Enzymatic Degradation of Star Poly(ε-Caprolactone) with Different Central Units
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Measurements
2.3. Expression and Purification of Cutinase
2.4. Cutinase Enzyme Activity
2.5. Enzymatic Hydrolysis
3. Results and Discussion
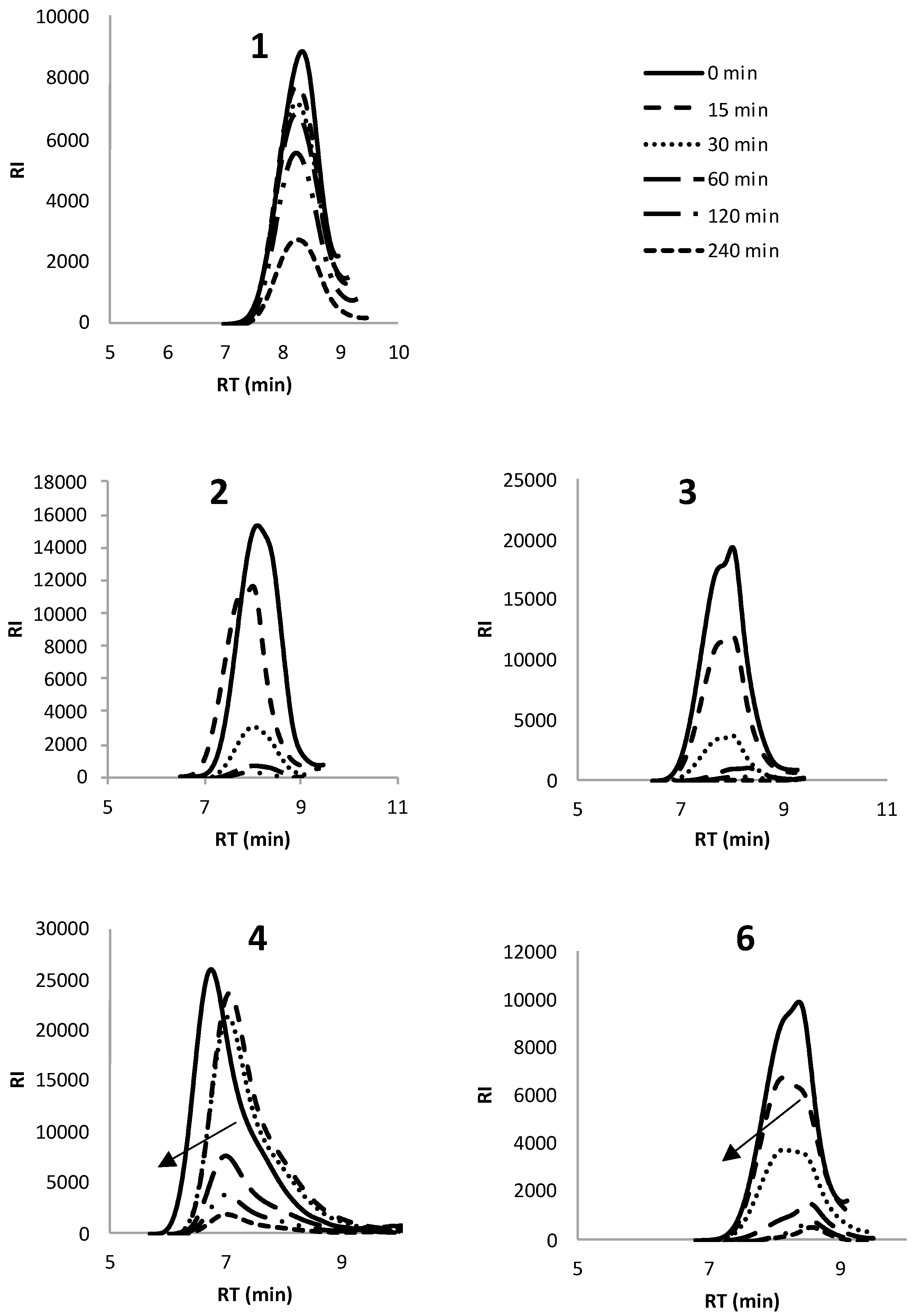
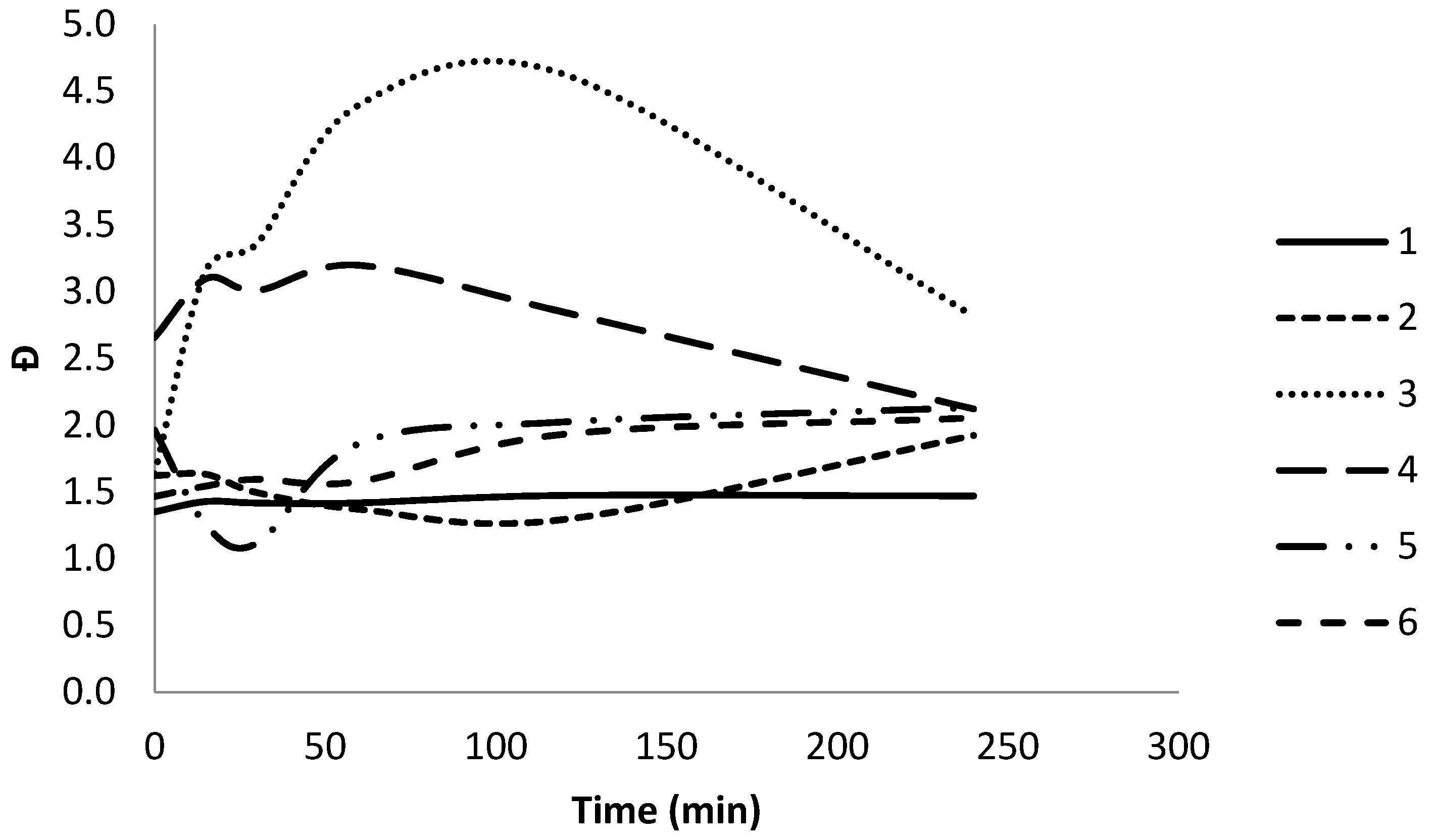
3.1. Enzymatic Degradation Using Lipase
3.2. Enzymatic Degradation Using Cutinase
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Geng, Y.; Discher, D.E. Hydrolytic Degradation of poly(ethylene oxide)-block-polycaprolactone worm micelles. J. Am. Chem. Soc. 2005, 127, 12780–12781. [Google Scholar] [CrossRef] [PubMed]
- Gref, R. Biodegradable long-circulating polymeric nanospheres. Science 1994, 263, 1600–1603. [Google Scholar] [CrossRef] [PubMed]
- Kumari, A. Biodegradable polymeric nanoparticles based drug delivery system. Colloids Surf. B 2010, 75, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Guillaume, S.M. Recent advances in ring-opening polymerization strategies toward a,x-hydroxy telechelic polyesters and resulting copolymers. Eur. Polym. J. 2013, 49, 768–779. [Google Scholar] [CrossRef]
- Nuyken, O.P.; Pask, S.D. Ring-opening polymerization—An introductory review. Polymers 2013, 5, 361–403. [Google Scholar] [CrossRef]
- Ghoroghchian, P.P.; Li, G.; Levine, D.H.; Davis, K.P.; Bates, F.S.; Hammer, D.A.; Therien, M.J. Bioresorbable Vesicles Formed through Spontaneous Self-Assembly of Amphiphilic Poly(ethylene oxide)-block-polycaprolactone. Macromolecules 2006, 39, 1673–1675. