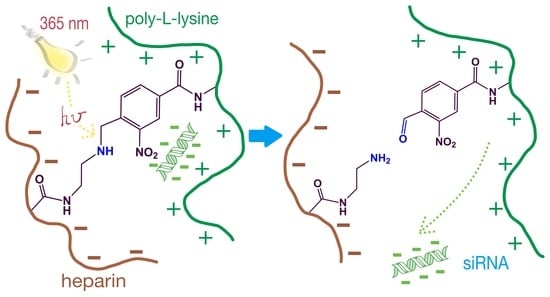
Photosensitive Poly-l-lysine/Heparin Interpolyelectrolyte Complexes for Delivery of Genetic Drugs
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Instruments
2.3. Cells
2.4. Methods
2.4.1. Poly-l-lysine Synthesis
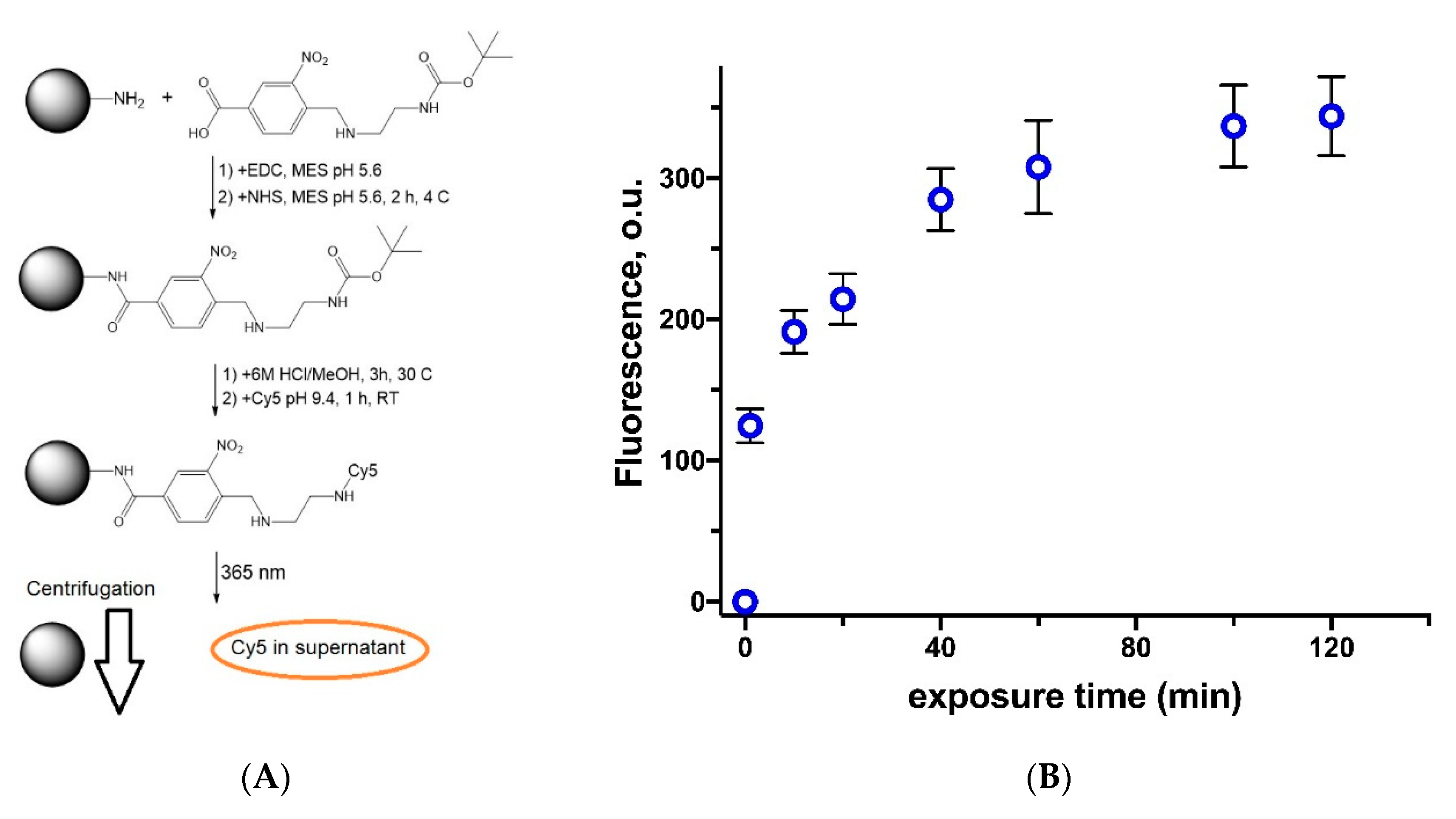
2.4.2. Photosensitive Linker Synthesis
2.4.3. IPECs Preparation
2.4.4. IPECs Characterization
2.4.5. Photo-Triggered Linker Decomposition
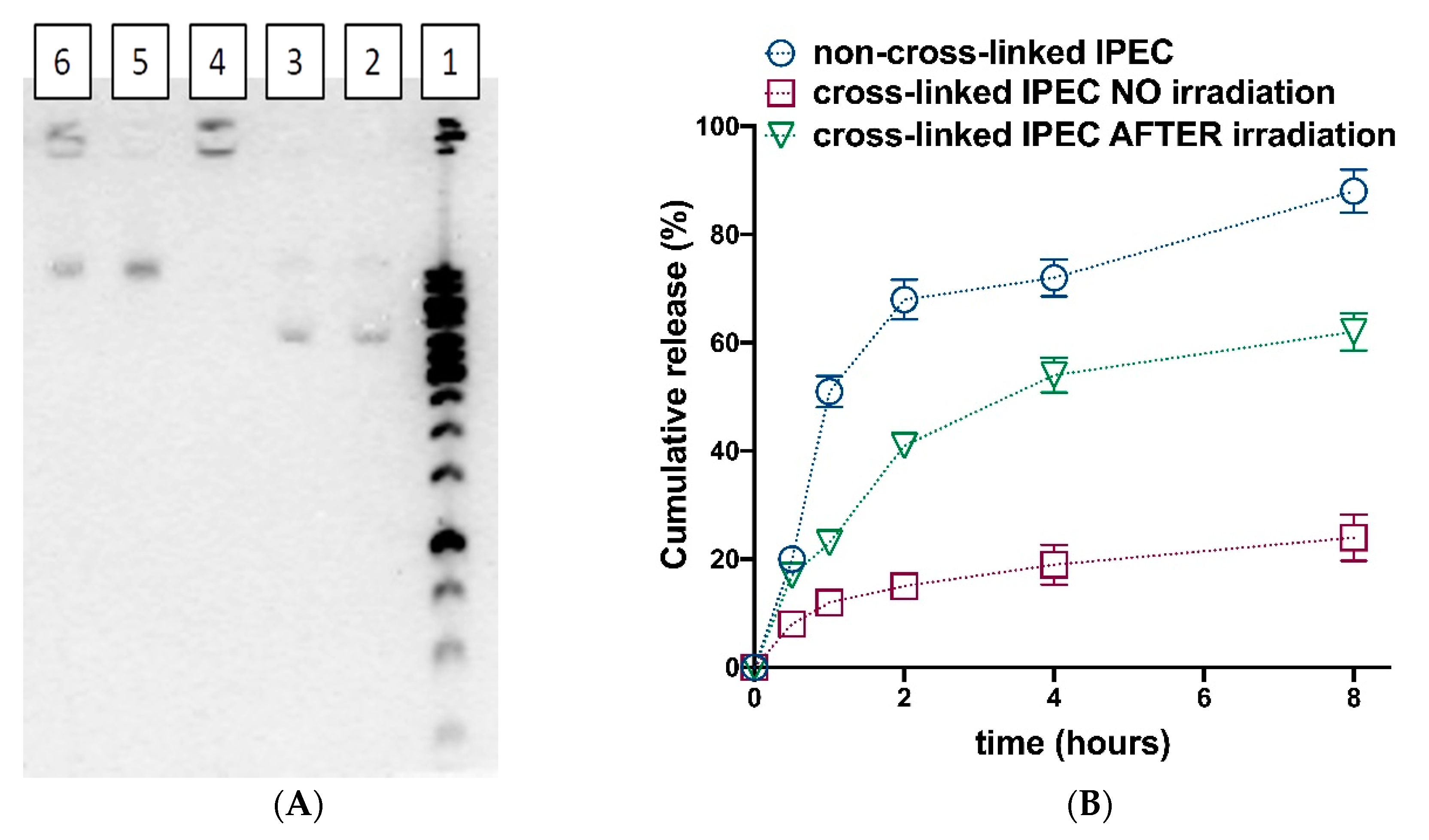
2.4.6. Photo-Triggered pDNA and Oligonucleotide Release
2.4.7. Cytotoxicity Assay
2.4.8. Particles Cellular Internalization
2.4.9. GFP Gene Silencing
2.4.10. Transfection
2.4.11. Statistics
3. Results and Discussion
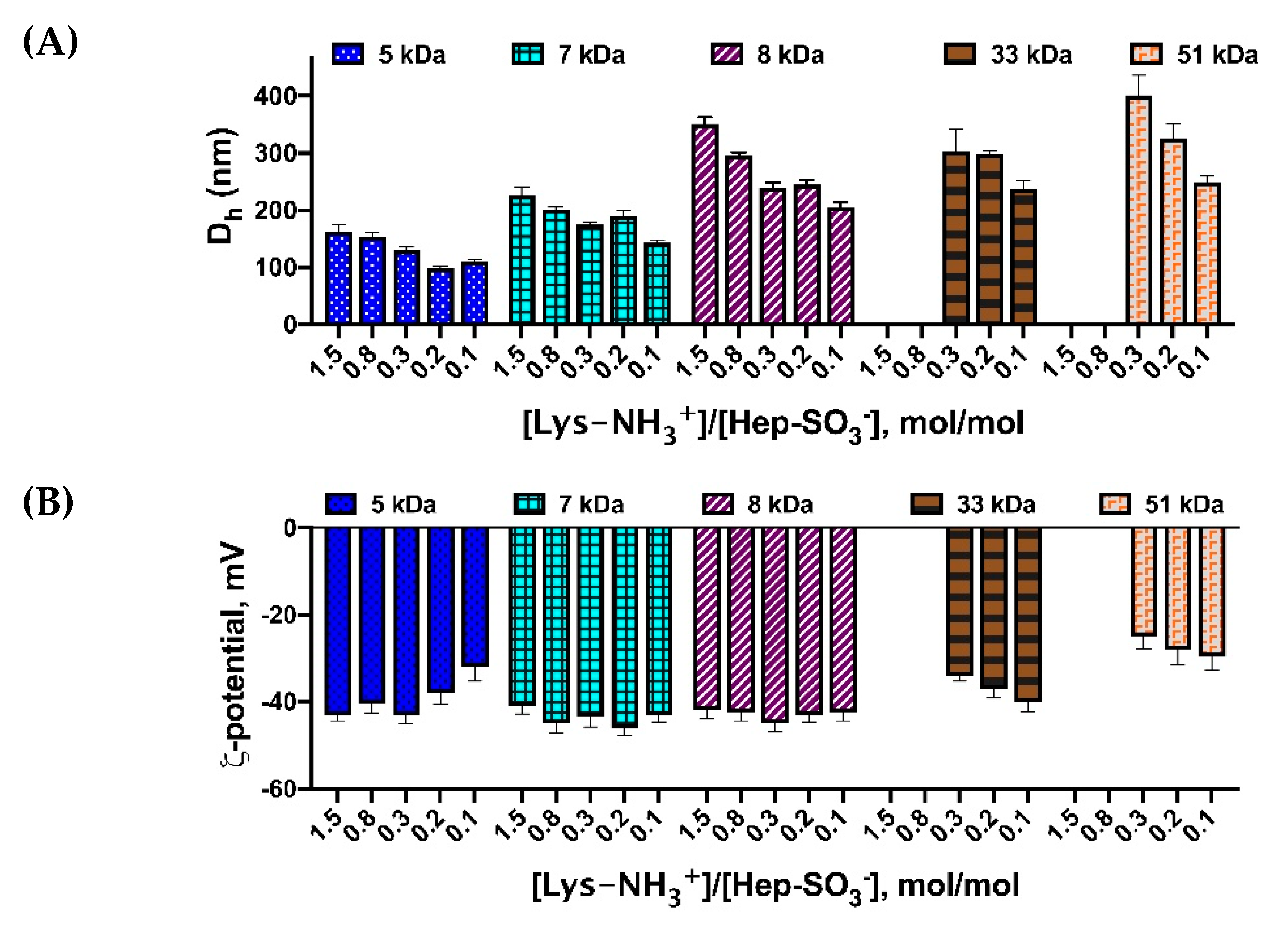
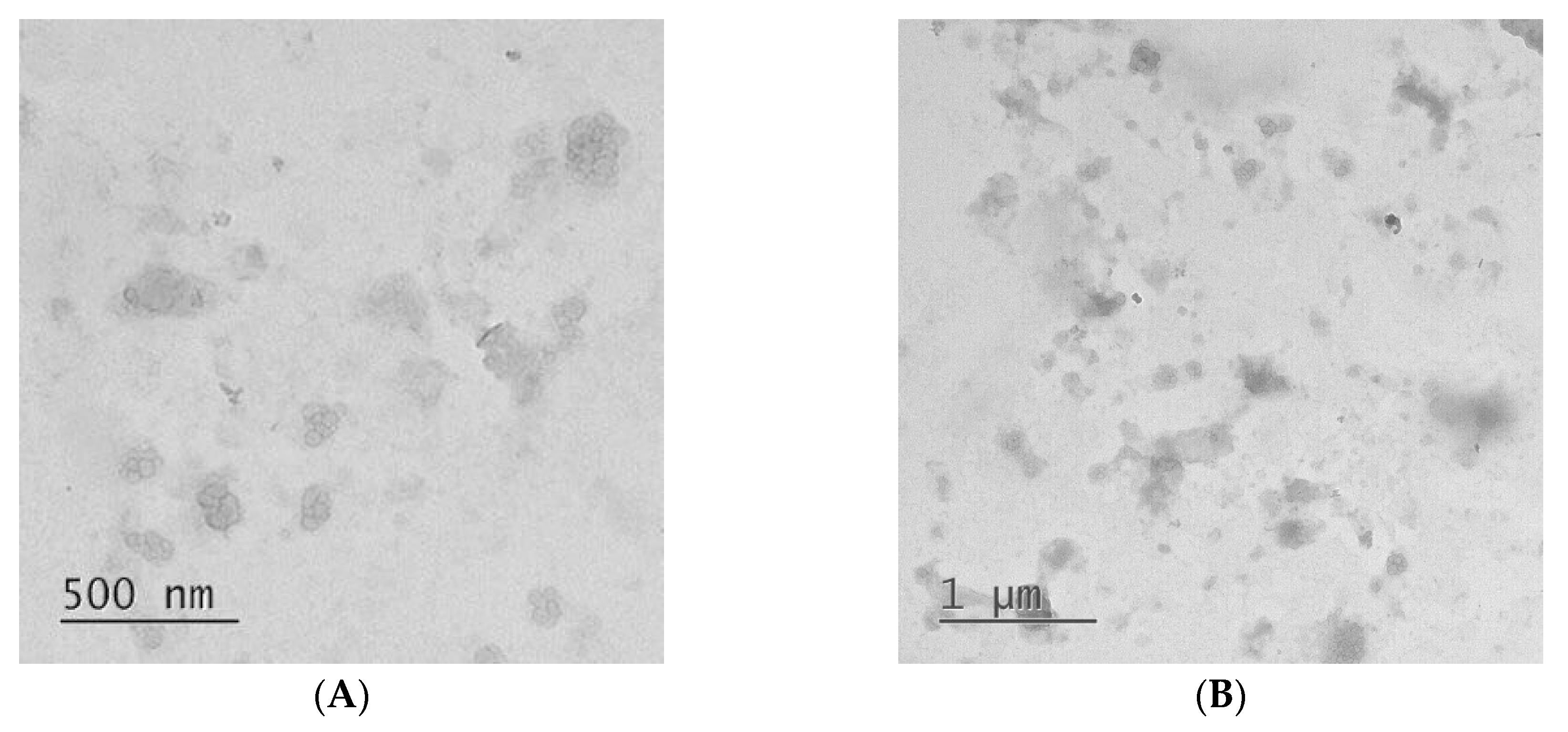
3.1. Preparation and Characterization of IPECs
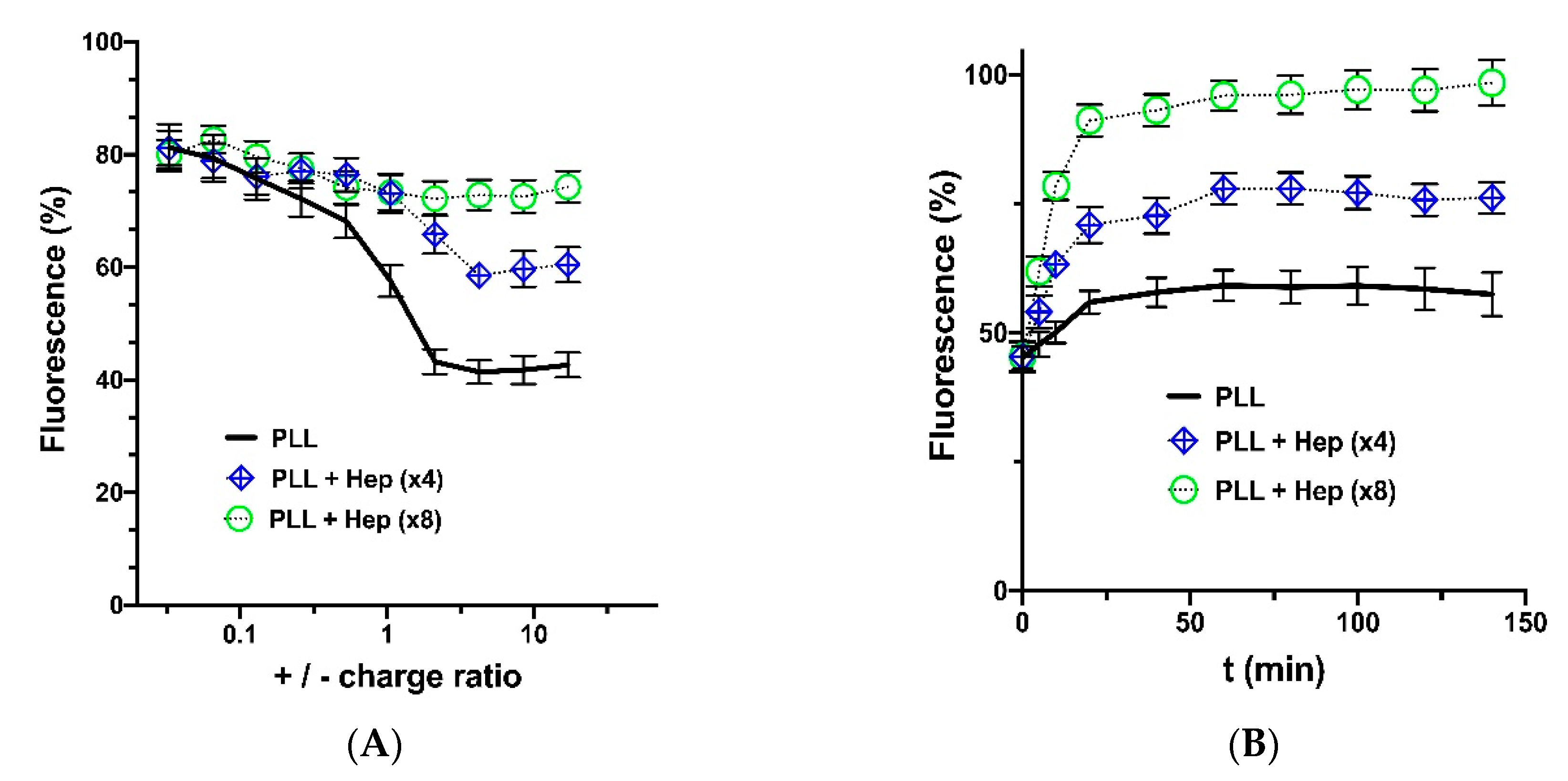
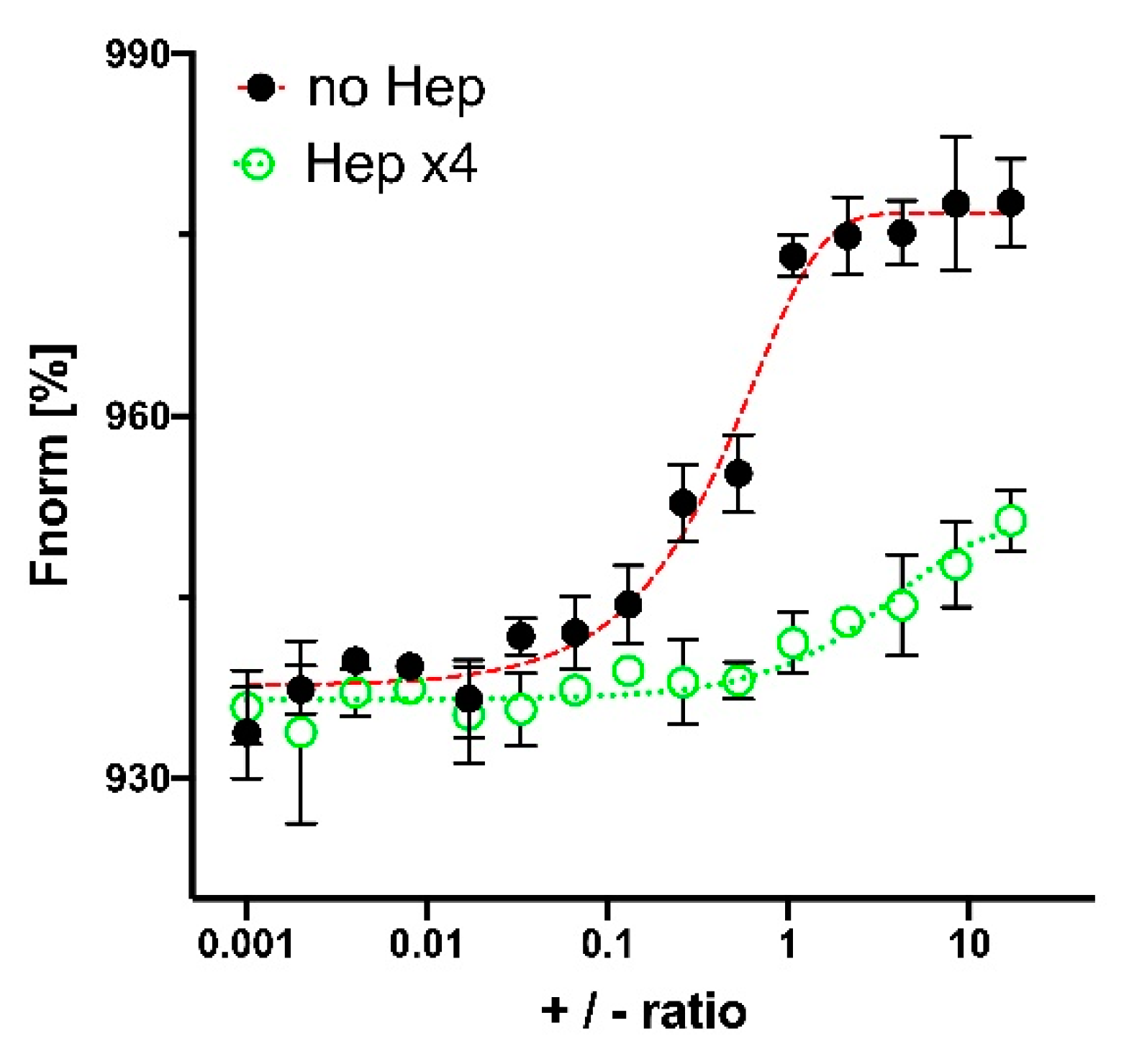
3.2. Hep Effect on PLL Interaction with Genetic Constructions
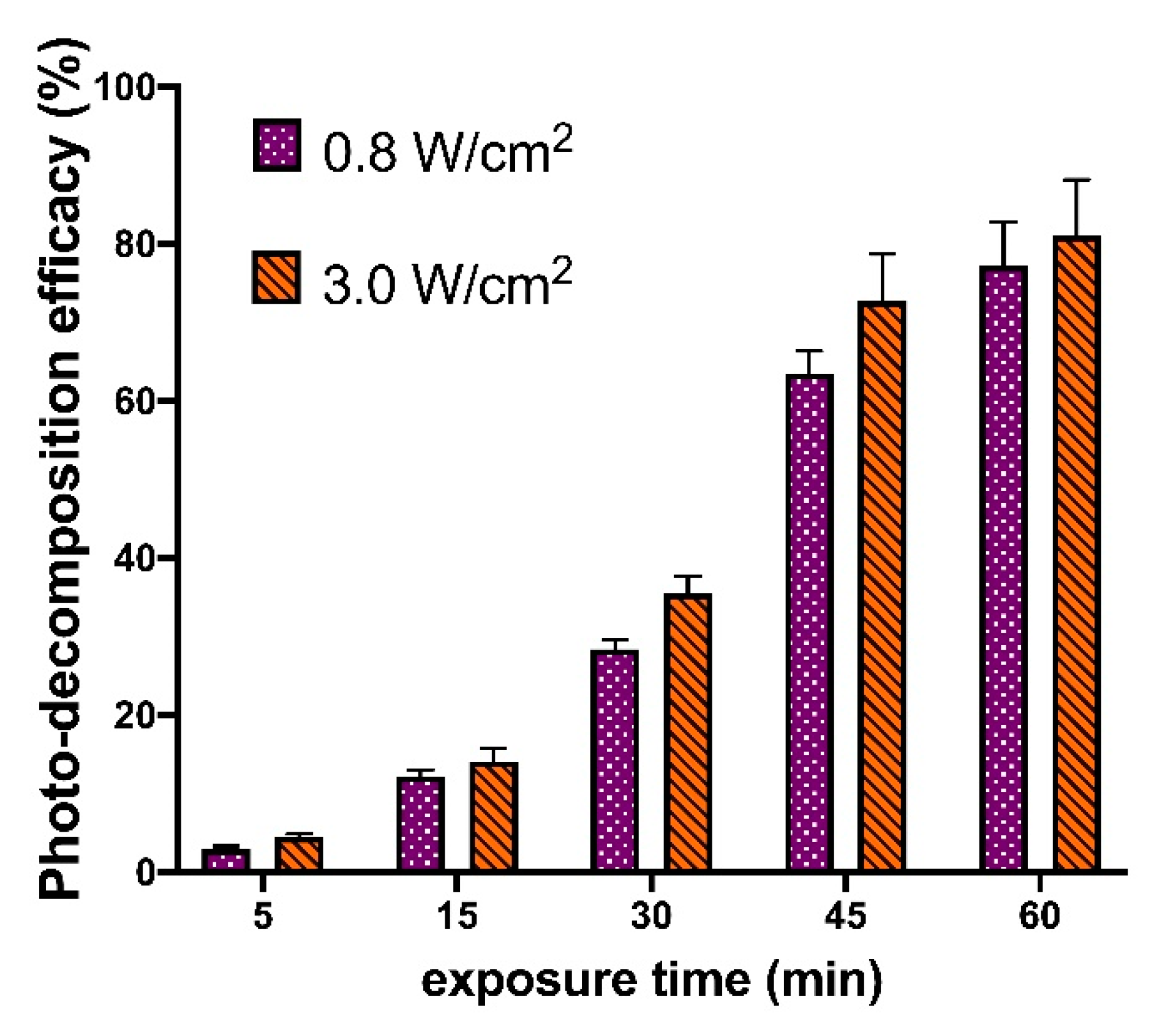
3.3. Assembling of Photosensitive IPECs and Linker Photo-Decomposition
3.4. Photosensitive Release
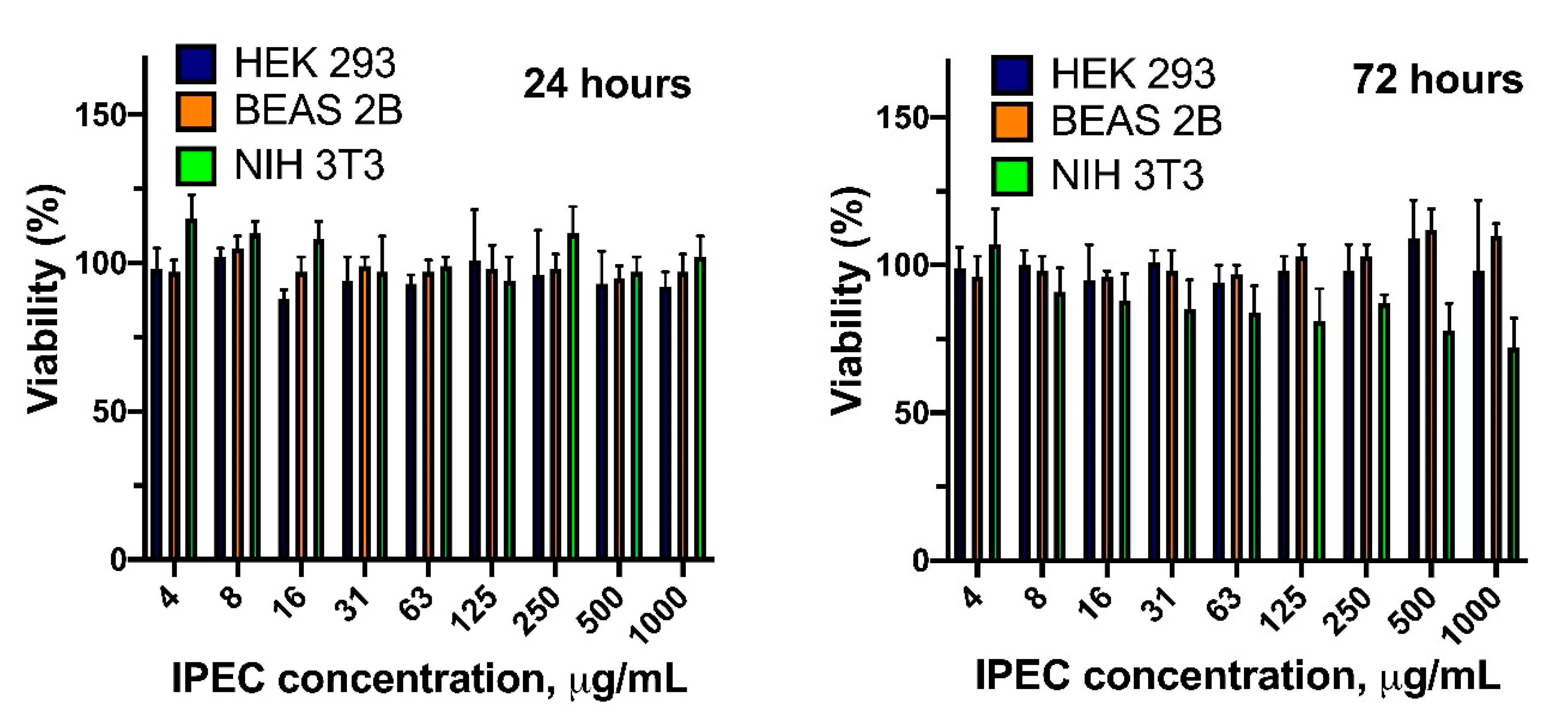
3.5. Cytotoxicity and Cellular Internalization
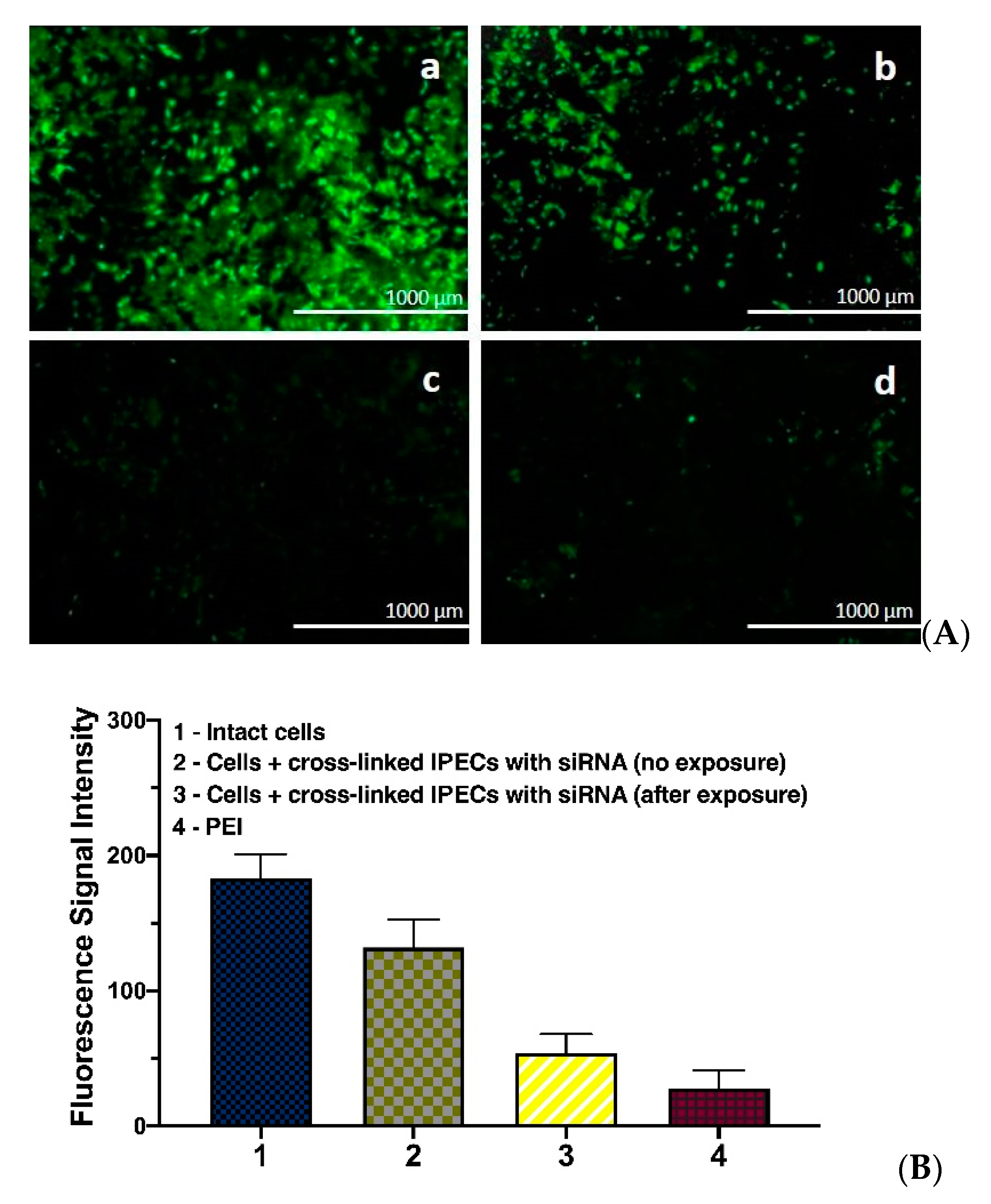
3.6. GFP Gene Expression Knockdown
3.7. Transfection Efficacy
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dunbar, C.E.; High, K.A.; Joung, J.K.; Kohn, D.B.; Ozawa, K.; Sadelain, M. Gene therapy comes of age. Science 2018, 359, eaan4672. [Google Scholar] [CrossRef] [PubMed]
