Crystal Structural Analysis of Oryza sativa SGT1-TPR Domain
Abstract
1. Introduction
2. Materials and Methods
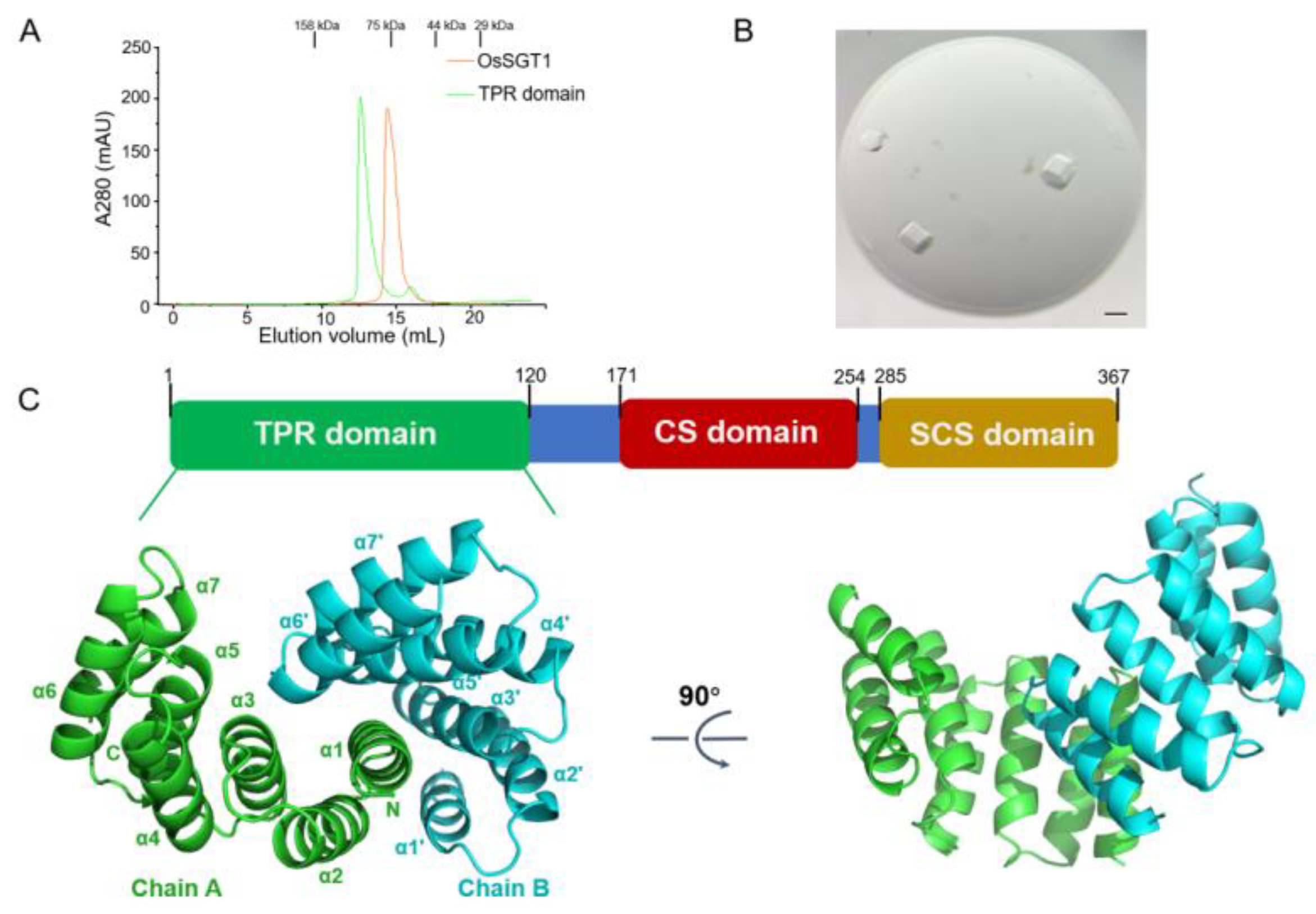
2.1. Protein Expression and Purification
2.2. Crystallization and X-Ray Diffraction Analysis
2.3. Structure Determination and Refinement
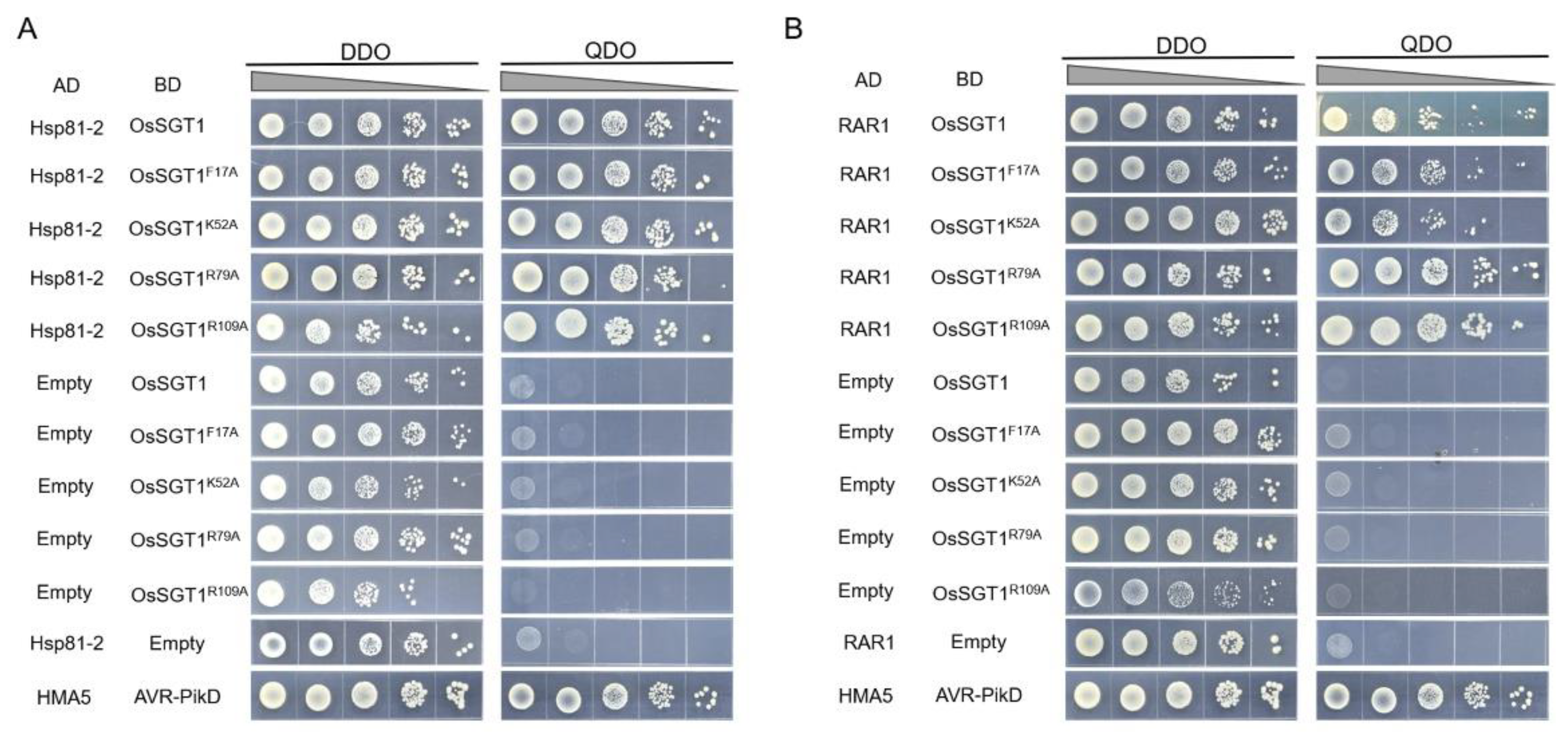
2.4. Yeast Two-Hybrid Assay
2.5. Circular Dichroism Spectroscopy
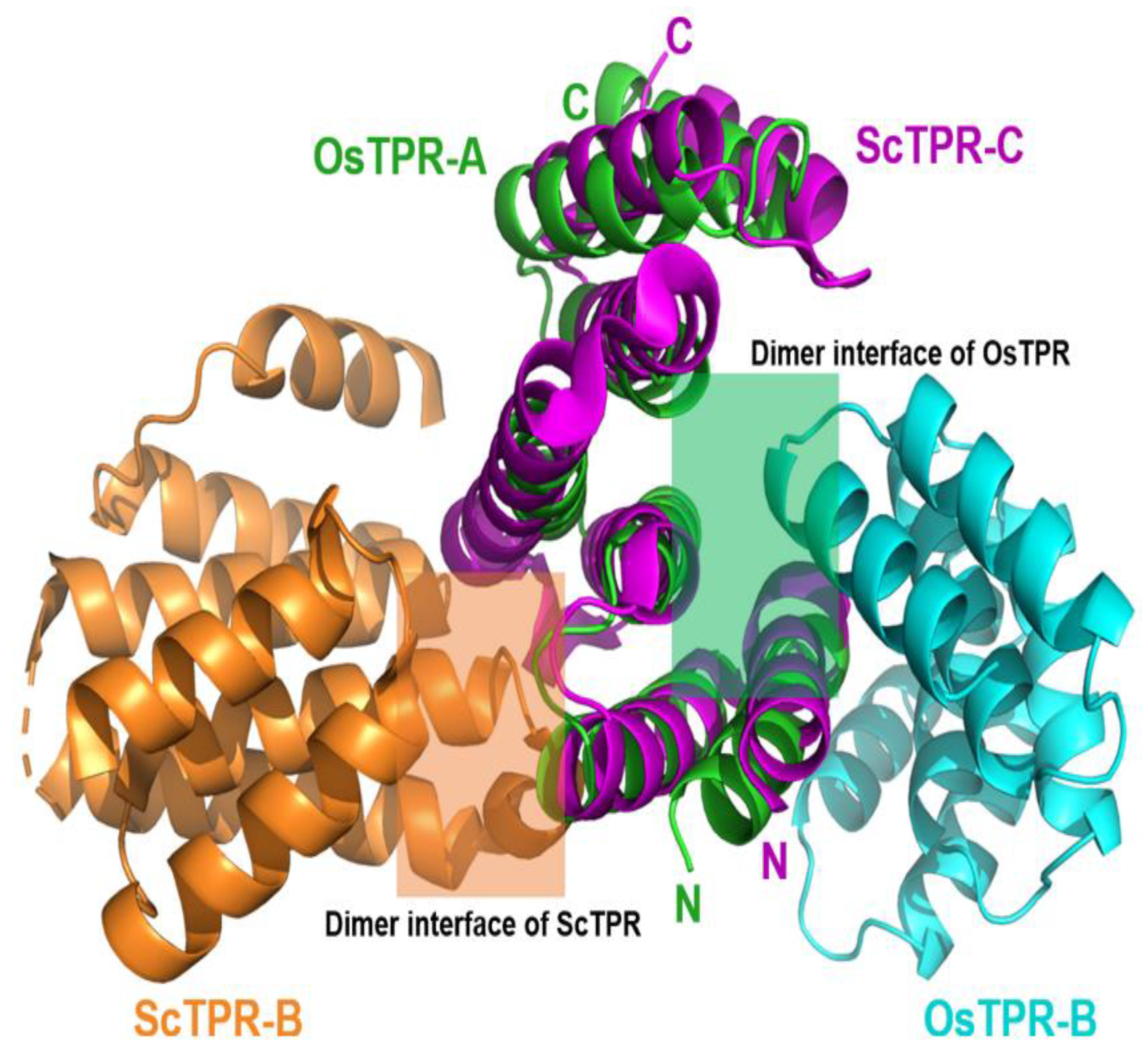
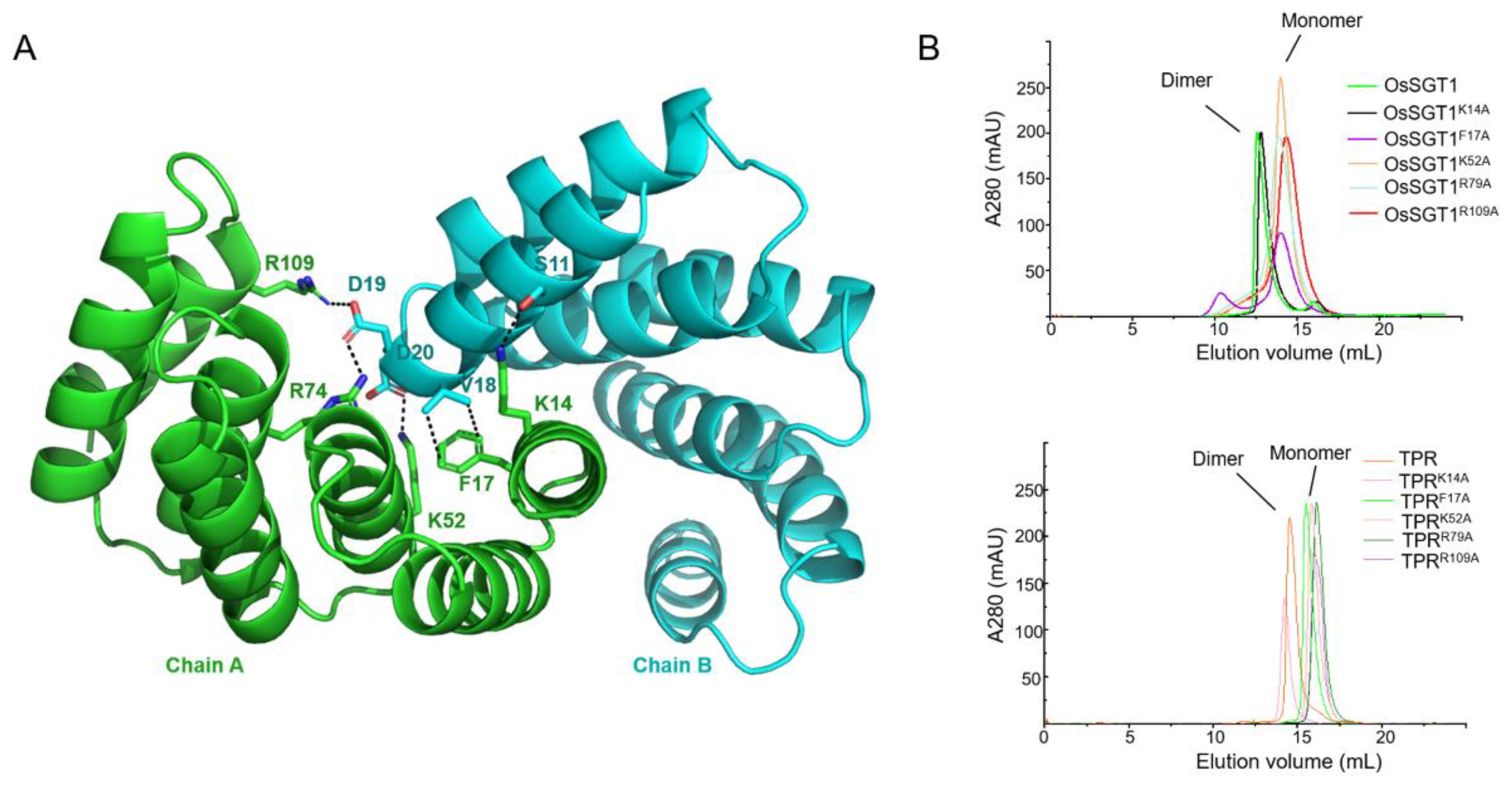
3. Results and Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jones, J.D.G.; Dangl, J.L. The Plant Immune System. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Chapman, A.V.E.; Elmore, J.M.; McReynolds, M.; Walley, J.W.; Wise, R.P. SGT1-Specific Domain Mutations Impair Interactions with the Barley MLA6 Immune Receptor in Association with Loss of NLR Protein. Mol. Plant-Microbe Interact. 2022, 35, 274–289. [Google Scholar] [CrossRef] [PubMed]
