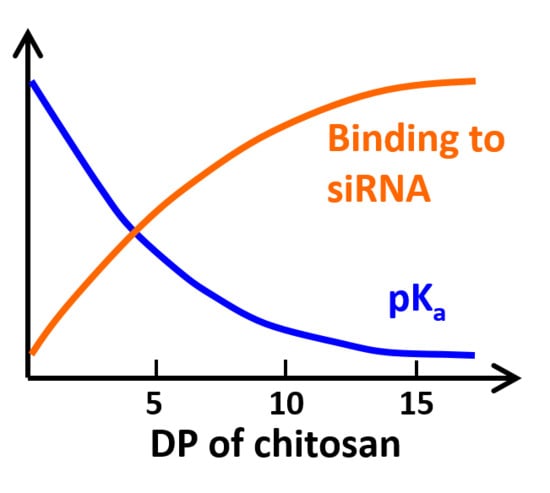
Effects of Chain Length of Chitosan Oligosaccharides on Solution Properties and Complexation with siRNA
Abstract
1. Introduction
2. Materials and Methods
3. Results
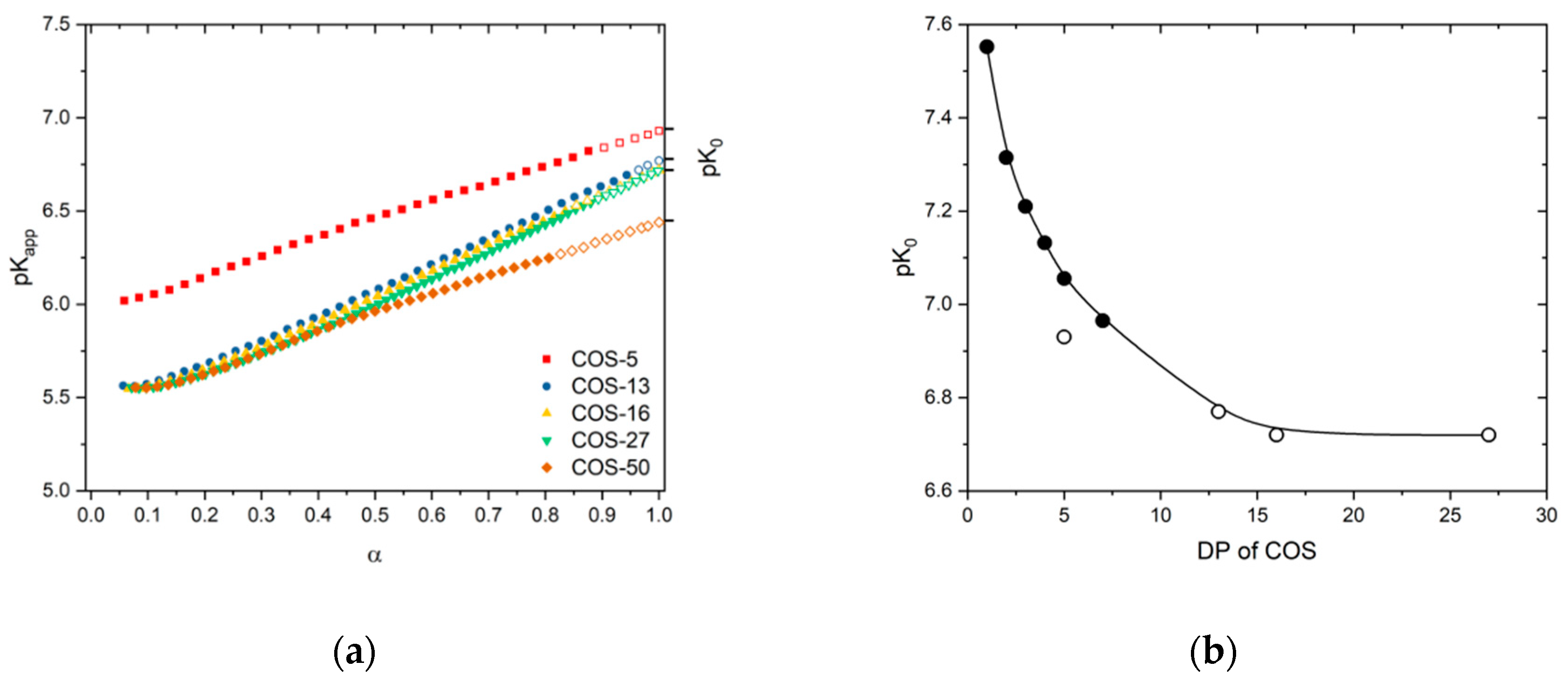
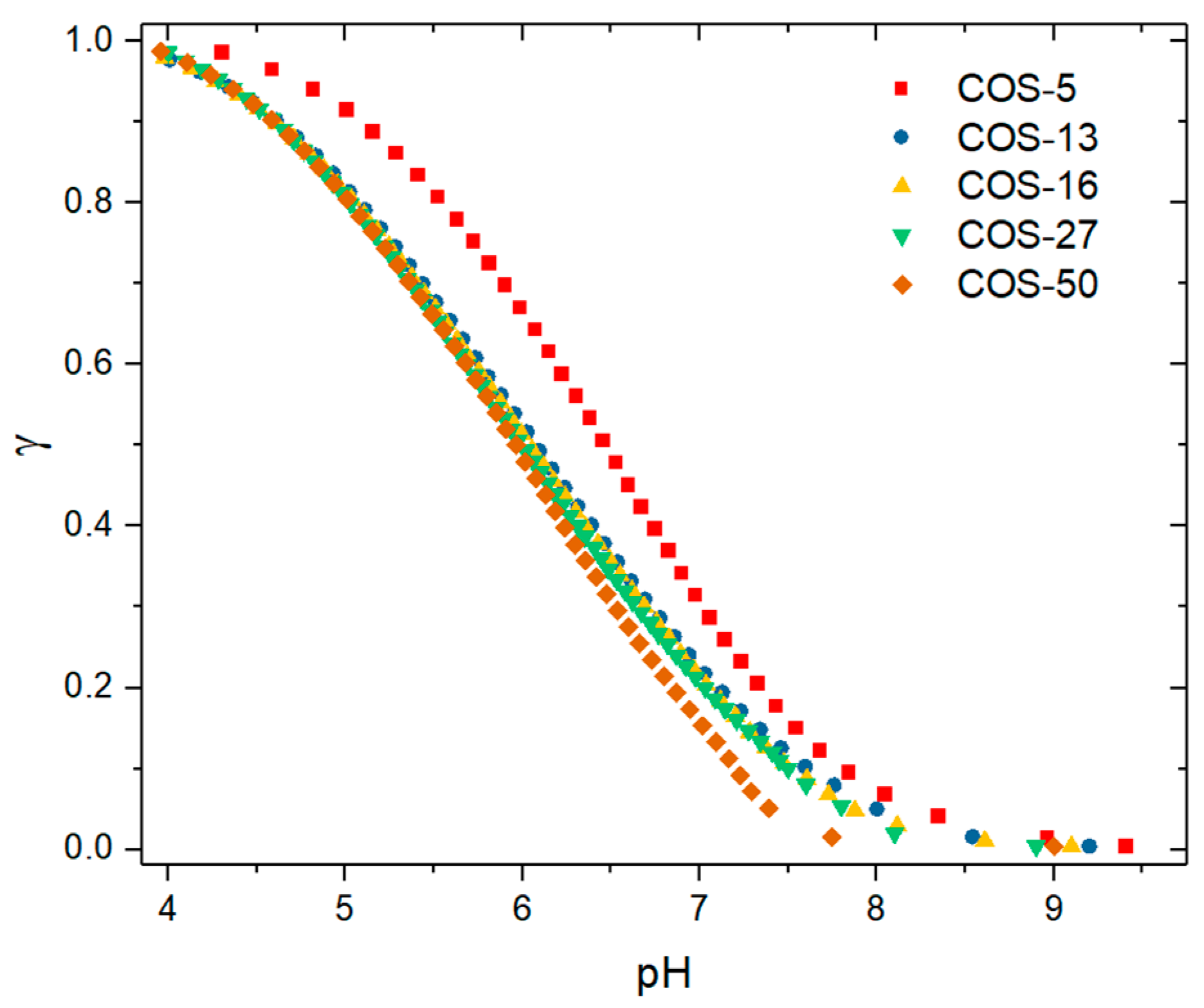
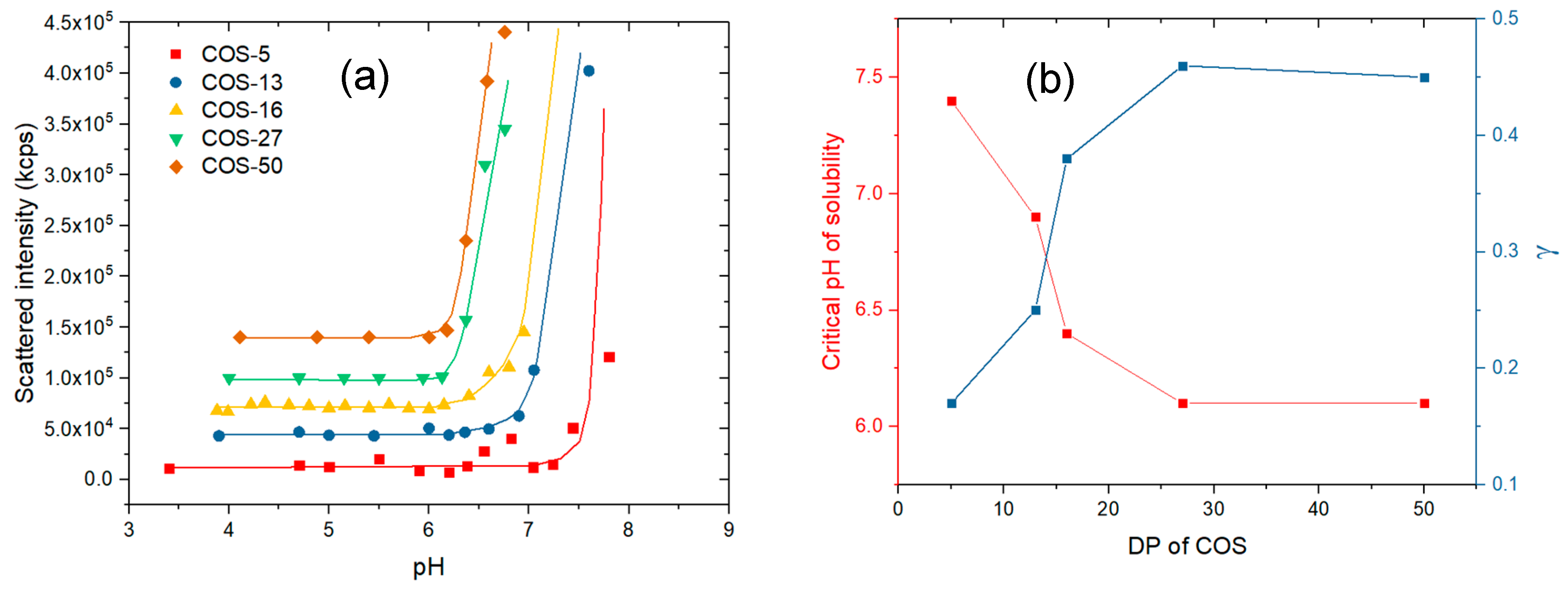
3.1. Solution Properties of Chitosan Oligosaccharides
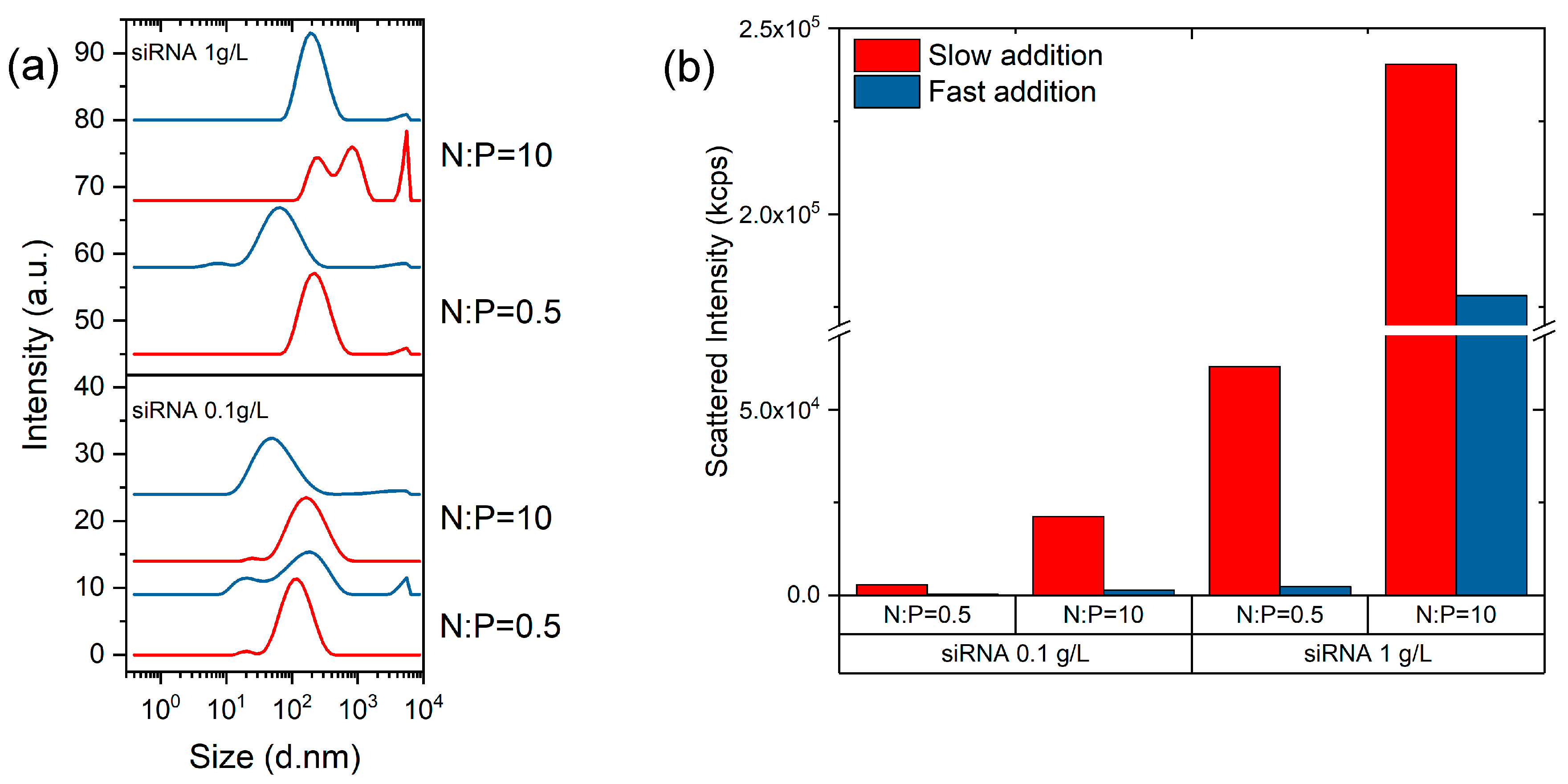
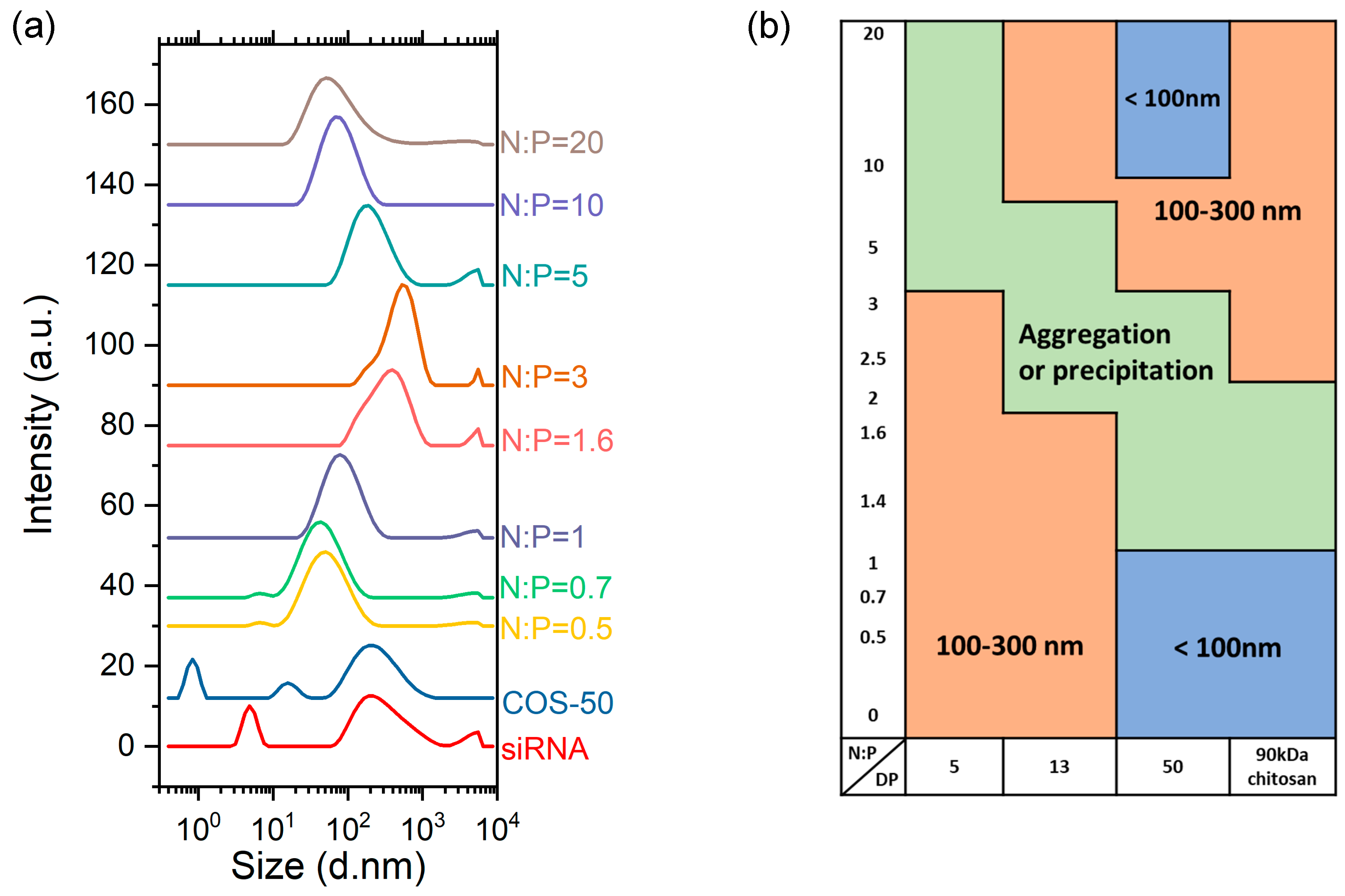
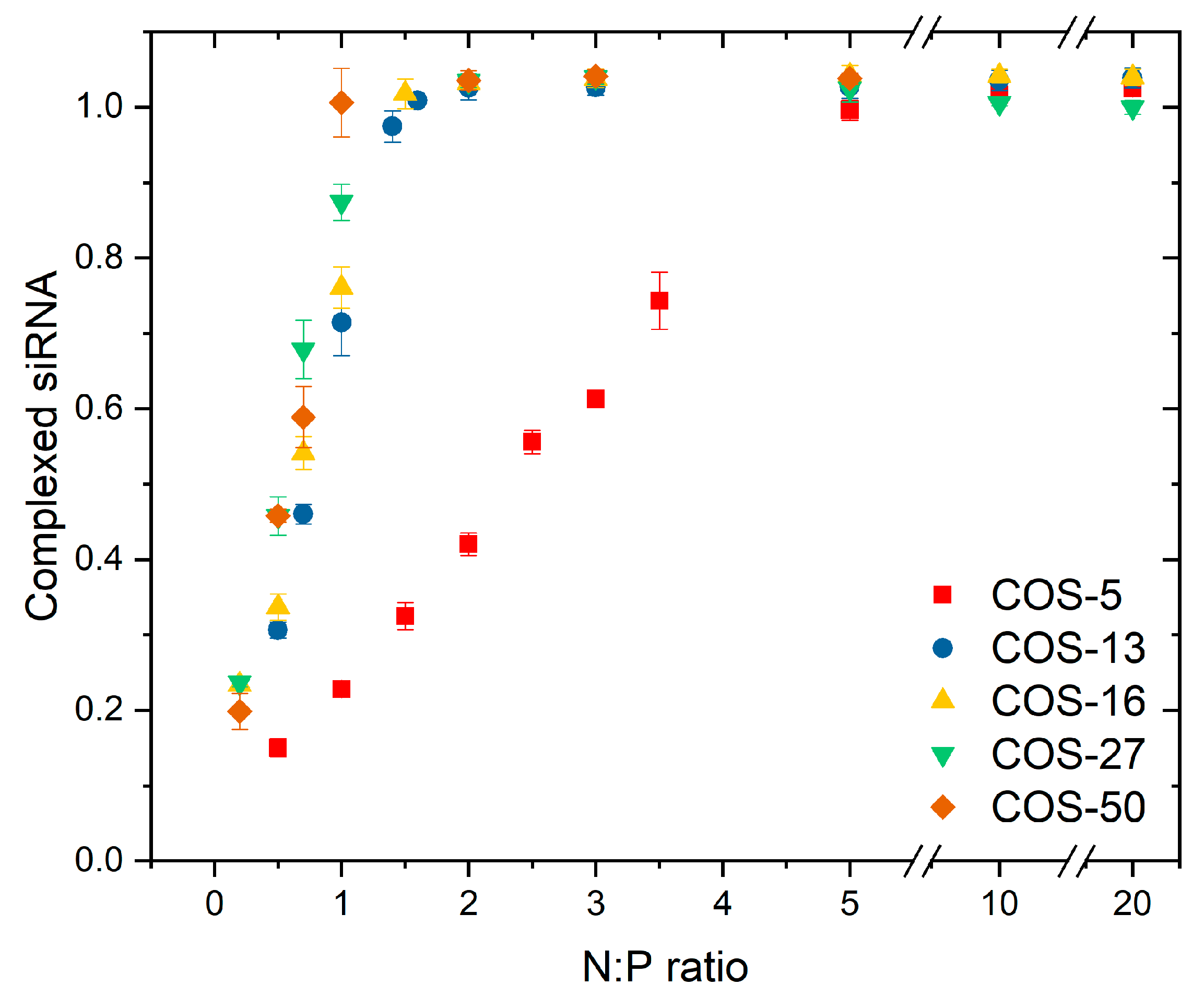
3.2. Complexation of COS with siRNA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zargar, V.; Asghari, M.; Dashti, A. A Review on Chitin and Chitosan Polymers: Structure, Chemistry, Solubility, Derivatives, and Applications. ChemBioEng Rev. 2015, 2, 204–226. [Google Scholar]
- Shariatinia, Z. Pharmaceutical applications of chitosan. Adv. Colloid Interface Sci. 2019, 263, 131–194. [Google Scholar]
- Holzerny, P.; Ajdini, B.; Heusermann, W.; Bruno, K.; Schuleit, M.; Meinel, L.; Keller, M. Biophysical properties of chitosan/siRNA polyplexes: Profiling the polymer/siRNA interactions and bioactivity. J. Control. Release 2012, 157, 297–304. [Google Scholar]
- Ragelle, H.; Riva, R.; Vandermeulen, G.; Naeye, B.; Pourcelle, V.; Le Duff, C.S.; D’Haese, C.; Nysten, B.; Braeckmans, K.; De Smedt, S.C.; et al. Chitosan nanoparticles for siRNA delivery: Optimizing formulation to increase stability and efficiency. J. Control. Release 2014, 176, 54–63. [Google Scholar]
- Vauthier, C.; Zandanel, C.; Ramon, A.L. Chitosan-based nanoparticles for in vivo delivery of interfering agents including siRNA. Curr. Opin. Colloid Interface Sci. 2013, 18, 406–418. [Google Scholar]
- Buschmann, M.D.; Merzouki, A.; Lavertu, M.; Thibault, M.; Jean, M.; Darras, V. Chitosans for delivery of nucleic acids. Adv. Drug Deliv. Rev. 2013, 65, 1234–1270. [Google Scholar]
- Lavertu, M.; Méthot, S.; Tran-Khanh, N.; Buschmann, M.D. High efficiency gene transfer using chitosan/DNA nanoparticles with specific combinations of molecular weight and degree of deacetylation. Biomaterials 2006, 27, 4815–4824. [Google Scholar]
- Esmaeilzadeh Gharehdaghi, E.; Amani, A.; Khoshayand, M.R.; Banan, M.; Esmaeilzadeh Gharehdaghi, E.; Amini, M.A.; Faramarzi, M.A. Chitosan nanoparticles for siRNA delivery: Optimization of processing/formulation parameters. Nucleic Acid Ther. 2014, 24, 420–427. [Google Scholar]