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.J.; Yuan, J.; Li, H.; Lei, Z. Synthesis and characterization of a series of biodegradable and biocompatible PEG-supportedpoly(lactic-ran-glycolic acid) amphiphilicbarbell-like copolymers. Polym. Sci. Part A Polym. Chem. 2008, 46, 3802–3812. [Google Scholar] [CrossRef]
- He, F.; Li, S.; Vert, M.; Zhuo, R. Enzymatric degradation of poly(-caprolactone)/poly(dl-lactide) blends in phosphate buffer solution. Polymer 2003, 44, 5145–5151. [Google Scholar] [CrossRef]
- Li, S.; Garreau, H.; Pauvert, B.; McGrath, J.; Toniolo, A.; Vert, M. Enzymatic degradation of block copolymers prepared from epsilon-caprolactone and poly(ethylene glycol). Biomacromolecules 2002, 3, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Gan, Z.; Yu, D.; Zhong, Z.; Liang, Q.; Jing, X. Enzymatric degradation of poly(-caprolactone)/poly(dl-lactide) blends in phosphate buffer solution. Polymer 1999, 40, 2859–2862. [Google Scholar] [CrossRef]
- Castilla-Cortázar, I.; Más-Estellés, J.; Meseguer-Dueñas, J.M.; Escobar Ivirico, J.L.; Marí, B.; Vidaurre, A. Hydrolytic and enzymatic degradation of a poly(ε-caprolactone) network. Polym. Degrad. Stab. 2012, 97, 1241–1248. [Google Scholar] [CrossRef]
- He, F. Enzyme-catalyzed polymerization and degradation of copolymers prepared from ϵ-caprolactone and poly(ethylene glycol). Polymer 2003, 44, 5145–5151. [Google Scholar] [CrossRef]
- Lee, C.; Kimura, J.; Chung, J.D. Mechanism of enzymatic degradation of poly(butylene succinate). Macromol. Res. 2008, 16, 651–658. [Google Scholar] [CrossRef]