- Shirasu, K. The HSP90-SGT1 Chaperone Complex for NLR Immune Sensors. Annu. Rev. Plant Biol. 2009, 60, 139–164. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wu, Q.; Tong, C.; Chen, H.; Miao, D.; Qian, X.; Zhao, X.; Jiang, L.; Tao, X. Characterization of the Roles of SGT1/RAR1, EDS1/NDR1, NPR1, and NRC/ADR1/NRG1 in Sw-5b-Mediated Resistance to Tomato Spotted Wilt Virus. Viruses 2021, 13, 1447. [Google Scholar] [CrossRef]
- Yu, G.; Xian, L.; Xue, H.; Yu, W.; Rufian, J.S.; Sang, Y.; Morcillo, R.J.L.; Wang, Y.; Macho, A.P. A Bacterial Effector Protein Prevents MAPK-Mediated Phosphorylation of SGT1 to Suppress Plant Immunity. PLoS Pathog. 2020, 16, e1008933. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, C.; Du, Y.; Cai, K.; Wang, Y.; Guo, J.; Bai, X.; Kang, Z.; Guo, J. A Stripe Rust Fungal Effector PstSIE1 Targets TaSGT1 to Facilitate Pathogen Infection. Plant J. 2022, 112, 1413–1428. [Google Scholar] [CrossRef]
- Yu, G.; Xian, L.; Zhuang, H.; Macho, A.P. SGT1 Is Not Required for Plant LRR-RLK-Mediated Immunity. Mol. Plant Pathol. 2021, 22, 145–150. [Google Scholar] [CrossRef]
- Siligardi, G.; Zhang, M.; Prodromou, C. The Stoichiometric Interaction of the Hsp90-Sgt1-Rar1 Complex by CD and SRCD Spectroscopy. Front. Mol. Biosci. 2018, 4, 95. [Google Scholar] [CrossRef]