- Sorlier, P.; Denuzière, A.; Viton, C.; Domard, A. Relation between the Degree of Acetylation and the Electrostatic Properties of Chitin and Chitosan. Biomacromolecules 2001, 2, 765–772. [Google Scholar]
- Huang, M.; Fong, C.-W.; Khor, E.; Lim, L.-Y. Transfection efficiency of chitosan vectors: Effect of polymer molecular weight and degree of deacetylation. J. Control. Release 2005, 106, 391–406. [Google Scholar]
- Liu, X.; Howard, K.A.; Dong, M.; Andersen, M.Ø.; Rahbek, U.L.; Johnsen, M.G.; Hansen, O.C.; Besenbacher, F.; Kjems, J. The influence of polymeric properties on chitosan/siRNA nanoparticle formulation and gene silencing. Biomaterials 2007, 28, 1280–1288. [Google Scholar]
- Malmo, J.; Sørgård, H.; Vårum, K.M.; Strand, S.P. siRNA delivery with chitosan nanoparticles: Molecular properties favoring efficient gene silencing. J. Control. Release 2012, 158, 261–268. [Google Scholar]
- Fernandes, J.C.; Qiu, X.; Winnik, F.M.; Benderdour, M.; Zhang, X.; Dai, K.; Shi, Q. Low molecular weight chitosan conjugated with folate for siRNA delivery in vitro: Optimization studies. Int. J. Nanomed. 2012, 7, 5833–5845. [Google Scholar]
- Alameh, M.; Lavertu, M.; Tran-Khanh, N.; Chang, C.-Y.; Lesage, F.; Bail, M.; Darras, V.; Chevrier, A.; Buschmann, M.D. siRNA Delivery with Chitosan: Influence of Chitosan Molecular Weight, Degree of Deacetylation, and Amine to Phosphate Ratio on in Vitro Silencing Efficiency, Hemocompatibility, Biodistribution, and in Vivo Efficacy. Biomacromolecules 2018, 19, 112–131. [Google Scholar]
- Strand, S.P.; Danielsen, S.; Christensen, B.E.; Vårum, K.M. Influence of Chitosan Structure on the Formation and Stability of DNA−Chitosan Polyelectrolyte Complexes. Biomacromolecules 2005, 6, 3357–3366. [Google Scholar]
- Allan, G.G.; Peyron, M. Molecular weight manipulation of chitosan I: Kinetics of depolymerization by nitrous acid. Carbohydr. Res. 1995, 277, 257–272. [Google Scholar]
- Allan, G.G.; Peyron, M. Molecular weight manipulation of chitosan II: Prediction and control of extent of depolymerization by nitrous acid. Carbohydr. Res. 1995, 277, 273–282. [Google Scholar]
- Fulmer, G.R.; Miller, A.J.M.; Sherden, N.H.; Gottlieb, H.E.; Nudelman, A.; Stoltz, B.M.; Bercaw, J.E.; Goldberg, K.I. NMR Chemical Shifts of Trace Impurities: Common Laboratory Solvents, Organics, and Gases in Deuterated Solvents Relevant to the Organometallic Chemist. Organometallics 2010, 29, 2176–2179. [Google Scholar]