- Fukuzaki, H.; Yoshida, M.; Asano, M.; Kumakura, M.; Mashimo, T.; Yuasa, H.; Imai, K.; Hidetoshi, Y. Synthesis of low-molecular-weight copoly(l-lactic acid/ɛ-caprolactone) by direct copolycondensation in the absence of catalysts, and enzymatic degradation of the polymers. Polymer 1990, 31, 2006–2014. [Google Scholar] [CrossRef]
- Mochizuki, M.; Hirano, M.; Kanmuri, Y.; Kudo, K.; Tokiwa, Y. Hydrolysis of polycaprolactone fibers by lipase: Effects of draw ratio on enzymatic degradation. J. Appl. Polym. Sci. 1995, 55, 289–296. [Google Scholar] [CrossRef]
- Gan, Z.; Liang, Q.; Zhang, J.; Jing, X. Enzymatic degradation of poly(ε-caprolactone) film in phosphate buffer solution containing lipases. Polym. Degrad. Stab. 1997, 56, 209–213. [Google Scholar] [CrossRef]
- Li, S.M.; Pignol, M.; Gasc, F.; Vert, M. Synthesis, Characterization, and enzymatic degradation of copolymers prepared from ε-caprolactone and β-butyrolactone. Macromolecules 2004, 37, 9798–9803. [Google Scholar] [CrossRef]
- Meseguer-Duenas, J.M.; Mas-Estelles, J.; Castilla-Cortazar, I.; Escobar Ivirico, J.L.; Vidaurre, A. Alkaline degradation study of linear and network poly(ε-caprolactone). J. Mater. Sci. Mater. Med. 2011, 22, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Ponsart, S.; Coudane, J.; Saulnier, B.; Morgat, J.-L.; Vert, M. Biodegradation of [(3)H]poly(epsilon-caprolactone) in the presence of active sludge extracts. Biomacromolecules 2001, 2, 373–377. [Google Scholar] [CrossRef] [PubMed]
- Khatiwala, V.K.; Shekhar, N.; Aggarwal, S.; Mandal, U.K. Biodegradation of poly(ecaprolactone)(PCL) film by alcaligenes faecalis. J. Polym. Environ. 2008, 16, 61–67. [Google Scholar] [CrossRef]