- Kohn, D.B.; Sadelain, M.; Glorioso, J.C. Occurrence of leukaemia following gene therapy of X-linked SCID. Nat. Rev. Cancer 2003, 3, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.E.; Ehrhardt, A.; Kay, M.A. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 2003, 4, 346–358. [Google Scholar] [CrossRef] [PubMed]
- Marshall, E. CLINICAL TRIALS:Gene Therapy Death Prompts Review of Adenovirus Vector. Science 1999, 286, 2244–2245. [Google Scholar] [CrossRef]
- Schaffert, D.; Wagner, E. Gene therapy progress and prospects: Synthetic polymer-based systems. Gene Ther. 2008, 15, 1131–1138. [Google Scholar] [CrossRef]
- Kotterman, M.A.; Chalberg, T.W.; Schaffer, D.V. Viral Vectors for Gene Therapy: Translational and Clinical Outlook. Annu. Rev. Biomed. Eng. 2015, 17, 63–89. [Google Scholar] [CrossRef]
- Little, S.R.; Langer, R. Nonviral delivery of cancer genetic vaccines. Adv. Biochem. Eng. Biotechnol. 2005, 99, 93–118. [Google Scholar] [CrossRef]
- Munkonge, F.M.; Dean, D.A.; Hillery, E.; Griesenbach, U.; Alton, E.W.F.W. Emerging significance of plasmid DNA nuclear import in gene therapy. Adv. Drug Deliv. Rev. 2003, 55, 749–760. [Google Scholar] [CrossRef]
- Gaspar, V.; de Melo-Diogo, D.; Costa, E.; Moreira, A.; Queiroz, J.; Pichon, C.; Correia, I.; Sousa, F. Minicircle DNA vectors for gene therapy: Advances and applications. Expert Opin. Biol. Ther. 2015, 15, 353–379. [Google Scholar] [CrossRef]
- Chang, C.-W.; Christensen, L.V.; Lee, M.; Kim, S.W. Efficient expression of vascular endothelial growth factor using minicircle DNA for angiogenic gene therapy. J. Control. Release 2008, 125, 155–163. [Google Scholar] [CrossRef]