- Tømmeraas, K.; Vårum, K.M.; Christensen, B.E.; Smidsrød, O. Preparation and characterisation of oligosaccharides produced by nitrous acid depolymerisation of chitosans. Carbohydr. Res. 2001, 333, 137–144. [Google Scholar]
- Salim, E.; Ailincai, D.; Trombotto, S. Chitooligosaccharide-2,5-anhydro-D-mannonic Acid. Molbank 2014, 2014, M832. [Google Scholar]
- Müller, M.; Keßler, B.; Fröhlich, J.; Poeschla, S.; Torger, B. Polyelectrolyte Complex Nanoparticles of Poly(ethyleneimine) and Poly(acrylic acid): Preparation and Applications. Polymers 2011, 3, 762–778. [Google Scholar]
- Schatz, C.; Domard, A.; Viton, C.; Pichot, C.; Delair, T. Versatile and Efficient Formation of Colloids of Biopolymer-Based Polyelectrolyte Complexes. Biomacromolecules 2004, 5, 1882–1892. [Google Scholar]
- Vitorazi, L.; Ould-Moussa, N.; Sekar, S.; Fresnais, J.; Loh, W.; Chapel, J.P.; Berret, J.F. Evidence of a two-step process and pathway dependency in the thermodynamics of poly(diallyldimethylammonium chloride)/poly(sodium acrylate) complexation. Soft Matter 2014, 10, 9496–9505. [Google Scholar]
- Bezrodnykh, E.A.; Blagodatskikh, I.V.; Kulikov, S.N.; Zelenikhin, P.V.; Yamskov, I.A.; Tikhonov, V.E. Consequences of chitosan decomposition by nitrous acid: Approach to non-branched oligochitosan oxime. Carbohydr. Polym. 2018, 195, 551–557. [Google Scholar]
- Katchalsky, A.; Shavit, N.; Eisenberg, H. Dissociation of weak polymeric acids and bases. J. Polym. Sci. 1954, 13, 69–84. [Google Scholar]
- Tsukada, S.; Inoue, Y. Conformational properties of chito-oligosaccharides: Titration, optical rotation, and carbon-13 N.M.R. studies of chito-oligosaccharides. Carbohydr. Res. 1981, 88, 19–38. [Google Scholar]
- Zentz, F.; Bédouet, L.; Almeida, M.J.; Milet, C.; Lopez, E.; Giraud, M. Characterization and Quantification of Chitosan Extracted from Nacre of the Abalone Haliotis tuberculata and the Oyster Pinctada maxima. Mar. Biotechnol. 2001, 3, 36–44. [Google Scholar]
- Filion, D.; Lavertu, M.; Buschmann, M.D. Ionization and Solubility of Chitosan Solutions Related to Thermosensitive Chitosan/Glycerol-Phosphate Systems. Biomacromolecules 2007, 8, 3224–3234. [Google Scholar]
- Domard, A. pH and c.d. measurements on a fully deacetylated chitosan: Application to CuII—polymer interactions. Int. J. Biol. Macromol. 1987, 9, 98–104. [Google Scholar]
- Domard, A.; Gey, C.; Taravel, F. Glucosamine oligomers: 2. N.m.r. studies on a DP3. Int. J. Biol. Macromol. 1991, 13, 105–109. [Google Scholar]