- Zhang, D.; Yang, X.; Wen, Z.; Li, Z.; Zhang, X.; Zhong, C.; She, J.; Zhang, Q.; Zhang, H.; Li, W.; et al. Proxitome Profiling Reveals a Conserved SGT1-NSL1 Signaling Module That Activates NLR-Mediated Immunity. Mol. Plant 2024, 17, 1369–1391. [Google Scholar] [CrossRef]
- Zhang, M.; Kadota, Y.; Prodromou, C.; Shirasu, K.; Pearl, L.H. Structural Basis for Assembly of Hsp90-Sgt1-CHORD Protein Complexes: Implications for Chaperoning of NLR Innate Immunity Receptors. Mol. Cell 2010, 39, 269–281. [Google Scholar] [CrossRef]
- Li, D.; Zhou, J.; Zheng, C.; Zheng, E.; Liang, W.; Tan, X.; Xu, R.; Yan, C.; Yang, Y.; Yi, K.; et al. OsTGAL1 Suppresses the Resistance of Rice to Bacterial Blight Disease by Regulating the Expression of Salicylic Acid Glucosyltransferase OsSGT1. Plant Cell Environ. 2022, 45, 1584–1602. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Zhang, Y.; Ding, Y. Binding Mechanism between Hsp90 and Sgt1 Explored by Homology Modeling and Molecular Dynamics Simulations in Rice. J. Mol. Model. 2012, 18, 4665–4673. [Google Scholar] [CrossRef] [PubMed]