- Bi, F.; Liu, N.; Fan, D. Small Interfering RNA: A New Tool for Gene Therapy. Curr. Gene Ther. 2005, 3, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Nimjee, S.M.; White, R.R.; Becker, R.C.; Sullenger, B.A. Aptamers as Therapeutics. Annu. Rev. Pharmacol. Toxicol. 2017, 57, 61–79. [Google Scholar] [CrossRef] [PubMed]
- Steger, C.M.; Bonaros, N.; Rieker, R.J.; Bonatti, J.; Schachner, T. Gene therapy with antisense oligonucleotides silencing c-myc reduces neointima formation and vessel wall thickness in a mouse model of vein graft disease. Exp. Mol. Pathol. 2018, 105, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Daya, S.; Berns, K.I. Gene Therapy Using Adeno-Associated Virus Vectors. Clin. Microbiol. Rev. 2008, 21, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Shirley, J.L.; de Jong, Y.P.; Terhorst, C.; Herzog, R.W. Immune Responses to Viral Gene Therapy Vectors. Mol. Ther. 2020, 28, 709–722. [Google Scholar] [CrossRef]
- Chen, Y.H.; Keiser, M.S.; Davidson, B.L. Viral Vectors for Gene Transfer. Curr. Protoc. Mouse Biol. 2018, 8, e58. [Google Scholar] [CrossRef]
- Allon, N.; Saxena, A.; Chambers, C.; Doctor, B.P. A new liposome-based gene delivery system targeting lung epithelial cells using endothelin antagonist. J. Control. Release 2012, 160, 217–224. [Google Scholar] [CrossRef]
- AL Qtaish, N.; Gallego, I.; Villate-Beitia, I.; Sainz-Ramos, M.; López-Méndez, T.B.; Grijalvo, S.; Eritja, R.; Soto-Sánchez, C.; Martínez-Navarrete, G.; Fernández, E.; et al. Niosome-Based Approach for In Situ Gene Delivery to Retina and Brain Cortex as Immune-Privileged Tissues. Pharmaceutics 2020, 12, 198. [Google Scholar] [CrossRef]