- Yoshioka, T.; Kamada, F.; Kawazoe, N.; Tateishi, T.; Chen, G. Structural changes and biodegradation of PLLA, PCL, and PLGA sponges during in vitro incubation. Polym. Eng. Sci. 2010, 50, 1895–1903. [Google Scholar] [CrossRef]
- Sekosan, G.; Vasanthan, N. Morphological changes of annealed poly-ε-caprolactone by enzymatic degradation with lipase. J. Polym. Sci. B Polym. Phys. 2010, 48, 202–211. [Google Scholar] [CrossRef]
- Inglis, G.D.; Yanke, L.J.; Selinger, L.B. Cutinolytic esterase activity of bacteria isolated from mixed-plant compost and characterization of a cutinase gene from Pseudomonas pseudoalcaligenes. Can. J. Microbiol. 2011, 57, 902–913. [Google Scholar] [CrossRef] [PubMed]
- Murphy, C.A.; Cameron, J.A.; Huang, S.J.; Vinopal, R.T. Fusarium polycaprolactone depolymerase is cutinase. Appl. Environ. Microbiol. 1996, 62, 456–460. [Google Scholar] [PubMed]
- Nishida, H.; Tokiwa, Y. Degradation of poly(2-oxepanone) by phytopathogens. Chem. Lett. 1994, 23, 1547–1550. [Google Scholar] [CrossRef]
- Perz, V.; Zumstein, M.T.; Sander, M.; Zitzenbacher, S.; Ribitsch, D.; Guebitz, G.M. Biomimetic approach to enhance enzymatic hydrolysis of the synthetic polyester poly(1,4-butylene adipate): Fusing binding modules to esterases. Biomacromolecules 2015, 16, 3889–3896. [Google Scholar] [CrossRef] [PubMed]
- Ribitsch, D.; Yebra, A.O.; Zitzenbacher, S.; Wu, J.; Nowitsch, S.; Steinkellner, G.; Greimel, K.; Doliska, A.; Oberdorfer, G.; Gruber, C.C.; et al. Fusion of binding domains to Thermobifida cellulosilytica cutinase to tune sorption characteristics and enhancing PET hydrolysis. Biomacromolecules 2013, 14, 1769–1776. [Google Scholar] [CrossRef] [PubMed]