- Umemura, K.; Satou, J.; Iwata, M.; Uozumi, N.; Koga, J.; Kawano, T.; Koshiba, T.; Anzai, H.; Mitomi, M. Contribution of Salicylic Acid Glucosyltransferase, OsSGT1, to Chemically Induced Disease Resistance in Rice Plants. Plant J. 2009, 57, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gao, M.; Li, Q.; Wang, L.; Wang, J.; Jeon, J.-S.; Qu, N.; Zhang, Y.; He, Z. OsRAR1 and OsSGT1 Physically Interact and Function in Rice Basal Disease Resistance. Mol. Plant-Microbe Interact. 2008, 21, 294–303. [Google Scholar] [CrossRef]
- Li, R.; Zheng, W.; Yang, R.; Hu, Q.; Ma, L.; Zhang, H. OsSGT1 Promotes Melatonin-Ameliorated Seed Tolerance to Chromium Stress by Affecting the OsABI5–OsAPX1 Transcriptional Module in Rice. Plant J. 2022, 112, 151–171. [Google Scholar] [CrossRef]
- Kang, B.; Zhang, Z.; Wang, L.; Zheng, L.; Mao, W.; Li, M.; Wu, Y.; Wu, P.; Mo, X. OsCYP2, a Chaperone Involved in Degradation of Auxin-Responsive Proteins, Plays Crucial Roles in Rice Lateral Root Initiation. Plant J. 2013, 74, 86–97. [Google Scholar] [CrossRef]