- Kumar, V.; Singh, R.; Chaudhuri, A. Cationic Transfection Lipids in Gene Therapy: Successes, Set-backs, Challenges and Promises. Curr. Med. Chem. 2003, 10, 1297–1306. [Google Scholar] [CrossRef]
- Thomas, M.; Klibanov, A.M. Non-viral gene therapy: Polycation-mediated DNA delivery. Appl. Microbiol. Biotechnol. 2003, 62, 27–34. [Google Scholar] [CrossRef]
- Gebhart, C.L.; Kabanov, A.V. Evaluation of polyplexes as gene transfer agents. J. Control. Release 2001, 73, 401–416. [Google Scholar] [CrossRef]
- Osipova, O.; Sharoyko, V.; Zashikhina, N.; Zakharova, N.; Tennikova, T.; Urtti, A.; Korzhikova-Vlakh, E. Amphiphilic polypeptides for VEGF siRNA delivery into retinal epithelial cells. Pharmaceutics 2020, 12, 39. [Google Scholar] [CrossRef] [PubMed]
- Kabanov, A.V. Taking polycation gene delivery systems from in vitro to in vivo. Pharm. Sci. Technol. Today 1999, 2, 365–372. [Google Scholar] [CrossRef]
- Erbacher, P.; Remy, J.S.; Behr, J.P. Gene transfer with synthetic virus-like particles via the integrin-mediated endocytosis pathway. Gene Ther. 1999, 6, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Wang, S.; Li, C.; Qian, M.; Yan, X.; Yao, S.; Peng, X.; Wang, Y.; Huang, R. Photothermal combined gene therapy achieved by polyethyleneimine-grafted oxidized mesoporous carbon nanospheres. Biomaterials 2016, 100, 134–142. [Google Scholar] [CrossRef]