- Perz, V.; Baumschlager, A.; Bleymaier, K.; Zitzenbacher, S.; Hromic, A.; Steinkellner, G.; Pairitsch, A.; Łyskowski, A.; Gruber, K.; Sinkel, C.; et al. Hydrolysis of synthetic polyesters by Clostridium botulinum esterases. Biotechnol. Bioeng. 2016, 113, 102. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.L.; Wang, L.; Dong, C.M. Synthesis, crystallization, and morphology of star-shaped poly(ɛ-caprolactone). Polym. Sci. Polym. Chem. 2005, 43, 5449–5457. [Google Scholar] [CrossRef]
- Li, Q.B.; Li, F.X.; Jia, L.; Li, Y.; Liu, Y.C.; Yu, J.Y.; Fang, Q.; Cao, A.M. New asymmetric AB(n)-shaped amphiphilic poly(ethylene glycol)-b-[poly(l-lactide)](n) (n = 2, 4, 8) bridged with dendritic ester linkages: I. Syntheses and their characterization. Biomacromolecules 2006, 7, 2377–2387. [Google Scholar] [CrossRef] [PubMed]
- Nederberg, F.; Appel, E.; Tan, J.P.K.; Kim, S.H.; Fukushima, K.; Sly, J.; Miller, R.D.; Waymouth, R.M.; Yang, Y.Y.; Hedrick, J.L. Simple approach to stabilized micelles employing miktoarm terpolymers and stereocomplexes with application in paclitaxel delivery. Biomacromolecules 2009, 10, 1460–1468. [Google Scholar] [CrossRef] [PubMed]
- Cohn, D.; Stern, T.; Gonzalez, M.F.; Epstein, J. Biodegradable poly(ethylene oxide)/poly(epsilon-caprolactone) multiblock copolymers. J. Biomed. Mater. Res. 2002, 59, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Herrero Acero, E.; Ribitsch, D.; Steinkellner, G.; Gruber, K.; Greimel, K.; Eiteljoerg, I.; Trotscha, E.; Wei, R.; Zimmermann, W.; Zinn, M.; et al. Enzymatic surface hydrolysis of PET: Effect of structural diversity on kinetic properties of cutinases from