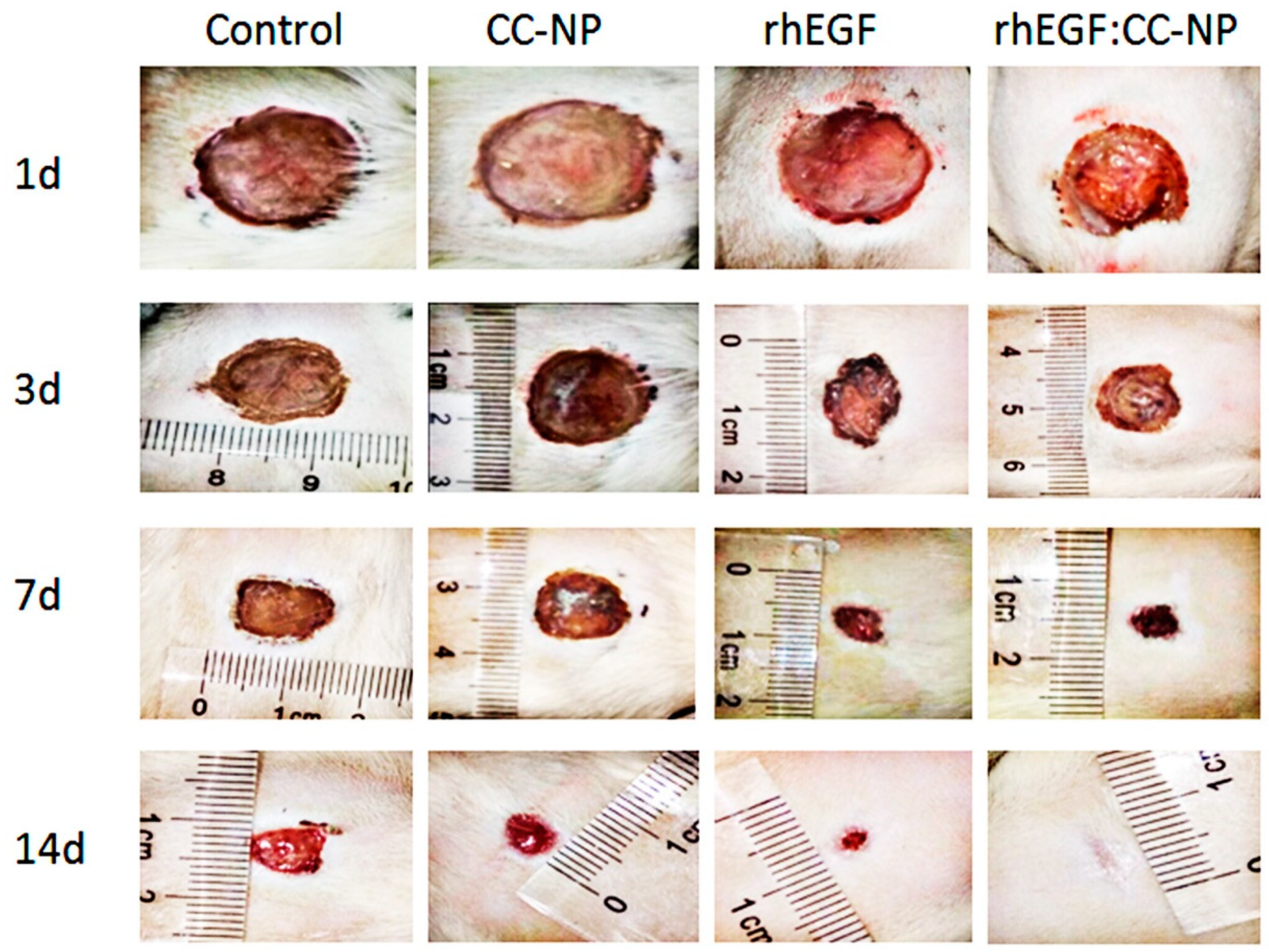
3.4.1. Change of Body Weight and Promotion of Wound Contraction
3.4.2. Results of H&E Staining
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sabine, W.; Richard, G. Regulation of Wound Healing by Growth Factors and Cytokines. Physiol. Rev. 2003, 83, 835–870. [Google Scholar]
- Horst, B.T.; Chouhan, G.; Moiemen, N.S.; Grover, L.M. Advances in keratinocyte delivery in burn wound care. Adv. Drug Deliv. Rev. 2017, 123, 18–32. [Google Scholar] [CrossRef] [PubMed]
- Morey, M.; Pandit, A. Responsive triggering systems for delivery in chronic wound healing. Adv. Drug Deliv. Rev. 2018, 129, 169–193. [Google Scholar] [CrossRef]
- Schultz, G.; Rotatori, D.S.; Clark, W. EGF and TGF-alpha in wound healing and repair. J. Cell. Biochem. 1991, 45, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Behm, B.; Babilas, P.; Landthaler, M.; Schreml, S. Cytokines, chemokines and growth factors in wound healing. J. Eur. Acad. Dermatol. Venereol. 2012, 26, 812–820. [Google Scholar] [CrossRef] [PubMed]
- Pessa, M.E.; Bland, K.I.; Copeland, E.M. Growth factors and determinants of wound repair. J. Surg. Res. 1987, 42, 207–217. [Google Scholar] [CrossRef]
- Gainza, G.; Pastor, M.; Aguirre, J.J.; Villullas, S.; Pedraz, J.L.; Hernandez, R.M.; Igartua, M. A novel strategy for the treatment of chronic wounds based on the topical administration of rhEGF-loaded lipid nanoparticles: In vitro bioactivity and in vivo effectiveness in healing-impaired db/db mice. J. Control. Release 2014, 185, 51–61. [Google Scholar] [CrossRef]
- Lao, G.; Yan, L.; Yang, C.; Zhang, L.; Zhang, S.; Zhou, Y. Controlled Release of Epidermal Growth Factor from Hydrogels Accelerates Wound Healing in Diabetic Rats. J. Am. Podiatr. Med. Assoc. 2012, 102, 89–98. [Google Scholar] [CrossRef]
- Ulubayram, K.; Cakar, A.N.; Korkusuz, P.; Ertan, C.; Hasirci, N. EGF containing gelatin-based wound dressing. Biomaterials 2001, 22, 1345–1356. [Google Scholar] [CrossRef]
- Ji, W.; Sun, Y.; Yang, F.; van den Beucken, J.J.; Fan, M.; Chen, Z.; Jansen, J.A. Bioactive Electrospun Scaffolds Delivering Growth Factors and Genes for Tissue Engineering Applications. Pharm. Res. 2011, 28, 1259–1272. [Google Scholar] [CrossRef]
- Kim, H.; Kong, W.H.; Seong, K.Y.; Sung, D.K.; Jeong, H.; Kim, J.K.; Yang, S.Y.; Hahn, S.K. Hyaluronate—Epidermal Growth Factor Conjugate for Skin Wound Healing and Regeneration. Biomacromolecules 2016, 17, 3694–3705. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhang, Y.; Liu, C.G. Polymeric nanoparticles based on carboxymethyl chitosan in combination with painless microneedle therapy systems for enhancing transdermal insulin delivery. RCS Adv. 2020, 10, 24319–24329. [Google Scholar] [CrossRef]
- Müller, R.H.; Mäder, K.; Gohla, S. Solid lipid nanoparticles (SLN) for controlled drug delivery—A review of the state of the art. Eur. J. Pharm. Biopharm. 1998, 45, 149–155. [Google Scholar]