- Kodama, H.; Miyajima, T.; Mori, M.; Takahashi, M.; Nishimura, H.; Ishiguro, S. A unified analytical treatment of the acid-dissociation equilibria of weakly acidic linear polyelectrolytes and the conjugate acids of weakly basic linear polyelectrolytes. Colloid Polym. Sci. 1997, 275, 938–945. [Google Scholar]
- Kulikov, S.; Tikhonov, V.; Blagodatskikh, I.; Bezrodnykh, E.; Lopatin, S.; Khairullin, R.; Philippova, Y.; Abramchuk, S. Molecular weight and pH aspects of the efficacy of oligochitosan against methicillin-resistant Staphylococcus aureus (MRSA). Carbohydr. Polym. 2012, 87, 545–550. [Google Scholar]
- Katchalsky, A.; Spitnik, P. Potentiometric titrations of polymethacrylic acid. J. Polym. Sci. 1947, 2, 432–446. [Google Scholar]
- Tsuchida, E.; Osada, Y.; Abe, K. Formation of polyion complexes between polycarboxylic acids and polycations carrying charges in the chain backbone. Die Makromol. Chem. 1974, 175, 583–592. [Google Scholar]
- Rinaudo, M.; Milas, M.; Dung, P.L. Characterization of chitosan. Influence of ionic strength and degree of acetylation on chain expansion. Int. J. Biol. Macromol. 1993, 15, 281–285. [Google Scholar]
- Qin, C.; Li, H.; Xiao, Q.; Liu, Y.; Zhu, J.; Du, Y. Water-solubility of chitosan and its antimicrobial activity. Carbohydr. Polym. 2006, 63, 367–374. [Google Scholar]
- Vårum, K.M.; Ottøy, M.H.; Smidsrød, O. Water-solubility of partially N-acetylated chitosans as a function of pH: Effect of chemical composition and depolymerisation. Carbohydr. Polym. 1994, 25, 65–70. [Google Scholar]
- Liu, X.; Chapel, J.P.; Schatz, C. Structure, thermodynamic and kinetic signatures of a synthetic polyelectrolyte coacervating system. Adv. Colloid Interface Sci. 2017, 239, 178–186. [Google Scholar]
- Bakeev, K.N.; Izumrudov, V.A.; Kuchanov, S.I.; Zezin, A.B.; Kabanov, V.A. Kinetics and mechanism of interpolyelectrolyte exchange and addition reactions. Macromolecules 1992, 25, 4249–4254. [Google Scholar]
- Kabanov, V.A. Basic Properties of Soluble Interpolyelectrolyte Complexes Applied to Bioengineering and Cell Transformations. In Macromolecular Complexes in Chemistry and Biology; Dubin, P., Bock, J., Davis, R., Schulz, D.N., Thies, C., Eds.; Springer: Berlin/Heidelberg, Germany, 1994; pp. 151–174. [Google Scholar]
- Majedi, F.S.; Hasani-Sadrabadi, M.M.; Hojjati Emami, S.; Shokrgozar, M.A.; VanDersarl, J.J.; Dashtimoghadam, E.; Bertsch, A.; Renaud, P. Microfluidic assisted self-assembly of chitosan based nanoparticles as drug delivery agents. Lab Chip 2013, 13, 204–207. [Google Scholar]