- Song, L.; Liang, X.; Yang, S.; Wang, N.; He, T.; Wang, Y.; Zhang, L.; Wu, Q.; Gong, C. Novel polyethyleneimine-R8-heparin nanogel for high-efficiency gene delivery in vitro and in vivo. Drug Deliv. 2018, 25, 122–131. [Google Scholar] [CrossRef]
- Kim, S.W. In Vitro Transfection with Oligonucleotide DNA Using Polylysine Copolymers. Cold Spring Harb. Protoc. 2012, 5, pdb.prot068635. [Google Scholar] [CrossRef]
- Chen, B.; Yu, L.; Li, Z.; Wu, C. Design of Free Triblock Polylysine-b-Polyleucine-b-Polylysine Chains for Gene Delivery. Biomacromolecules 2018, 19, 1347–1357. [Google Scholar] [CrossRef]
- Pilipenko, I.; Korzhikov-Vlakh, V.; Sharoyko, V.; Zhang, N.; Schäfer-Korting, M.; Rühl, E.; Zoschke, C.; Tennikova, T. pH-sensitive chitosan–heparin nanoparticles for effective delivery of genetic drugs into epithelial cells. Pharmaceutics 2019, 11, 317. [Google Scholar] [CrossRef]
- Strand, S.P.; Lelu, S.; Reitan, N.K.; de Lange Davies, C.; Artursson, P.; Vårum, K.M. Molecular design of chitosan gene delivery systems with an optimized balance between polyplex stability and polyplex unpacking. Biomaterials 2010, 31, 975–987. [Google Scholar] [CrossRef]
- Chen, H.H.; Ho, Y.-P.; Jiang, X.; Mao, H.-Q.; Wang, T.-H.; Leong, K.W. Quantitative Comparison of Intracellular Unpacking Kinetics of Polyplexes by a Model Constructed From Quantum Dot-FRET. Mol. Ther. 2008, 16, 324–332. [Google Scholar] [CrossRef]
- Douglas, K.L.; Piccirillo, C.A.; Tabrizian, M. Effects of alginate inclusion on the vector properties of chitosan-based nanoparticles. J. Control. Release 2006, 115, 354–361. [Google Scholar] [CrossRef] [PubMed]