- Sonin, D.; Pochkaeva, E.; Zhuravskii, S.; Postnov, V.; Korolev, D.; Vasina, L.; Kostina, D.; Mukhametdinova, D.; Zelinskaya, I.; Skorik, Y.; et al. Biological Safety and Biodistribution of Chitosan Nanoparticles. Nanomaterials 2020, 10, 810. [Google Scholar] [CrossRef]
- Zhang, P.; Zhao, S.; Yu, Y.; Wang, H.; Yang, Y.; Liu, C. Biocompatibility Profile and In Vitro Cellular Uptake of Self-assembled Alginate Nanoparticles. Molecules 2019, 24, 555. [Google Scholar] [CrossRef]
- Hong, L.; Han, D.; Li, M.X.; Zhang, P.; Liu, C.G. Development and validation of an ultraviolet-visible (uv-vis) spectrophotometric method for determination of phenylethyl resorcinol in new topical nanoemulsions. Int. J. Cosmet. Sci. 2017, 39, 337–343. [Google Scholar] [CrossRef]
- Aditya, A.; Chattopadhyay, S.; Jha, D.; Gautam, H.K.; Maiti, S.; Ganguli, M. Zinc Oxide nanoparticles dispersed in ionic liquids show high antimicrobial efficacy to skin-specific bacteria. ACS Appl. Mater. Interfaces 2018, 10, 15401–15411. [Google Scholar] [CrossRef] [PubMed]
- Vilela, P.; Heuer-Jungemann, A.; El-Sagheer, A.; Brown, T.; Muskens, O.L.; Smyth, N.R.; Kanaras, A.G. Sensing of vimentin mRNA in 2D and 3D models of wounded skin using DNA-coated gold nanoparticles. Small 2018, 14, 1703489. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Guo, H.; Liu, C.G. Fabrication of Carboxylmethyl Chitosan Nanocarrier via Self-Assembly for Efficient Delivery of Phenylethyl Resorcinol in B16 Cells. Polymers 2020, 12, 408. [Google Scholar] [CrossRef]
- Jiang, Z.; Han, B.; Li, H.; Yang, Y.; Liu, W. Carboxymethyl chitosan represses tumor angiogenesis in vitro and in vivo. Carbohydr. Polym. 2015, 129, 1–8. [Google Scholar] [CrossRef]
- Choi, J.K.; Jang, J.H.; Jang, W.H.; Kim, J.; Bae, I.H.; Bae, J.; Park, Y.H.; Kim, B.J.; Lim, K.M.; Park, J.W. The effect of epidermal growth factor (EGF) conjugated with low-molecular-weight protamine (LMWP) on wound healing of the skin. Biomaterials 2012, 33, 8579–8590. [Google Scholar] [CrossRef] [PubMed]
- Sapru, S.; Das, S.; Mandal, M.; Ghosh, A.K.; Kundu, S.C. Prospects of nonmulberry silk protein sericin-based nanofibrous matrices for wound healing—In vitro and in vivo investigations. Acta Biomater. 2018, 78, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Nian, S.; Huang, Q. Assembly of kafirin/carboxymethyl chitosan nanoparticles to enhance the cellular uptake of curcumin. Food Hydrocoll. 2015, 51, 166–175. [Google Scholar] [CrossRef]
- Shi, L.; Tang, C.; Yin, C. Glycyrrhizin-modified O-carboxymethyl chitosan nanoparticles as drug vehicles targeting hepatocellular carcinoma. Biomaterials 2012, 33, 7594–7604. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Liu, C.G.; Huang, Z.H.; Xue, F.F. Preparation and Characterization of Nanoparticles Based on Hydrophobic Alginate Derivative as Carriers for Sustained Release of Vitamin D-3. J. Agric. Food Chem. 2011, 59, 1962–1967. [Google Scholar] [CrossRef]