thermobifida. Macromolecules 2011, 44, 4632–4640. [Google Scholar] [CrossRef] [Green Version]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Carrez, C. Le Ferrocyanure de Potassium et l’acétate de Zinc comme Agents de Défécation des Urines. Ann. Chim. Anal. 1908, 13, 97–101. [Google Scholar]
- Culhaoglu, T.; Zheng, D.; Méchin, V.; Baumberger, S. Adaptation of the Carrez procedure for the purification of ferulic and p-coumaric acids released from lignocellulosic biomass prior to LC/MS analysis. J. Chromatogr. B 2011, 879, 3017–3021. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Wu, L.; Sun, Y.; Zhang, M. Rheological properties and crystallization behavior of multi-walled carbon nanotube/poly(ε-caprolactone) composites. J. Polym. Sci. B Polym. Phys. 2007, 45, 3137–3147. [Google Scholar] [CrossRef]
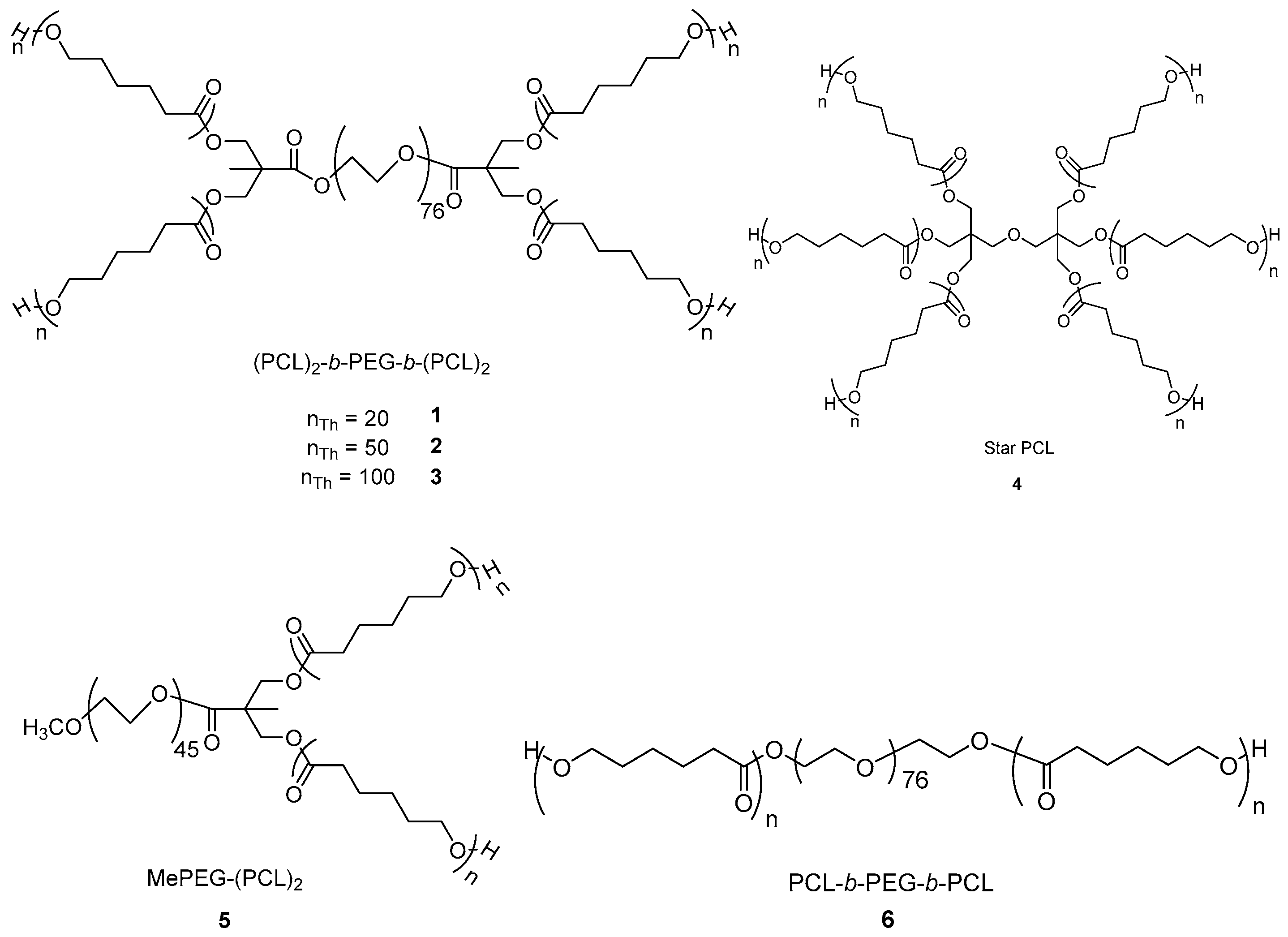
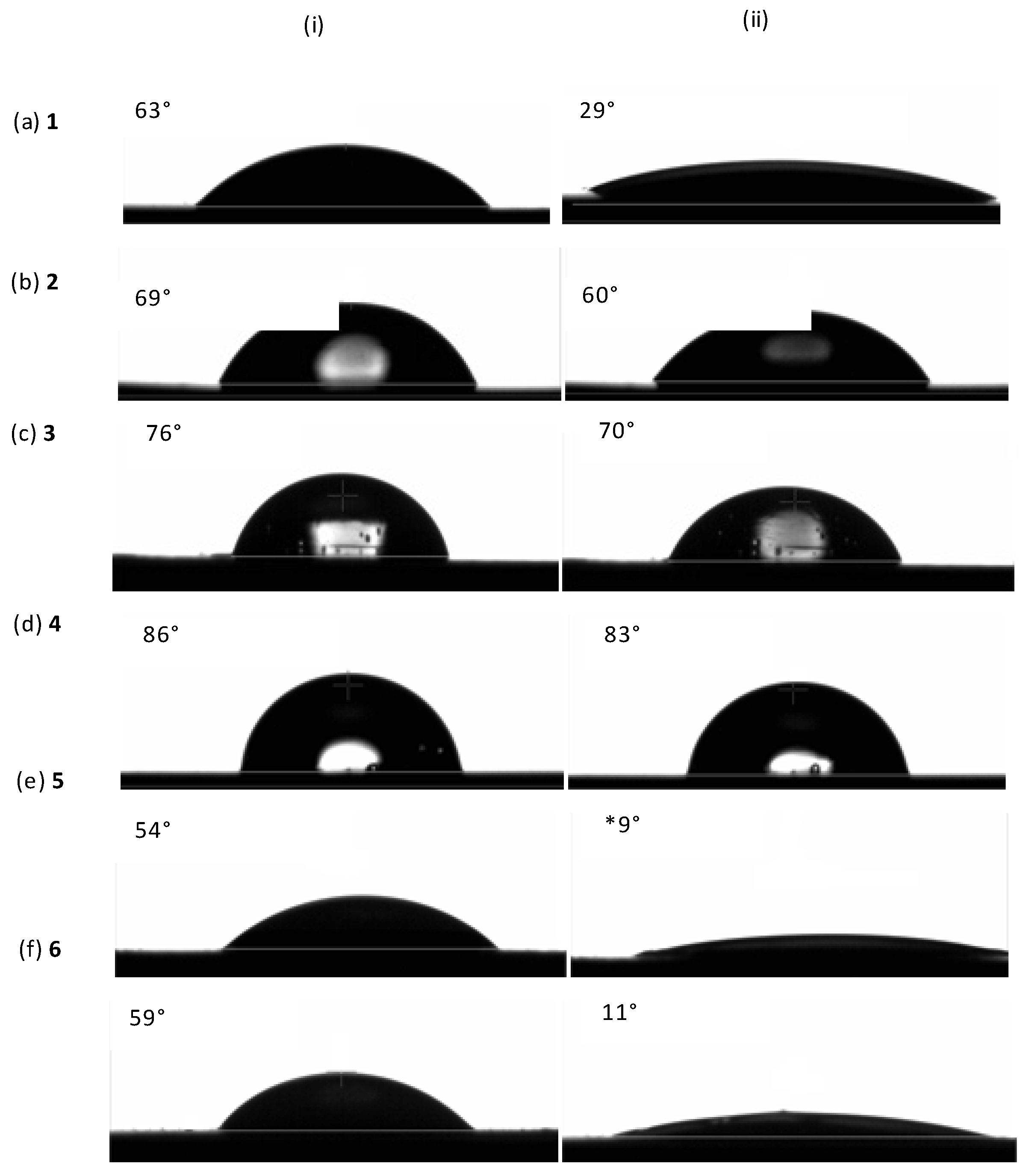


| Sample | Shape | a nTh | b nNMR | cMnTh | dMnNMR | eMnGPC | Ð | f %PEG |
|---|---|---|---|---|---|---|---|---|
| (×10−4 g mol−1) | ||||||||
| 1 | Star | 20 | 19 | 1.28 | 1.20 | 0.96 | 1.47 | 28 |
| 2 | Star | 50 | 52 | 2.62 | 2.71 | 0.96 | 1.75 | 14 |
| 3 | Star | 100 | 114 | 4.93 | 5.54 | 1.35 | 1.78 | 6 |
| 4 | Star | 100 | 67 | 6.87 | 4.61 | 2.80 | 2.20 | 0 |
| 5 | Y-shaped | 20 | 22 | 0.67 | 0.70 | 0.68 | 1.36 | 30 |
| 6 | Linear | 40 | 40 | 1.25 | 1.25 | 0.89 | 1.49 | 27 |
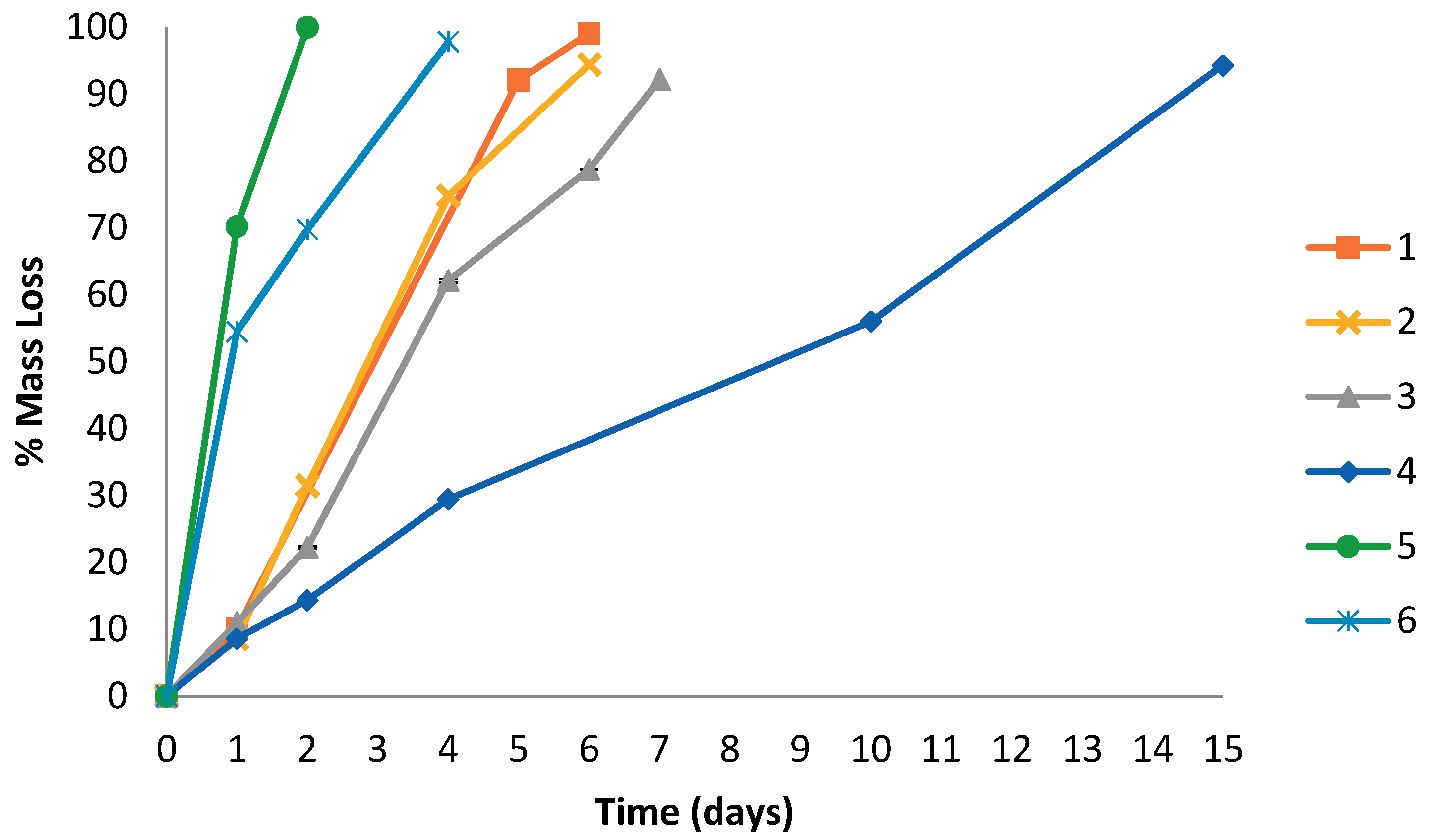
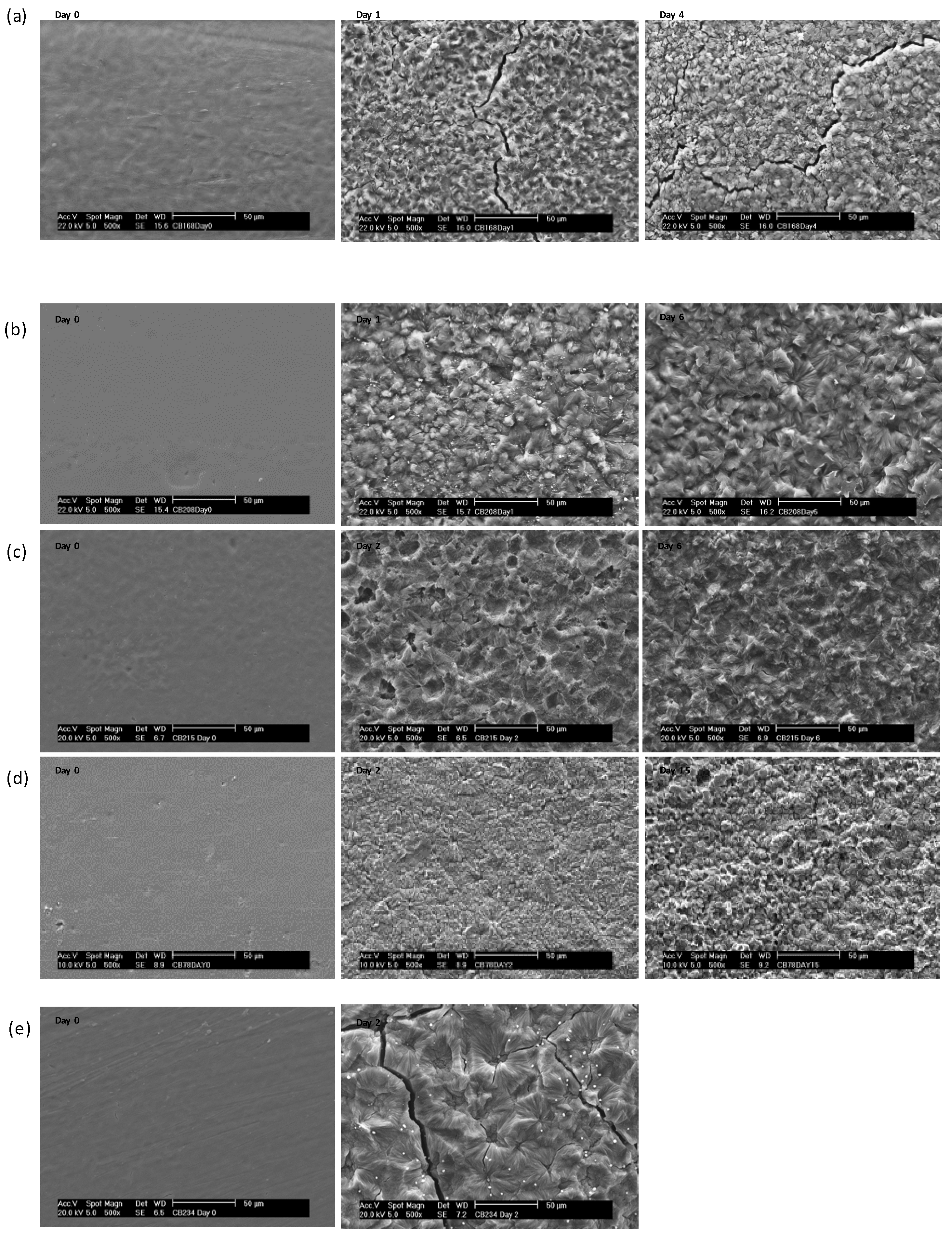
| Sample | PEG | Time | Mass loss | ΔHm | ΔHc | χc | Tm | Tc |
|---|---|---|---|---|---|---|---|---|
| % | day | % | J g−1 | % | °C | |||
| 1 | 28 | 0 | - | 26 | 18 | 12 | 54 | 28 |