- De la Fuente, M.; Seijo, B.; Alonso, M.J. Novel Hyaluronic Acid-Chitosan Nanoparticles for Ocular Gene Therapy. Investig. Opthalmol. Vis. Sci. 2008, 49, 2016. [Google Scholar] [CrossRef] [PubMed]
- Layek, B.; Singh, J. Caproic acid grafted chitosan cationic nanocomplexes for enhanced gene delivery: Effect of degree of substitution. Int. J. Pharm. 2013, 447, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Kontturi, L.S.; Van Den Dikkenberg, J.; Urtti, A.; Hennink, W.E.; Mastrobattista, E. Light-triggered cellular delivery of oligonucleotides. Pharmaceutics 2019, 11, 90. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Deng, W.; Xu, X.; Zhao, X.; Vo, J.N.; Anwer, A.G.; Williams, T.C.; Cui, H.; Goldys, E.M. Photoresponsive endosomal escape enhances gene delivery using liposome-polycation-DNA (LPD) nanovectors. J. Mater. Chem. B 2018, 6, 5269–5281. [Google Scholar] [CrossRef]
- Li, H.-J.; Wang, H.-X.; Sun, C.-Y.; Du, J.-Z.; Wang, J. Shell-detachable nanoparticles based on a light-responsive amphiphile for enhanced siRNA delivery. RSC Adv. 2014, 4, 1961–1964. [Google Scholar] [CrossRef]
- Foster, A.A.; Greco, C.T.; Green, M.D.; Epps, T.H.; Sullivan, M.O. Light-mediated activation of siRNA release in diblock copolymer assemblies for controlled gene silencing. Adv. Healthc. Mater. 2015, 4, 760–770. [Google Scholar] [CrossRef]
- Wang, J.; He, X.; Shen, S.; Cao, Z.; Yang, X. ROS-Sensitive Cross-Linked Polyethylenimine for Red-Light-Activated siRNA Therapy. ACS Appl. Mater. Interfaces 2019, 11, 1855–1863. [Google Scholar] [CrossRef]