- Debus, H.; Beck-Broichsitter, M.; Kissel, T. Optimized preparation of pDNA/poly(ethylene imine) polyplexes using a microfluidic system. Lab Chip 2012, 12, 2498–2506. [Google Scholar]
- Iliescu, C.; Tresset, G. Microfluidics-Driven Strategy for Size-Controlled DNA Compaction by Slow Diffusion through Water Stream. Chem. Mater. 2015, 27, 8193–8197. [Google Scholar]
- Eimer, W.; Pecora, R. Rotational and translational diffusion of short rodlike molecules in solution: Oligonucleotides. J. Chem. Phys. 1991, 94, 2324–2329. [Google Scholar]
- Sedlák, M. What Can Be Seen by Static and Dynamic Light Scattering in Polyelectrolyte Solutions and Mixtures? Langmuir 1999, 15, 4045–4051. [Google Scholar]
- Philippova, O.E.; Volkov, E.V.; Sitnikova, N.L.; Khokhlov, A.R.; Desbrieres, J.; Rinaudo, M. Two Types of Hydrophobic Aggregates in Aqueous Solutions of Chitosan and Its Hydrophobic Derivative. Biomacromolecules 2001, 2, 483–490. [Google Scholar]
- Korchagina, E.V.; Philippova, O.E. Multichain Aggregates in Dilute Solutions of Associating Polyelectrolyte Keeping a Constant Size at the Increase in the Chain Length of Individual Macromolecules. Biomacromolecules 2010, 11, 3457–3466. [Google Scholar]
- Blagodatskikh, I.V.; Bezrodnykh, E.A.; Abramchuk, S.S.; Muranov, A.V.; Sinitsyna, O.V.; Khokhlov, A.R.; Tikhonov, V.E. Short chain chitosan solutions: Self-assembly and aggregates disruption effects. J. Polym. Res. 2013, 20, 73. [Google Scholar]
- Mende, M.; Schwarz, S.; Zschoche, S.; Petzold, G.; Janke, A. Influence of the Hydrophobicity of Polyelectrolytes on Polyelectrolyte Complex Formation and Complex Particle Structure and Shape. Polymers 2011, 3, 1363–1376. [Google Scholar]
- Cousin, F.; Gummel, J.; Ung, D.; Boué, F. Polyelectrolyte−Protein Complexes: Structure and Conformation of Each Specie Revealed by SANS. Langmuir 2005, 21, 9675–9688. [Google Scholar]
- Alatorre-Meda, M.; Taboada, P.; Hartl, F.; Wagner, T.; Freis, M.; Rodríguez, J.R. The influence of chitosan valence on the complexation and transfection of DNA: The weaker the DNA–chitosan binding the higher the transfection efficiency. Colloids Surf. B Biointerfaces 2011, 82, 54–62. [Google Scholar]
- Richards Grayson, A.C.; Doody, A.M.; Putnam, D. Biophysical and Structural Characterization of Polyethylenimine-Mediated siRNA Delivery in Vitro. Pharm. Res. 2006, 23, 1868–1876. [Google Scholar]