| 1 | 10.0 | 64 | 44 | 29 | 55 | 36 | ||
| 4 | 47.3 | 43 | 40 | 26 | 50 | 34 | ||
| 5 | 92.1 | 41 | 41 | 26 | 51 | 34 | ||
| 6 | 99.1 | - | - | - | - | - | ||
| 2 | 14 | 0 | - | 36 | 24 | 16 | 57 | 34 |
| 1 | 8.6 | 88 | 60 | 41 | 56 | 38 | ||
| 2 | 31.5 | 58 | 60 | 41 | 52 | 37 | ||
| 4 | 74.8 | 72 | 71 | 48 | 52 | 38 | ||
| 6 | 94.4 | 51 | 50 | 34 | 52 | 38 | ||
| 3 | 6 | 0 | - | 68 | 47 | 33 | 59 | 33 |
| 1 | 11.0 | 66 | 70 | 46 | 57 | 34 | ||
| 2 | 22.3 | 61 | 61 | 43 | 59 | 34 | ||
| 4 | 62.1 | 48 | 53 | 34 | 57 | 34 | ||
| 6 | 78.8 | 71 | 73 | 50 | 55 | 36 | ||
| 4 | 0 | 0 | - | 90 | 101 | 73 | 58 | 31 |
| 1 | 8.6 | 36 | 41 | 29 | 58 | 31 | ||
| 2 | 14.3 | 42 | 48 | 34 | 57 | 31 | ||
| 4 | 29.5 | 10 | 11 | 8 | 57 | 32 | ||
| 15 | 94.3 | 4 | 5 | 3 | 56 | 31 | ||
| 5 | 29 | 0 | - | 51 | 50 | 32 | 50 | 27 |
| 1 | 70.2 | 37 | 25 | 16 | 57 | 30 | ||
| 2 | 100 | - | - | - | - | - | ||
| 6 | 27 | 0 | - | 40 | 40 | 26 | 54 | 29 |
| 2 | 69.8 | 52 | 51 | 33 | 54 | 34 | ||
| 4 | 97.8 | - | - | - | - | - | ||
| Polymer | Enzymatic degradation | Mn | Mw | Đ |
|---|---|---|---|---|
| (min) | (×104 g mol−1) | |||
| 1 | 0 | 0.57 | 0.77 | 1.4 |
| 15 | 0.62 | 0.65 | 1.4 | |
| 30 | 0.55 | 0.77 | 1.4 | |
| 60 | 0.54 | 0.77 | 1.4 | |
| 120 | 0.52 | 0.77 | 1.5 | |
| 240 | 0.51 | 0.75 | 1.5 | |
| 2 | 0 | 0.64 | 1.03 | 1.6 |
| 15 | 0.83 | 1.35 | 1.6 | |
| 30 | 0.75 | 1.12 | 1.5 | |
| 60 | 0.75 | 0.80 | 1.4 | |
| 120 | 0.93 | 1.29 | 1.3 | |
| 240 | 0.91 | 1.72 | 1.9 | |
| 3 | 0 | 0.98 | 1.61 | 1.6 |
| 15 | 0.39 | 1.24 | 3.2 | |
| 30 | 0.34 | 1.17 | 3.4 | |
| 60 | 0.13 | 0.55 | 4.4 | |
| 120 | 0.07 | 0.33 | 4.6 | |