- Liu, C.G.; Chen, X.G.; Park, H.J. Self-assembled nanoparticles based on linoleic-acid modified chitosan: Stability and adsorption of trypsin. Carbohydr. Polym. 2005, 62, 293–298. [Google Scholar] [CrossRef]
- Zhang, P.; Zhao, S.R.; Li, J.X.; Hong, L.; Raja, M.A.; Yu, L.J.; Liu, C.G. Nanoparticles based on phenylalanine ethyl ester-alginate conjugate as vitamin B2 delivery system. J. Biomater. Appl. 2016, 31, 13–22. [Google Scholar] [CrossRef]
- Xu, X.Q.; Shen, H.; Xu, J.R.; Xie, M.Q.; Li, X.J. The colloidal stability and core-shell structure of magnetite nanoparticles coated with alginate. Appl. Surf. Sci. 2006, 253, 2158–2164. [Google Scholar] [CrossRef]
- Ma, Z.; Bai, J.; Wang, Y.; Jiang, X. Impact of Shape and Pore Size of Mesoporous Silica Nanoparticles on Serum Protein Adsorption and RBCs Hemolysis. ACS Appl. Mater. Interfaces 2014, 6, 2431–2438. [Google Scholar] [CrossRef]
- Kumar, K.A.; John, J.; Sooraj, T.R.; Raj, S.A.; Unnikrishnan, N.V.; Selvaraj, N.B. Surface Plasmon response of silver nanoparticles doped silica synthesised via sol-gel route. Appl. Surf. Sci. 2018, 472, 40–45. [Google Scholar] [CrossRef]
- Li, H.; Xu, Y.; Sun, X.; Wang, S.; Wang, J.; Zhu, J.; Wang, D.; Zhao, L. Stability, bioactivity, and bioaccessibility of fucoxanthin in zein-caseinate composite nanoparticles fabricated at neutral pH by antisolvent precipitation. Food Hydrocoll. 2018, 84, 379–388. [Google Scholar] [CrossRef]
- Souto, E.B.; Wissing, S.A.; Barbosa, C.M.; Müller, R.H. Development of a controlled release formulation based on SLN and NLC for topical clotrimazole delivery. Int. J. Pharm. 2004, 278, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Akagi, T.; Kaneko, T.; Kida, T.; Akashi, M. Preparation and characterization of biodegradable nanoparticles based on poly(γ-glutamic acid) with l -phenylalanine as a protein carrier. J. Control. Release 2005, 108, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.Y.; Ma, G.H.; Su, Z.G. Preparation of uniform sized chitosan microspheres by membrane emulsification technique and application as a carrier of protein drug. J. Control. Release 2005, 106, 62–75. [Google Scholar] [CrossRef]
- Li, Q.; Liu, C.-G.; Yu, Y. Separation of monodisperse alginate nanoparticles and effect of particle size on transport of vitamin E. Carbohydr. Polym. 2015, 124, 274–279. [Google Scholar] [CrossRef]
- Zhang, J.; Song, H.; Ji, S.; Wang, X.; Huang, P.; Zhang, C.; Wang, W.; Kong, D. NO prodrug-conjugated, self-assembled, pH-responsive and galactose receptor targeted nanoparticles for co-delivery of nitric oxide and doxorubicin. Nanoscale 2018, 10, 4179–4188. [Google Scholar] [CrossRef]