- Terayama, H. Method of colloid titration (a new titration between polymer ions). J. Polym. Sci. 1952, 8, 243–253. [Google Scholar]
- Schatz, C.; Lucas, J.-M.; Viton, C.; Domard, A.; Pichot, C.; Delair, T. Formation and Properties of Positively Charged Colloids Based on Polyelectrolyte Complexes of Biopolymers. Langmuir 2004, 20, 7766–7778. [Google Scholar]
- Drogoz, A.; David, L.; Rochas, C.; Domard, A.; Delair, T. Polyelectrolyte Complexes from Polysaccharides: Formation and Stoichiometry Monitoring. Langmuir 2007, 23, 10950–10958. [Google Scholar]
- Vleugels, L.F.W.; Ricois, S.; Voets, I.K.; Tuinier, R. Reversal of metachromasy revisited; displacement of Toluidine-blue from alginate by surfactants. Colloids Surfaces A Physicochem. Eng. Asp. 2017, 529, 454–461. [Google Scholar]
- Sato, S.; Matsumoto, S.; Freivalds, T.; Erenpreisa, J. Consideration on the Metachromatic Spectra of Toluidine Blue Dimers Formed on DNA Oligomers. Bull. Chem. Soc. Jpn. 2010, 83, 1216–1222. [Google Scholar]
- Fukada, H.; Takahashi, K. Enthalpy and heat capacity changes for the proton dissociation of various buffer components in 0.1 M potassium chloride. Proteins Struct. Funct. Bioinform. 1998, 33, 159–166. [Google Scholar]
- Ma, P.L.; Lavertu, M.; Winnik, F.M.; Buschmann, M.D. New Insights into Chitosan−DNA Interactions Using Isothermal Titration Microcalorimetry. Biomacromolecules 2009, 10, 1490–1499. [Google Scholar]
- Petrov, A.I.; Antipov, A.A.; Sukhorukov, G.B. Base−Acid Equilibria in Polyelectrolyte Systems: From Weak Polyelectrolytes to Interpolyelectrolyte Complexes and Multilayered Polyelectrolyte Shells. Macromolecules 2003, 36, 10079–10086. [Google Scholar]
- Priftis, D.; Megley, K.; Laugel, N.; Tirrell, M. Complex coacervation of poly(ethylene-imine)/polypeptide aqueous solutions: Thermodynamic and rheological characterization. J. Colloid Interface Sci. 2013, 398, 39–50. [Google Scholar]
- Feng, X.; Leduc, M.; Pelton, R. Polyelectrolyte complex characterization with isothermal titration calorimetry and colloid titration. Colloids Surf. A Physicochem. Eng. Asp. 2008, 317, 535–542. [Google Scholar]
- Kim, W.; Yamasaki, Y.; Kataoka, K. Development of a Fitting Model Suitable for the Isothermal Titration Calorimetric Curve of DNA with Cationic Ligands. J. Phys. Chem. B 2006, 110, 10919–10925. [Google Scholar]
- Utsuno, K.; Uludag, H. Thermodynamics of polyethylenimine-DNA binding and DNA condensation. Biophys. J. 2010, 99, 201–207. [Google Scholar]