- Ruponen, M.; Ylä-Herttuala, S.; Urtti, A. Interactions of polymeric and liposomal gene delivery systems with extracellular glycosaminoglycans: Physicochemical and transfection studies. Biochim. Biophys. Acta Biomembr. 1999, 1415, 331–341. [Google Scholar] [CrossRef]
- Boyle, W.S.; Senger, K.; Tolar, J.; Reineke, T.M. Heparin Enhances Transfection in Concert with a Trehalose-Based Polycation with Challenging Cell Types. Biomacromolecules 2017, 18, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Guy, C.A.; Fields, G.B. Trifluoroacetic acid cleavage and deprotection of resin-bound peptides following synthesis by Fmoc chemistry. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 1997; Volume 289, pp. 67–83. [Google Scholar] [CrossRef]
- Garg, H.G.; Linhardt, R.J.; Hales, C.A. Chemistry and Biology of Heparin and Heparan Sulfate; Elsevier: Amsterdam, The Netherlands, 2005; ISBN 9780080448596. [Google Scholar]
- McCray, J.A.; Trentham, D.R. Properties and Uses of Photoreactive Caged Compounds. Annu. Rev. Biophys. Biophys. Chem. 1989, 18, 239–270. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, M.R.; San Román, J. Smart Polymers and Their Applications; Woodhead Publishing: Cambridge, UK, 2014; ISBN 9780857096951. [Google Scholar]
- Kritchenkov, I.S.; Zhukovsky, D.D.; Mohamed, A.; Korzhikov-Vlakh, V.A.; Tennikova, T.B.; Lavrentieva, A.; Scheper, T.; Pavlovskiy, V.V.; Porsev, V.V.; Evarestov, R.A.; et al. Functionalized Pt(II) and Ir(III) NIR Emitters and Their Covalent Conjugates with Polymer-Based Nanocarriers. Bioconj. Chem. 2020, 31. [Google Scholar] [CrossRef] [PubMed]