| 240 | 0.07 | 0.22 | 2.8 | |
| 4 | 0 | 2.30 | 6.11 | 2.7 |
| 15 | 1.26 | 3.89 | 3.1 | |
| 30 | 1.29 | 3.87 | 3.0 | |
| 60 | 1.20 | 4.85 | 3.2 | |
| 120 | 0.93 | 3.80 | 2.8 | |
| 240 | 1.92 | 4.06 | 2.1 | |
| 5 | 0 | 0.17 | 0.33 | 2.0 |
| 15 | 0.23 | 0.29 | 1.3 | |
| 30 | 0.69 | 0.79 | 1.1 | |
| 60 | 0.54 | 1.00 | 1.9 | |
| 120 | 0.39 | 0.79 | 2.0 | |
| 240 | 0.35 | 0.75 | 2.1 | |
| 6 | 0 | 0.58 | 0.86 | 1.5 |
| 15 | 0.54 | 0.83 | 1.5 | |
| 30 | 0.51 | 0.81 | 1.6 | |
| 60 | 0.38 | 0.60 | 1.6 | |
| 120 | 0.21 | 0.57 | 1.9 | |
| 240 | 0.20 | 0.40 | 2.1 | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blackwell, C.J.; Haernvall, K.; Guebitz, G.M.; Groombridge, M.; Gonzales, D.; Khosravi, E. Enzymatic Degradation of Star Poly(ε-Caprolactone) with Different Central Units. Polymers 2018, 10, 1266. https://doi.org/10.3390/polym10111266
Blackwell CJ, Haernvall K, Guebitz GM, Groombridge M, Gonzales D, Khosravi E. Enzymatic Degradation of Star Poly(ε-Caprolactone) with Different Central Units. Polymers. 2018; 10(11):1266. https://doi.org/10.3390/polym10111266
Chicago/Turabian StyleBlackwell, Catherine J., Karolina Haernvall, Georg M. Guebitz, Michael Groombridge, Denis Gonzales, and Ezat Khosravi. 2018. "Enzymatic Degradation of Star Poly(ε-Caprolactone) with Different Central Units" Polymers 10, no. 11: 1266. https://doi.org/10.3390/polym10111266
APA StyleBlackwell, C. J., Haernvall, K., Guebitz, G. M., Groombridge, M., Gonzales, D., & Khosravi, E. (2018). Enzymatic Degradation of Star Poly(ε-Caprolactone) with Different Central Units. Polymers, 10(11), 1266. https://doi.org/10.3390/polym10111266