- Alonso, T.; Irigoyen, J.; Iturri, J.J.; Larena, I.L.; Moya, S.E. Study of the multilayer assembly and complex formation of poly(diallyldimethylammonium chloride) (PDADMAC) and poly(acrylic acid) (PAA) as a function of pH. Soft Matter 2013, 9, 1920–1928. [Google Scholar]
- Tsuchida, E.; Abe, K. Interactions between macromolecules in solution and intermacromolecular complexes. In Interactions Between Macromolecules in Solution and Intermacromolecular Complexes; Tsuchida, E., Abe, K., Eds.; Springer: Berlin/Heidelberg, Germany, 1982; pp. 1–119. [Google Scholar]
- Kabanov, V.A.; Papisov, I.M. Formation of complexes between complementary synthetic polymers and oligomers in dilute solution review. Polym. Sci. USSR 1979, 21, 261–307. [Google Scholar]
- Pärnaste, L.; Arukuusk, P.; Langel, K.; Tenson, T.; Langel, Ü. The Formation of Nanoparticles between Small Interfering RNA and Amphipathic Cell-Penetrating Peptides. Mol. Ther. Nucleic Acids 2017, 7, 1–10. [Google Scholar]
- Schatz, C.; Viton, C.; Delair, T.; Pichot, C.; Domard, A. Typical Physicochemical Behaviors of Chitosan in Aqueous Solution. Biomacromolecules 2003, 4, 641–648. [Google Scholar]
- Kabanov, V.A. The Cooperative Interactions of Complementary Synthetic Macromolecules in Solutions. In Macromolecular Chemistry–8; Saarela, K., Ed.; Butterworth-Heinemann: Oxford, UK, 1973; pp. 121–145. [Google Scholar]
- Papisov, I.M.; Litmanovich, A.A. Specificity of cooperative interactions between simple synthetic macromolecules and its connection with chain length. Polym. Sci. USSR 1977, 19, 830–837. [Google Scholar]
- Plank, C.; Tang, M.X.; Wolfe, A.R.; Szoka, F.C. Branched Cationic Peptides for Gene Delivery: Role of Type and Number of Cationic Residues in Formation and in Vitro Activity of DNA Polyplexes. Hum. Gene Ther. 1999, 10, 319–332. [Google Scholar]
- Tsuchida, E.; Osada, Y. The rôle of the chain length in the stability of polyion complexes. Die Makromol. Chem. 1974, 175, 593–601. [Google Scholar]
- Kharenko, A.V.; Starikova, E.A.; Lutsenko, V.V.; Zezin, A.B. Study of cooperative reactions of oligophosphates and polyphosphates with polybases. Vysokomol. Soedin. Seriya A 1976, 18, 1604–1608. [Google Scholar]
- Ikawa, T.; Abe, K.; Honda, K.; Tsuchida, E. Interpolymer complex between poly(ethylene oxide) and poly(carboxylic acid). J. Polym. Sci. Polym. Chem. Ed. 1975, 13, 1505–1514. [Google Scholar]