| # | [Lys–NH3+]:[Hep–SO3−]:[Linker] mol/mol/mol | q (Bound Linker), mol% | Dh, nm | ζ-Potential, mV | PDI |
|---|---|---|---|---|---|
| 1 | 1:3:0.4 | 100 | 112 ± 5 | −38 ± 1 | 0.2 |
| 2 | 1:3:0.8 | 98 | 105 ± 2 | −31 ± 2 | 0.2 |
| 3 | 1:3:1.6 | 81 | 103 ± 1 | −38 ± 1 | 0.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Korzhikov-Vlakh, V.; Katernuk, I.; Pilipenko, I.; Lavrentieva, A.; Guryanov, I.; Sharoyko, V.; Manshina, A.A.; Tennikova, T.B. Photosensitive Poly-l-lysine/Heparin Interpolyelectrolyte Complexes for Delivery of Genetic Drugs. Polymers 2020, 12, 1077. https://doi.org/10.3390/polym12051077
Korzhikov-Vlakh V, Katernuk I, Pilipenko I, Lavrentieva A, Guryanov I, Sharoyko V, Manshina AA, Tennikova TB. Photosensitive Poly-l-lysine/Heparin Interpolyelectrolyte Complexes for Delivery of Genetic Drugs. Polymers. 2020; 12(5):1077. https://doi.org/10.3390/polym12051077
Chicago/Turabian StyleKorzhikov-Vlakh, Viktor, Iuliia Katernuk, Iuliia Pilipenko, Antonina Lavrentieva, Ivan Guryanov, Vladimir Sharoyko, Alina A. Manshina, and Tatiana B. Tennikova. 2020. "Photosensitive Poly-l-lysine/Heparin Interpolyelectrolyte Complexes for Delivery of Genetic Drugs" Polymers 12, no. 5: 1077. https://doi.org/10.3390/polym12051077
APA StyleKorzhikov-Vlakh, V., Katernuk, I., Pilipenko, I., Lavrentieva, A., Guryanov, I., Sharoyko, V., Manshina, A. A., & Tennikova, T. B. (2020). Photosensitive Poly-l-lysine/Heparin Interpolyelectrolyte Complexes for Delivery of Genetic Drugs. Polymers, 12(5), 1077. https://doi.org/10.3390/polym12051077