- Losi, P.; Briganti, E.; Errico, C.; Lisella, A.; Sanguinetti, E.; Chiellini, F.; Soldani, G. Fibrin-based scaffold incorporating VEGF- and bFGF-loaded nanoparticles stimulates wound healing in diabetic mice. Acta Biomater. 2013, 9, 7814–7821. [Google Scholar] [CrossRef]
- Park, J.H.; Choi, S.H.; Park, S.J.; Lee, Y.J.; Park, J.H.; Song, P.H.; Cho, C.M.; Ku, S.K.; Song, C.H. Promoting Wound Healing Using Low Molecular Weight Fucoidan in a Full-Thickness Dermal Excision Rat Model. Mar. Drugs 2017, 15, 112. [Google Scholar] [CrossRef]
- Chigurupati, S.; Mughal, M.R.; Okun, E.; Das, S.; Kumar, A.; McCaffery, M.; Seal, S.; Mattson, M.P. Effects of cerium oxide nanoparticles on the growth of keratinocytes, fibroblasts and vascular endothelial cells in cutaneous wound healing. Biomaterials 2013, 34, 2194–2201. [Google Scholar] [CrossRef]
- Chantre, C.O.; Campbell, P.H.; Golecki, H.M.; Buganza, A.T.; Capulli, A.K.; Deravi, L.F.; Dauth, S.; Sheehy, S.P.; Paten, J.A.; Gledhill, K.; et al. Production-scale fibronectin nanofibers promote wound closure and tissue repair in a dermal mouse model. Biomaterials 2018, 166, 96–108. [Google Scholar] [CrossRef]
- Hardwicke, J.; Moseley, R.; Stephens, P.; Harding, K.; Duncan, R.; Thomas, D.W. Bioresponsive Dextrin-rhEGF Conjugates: In Vitro Evaluation in Models Relevant to Its Proposed Use as a Treatment for Chronic Wounds. Mol. Pharm. 2010, 7, 699–707. [Google Scholar] [CrossRef] [PubMed]






| Formulation | DS (%) | Mean Size (nm) | PDI | Zeta Potential | Transmittance (%) | LE (%) | Hemolysis Rate (HR, %) |
|---|---|---|---|---|---|---|---|
| CC1 | 3.78 | 196.4 ± 5.49 | 0.272 | −19.8 ± 3.07 | 72 | 72.16 ± 2.69 | 2.83 |
| CC2 | 4.47 | 167.2 ± 11.38 | 0.239 | −20.1 ± 1.05 | 55 | 78.68 ± 1.83 | 2.65 |
| CC3 | 6.52 | 155.3 ± 4.62 | 0.202 | −23.3 ± 0.37 | 93 | 82.43 ± 3.14 | 1.48 |
| Groups | 3rd d (%) | 7th d (%) | 11th d (%) | 14th d (%) |
|---|---|---|---|---|
| Control | 32.24 ± 1.96 | 58.98 ± 3.02 | 71.04 ± 4.62 | 80.86 ± 3.29 |
| CC-NPs | 29.47 ± 3.14 | 61.67 ± 1.84 | 73.49 ± 5.17 | 84.61 ± 4.06 |
| rhEGF | 39.10 ± 2.37 | 73.53 ± 3.62 | 83.90 ± 3.24 | 92.34 ± 2.17 |
| rhEGF: CC-NPs | 46.20 ± 4.08 | 80.89 ± 4.07 | 91.27 ± 1.63 | 98.61 ± 1.64 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, P.; Liu, C. Enhancement of Skin Wound Healing by rhEGF-Loaded Carboxymethyl Chitosan Nanoparticles. Polymers 2020, 12, 1612. https://doi.org/10.3390/polym12071612
Zhang P, Liu C. Enhancement of Skin Wound Healing by rhEGF-Loaded Carboxymethyl Chitosan Nanoparticles. Polymers. 2020; 12(7):1612. https://doi.org/10.3390/polym12071612
Chicago/Turabian StyleZhang, Pei, and Chenguang Liu. 2020. "Enhancement of Skin Wound Healing by rhEGF-Loaded Carboxymethyl Chitosan Nanoparticles" Polymers 12, no. 7: 1612. https://doi.org/10.3390/polym12071612
APA StyleZhang, P., & Liu, C. (2020). Enhancement of Skin Wound Healing by rhEGF-Loaded Carboxymethyl Chitosan Nanoparticles. Polymers, 12(7), 1612. https://doi.org/10.3390/polym12071612