| SEC-MALLS | 1H NMR | |||||
|---|---|---|---|---|---|---|
| COS | Mn (g/mol) | Mw (g/mol) | Đ | dn/dc (mL/g) | DPn 1 | DPn 1 |
| COS-5 | 1030 | 1240 | 1.20 | 0.1660 | 5.4 | 4.5 |
| COS-13 | 2290 | 2900 | 1.26 | 0.1752 | 13.2 | 12.2 |
| COS-16 | 2710 | 3630 | 1.34 | 0.1860 | 15.8 | 20.2 |
| COS-27 | 4560 | 6420 | 1.41 | 0.1863 | 27.3 | 33.0 |
| COS-50 | 8190 | 11970 | 1.46 | 0.1801 | 49.8 | 69.7 |
| DPn | pK0 | pK1/2 | n |
|---|---|---|---|
| 5 | 6.93 | 6.46 | 1.53 |
| 13 | 6.77 | 6.08 | 1.74 |
| 16 | 6.72 | 6.04 | 1.73 |
| 27 | 6.72 | 6.00 | 1.73 |
| 50 | 6.44 | 5.95 | 1.56 |
| Ion Pairing | Aggregation | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| ΔH | TΔS | ΔG | Kb | n | ΔH | TΔS | ΔG | Kb | |
| COS-5 | −1.1 | 23.5 | −24.5 | 2.0 × 104 | 3.0 | 0.7 | 28.8 | −28.1 | 8.5 × 104 |
| COS-13 | −5.1 | 24.0 | −29.1 | 1.3 × 105 | 1.1 | 4.4 | 32.7 | −28.4 | 9.4 × 104 |
| COS-16 | −4.8 | 24.9 | −29.7 | 1.6 × 105 | 1.0 | 3.4 | 34.4 | −30.9 | 2.6 × 105 |
| COS-50 | −5.2 | 26.8 | −31.9 | 4.0 × 105 | 1.0 | 1.9 | 32.8 | −30.9 | 2.6 × 105 |
| Parent chitosan | −4.3 | 29.8 | −34.1 | 9.6 × 105 | 1.0 | ||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Delas, T.; Mock-Joubert, M.; Faivre, J.; Hofmaier, M.; Sandre, O.; Dole, F.; Chapel, J.P.; Crépet, A.; Trombotto, S.; Delair, T.; et al. Effects of Chain Length of Chitosan Oligosaccharides on Solution Properties and Complexation with siRNA. Polymers 2019, 11, 1236. https://doi.org/10.3390/polym11081236
Delas T, Mock-Joubert M, Faivre J, Hofmaier M, Sandre O, Dole F, Chapel JP, Crépet A, Trombotto S, Delair T, et al. Effects of Chain Length of Chitosan Oligosaccharides on Solution Properties and Complexation with siRNA. Polymers. 2019; 11(8):1236. https://doi.org/10.3390/polym11081236
Chicago/Turabian StyleDelas, Tim, Maxime Mock-Joubert, Jimmy Faivre, Mirjam Hofmaier, Olivier Sandre, François Dole, Jean Paul Chapel, Agnès Crépet, Stéphane Trombotto, Thierry Delair, and et al. 2019. "Effects of Chain Length of Chitosan Oligosaccharides on Solution Properties and Complexation with siRNA" Polymers 11, no. 8: 1236. https://doi.org/10.3390/polym11081236
APA StyleDelas, T., Mock-Joubert, M., Faivre, J., Hofmaier, M., Sandre, O., Dole, F., Chapel, J. P., Crépet, A., Trombotto, S., Delair, T., & Schatz, C. (2019). Effects of Chain Length of Chitosan Oligosaccharides on Solution Properties and Complexation with siRNA. Polymers, 11(8), 1236. https://doi.org/10.3390/polym11081236