- Zeng, G. Sticky-End PCR: New Method for Subcloning. BioTechniques 1998, 25, 206–208. [Google Scholar] [CrossRef]
- Otwinowski, Z.; Minor, W. [20] Processing of X-Ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar]
- Xtriage and Fest: Automatic Assessment of X-Ray Data and Substructure Structure Factor Estimation. Available online: https://www.researchgate.net/publication/228661841_Xtriage_and_Fest_automatic_assessment_of_X-ray_data_and_substructure_structure_factor_estimation (accessed on 30 May 2025).
- von Horsten, S.; Essen, L.-O. Conformational Change of Tetratricopeptide Repeats Region Triggers Activation of Phytochrome-Associated Protein Phosphatase 5. Front. Plant Sci. 2021, 12, 733069. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser Crystallographic Software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallogr. Sect. D-Struct. Biol. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- Schomaker, V.; Trueblood, K.N. On the Rigid-Body Motion of Molecules in Crystals. Acta Crystallogr. Sect. B Struct. Sci. Cryst. Eng. Mater. 1968, 24, 63–76. [Google Scholar] [CrossRef]
- Cantu, D.; Yang, B.; Ruan, R.; Li, K.; Menzo, V.; Fu, D.; Chern, M.; Ronald, P.C.; Dubcovsky, J. Comparative Analysis of Protein-Protein Interactions in the Defense Response of Rice and Wheat. BMC Genom. 2013, 14, 166. [Google Scholar] [CrossRef]
- Tai, Y.-S. Interactome of Signaling Networks in Wheat: The Protein–Protein Interaction between TaRAR1 and TaSGT1. Mol. Biol. Rep. 2008, 35, 337–343. [Google Scholar] [CrossRef]
- Seo, Y.-S.; Lee, S.-K.; Song, M.-Y.; Suh, J.-P.; Hahn, T.-R.; Ronald, P.; Jeon, J.-S. The HSP90-SGT1-RAR1 Molecular Chaperone Complex: A Core Modulator in Plant Immunity. J. Plant Biol. 2008, 51, 1–10. [Google Scholar] [CrossRef]
- Shen, Q.; Schulze-Lefert, P. Rumble in the Nuclear Jungle: Compartmentalization, Trafficking, and Nuclear Action of Plant Immune Receptors. EMBO J. 2007, 26, 4293–4301. [Google Scholar] [CrossRef]
- Kitagawa, K.; Skowyra, D.; Elledge, S.J.; Harper, J.W.; Hieter, P. SGT1 Encodes an Essential Component of the Yeast Kinetochore Assembly Pathway and a Novel Subunit of the SCF Ubiquitin Ligase Complex. Mol. Cell 1999, 4, 21–33. [Google Scholar] [CrossRef]
- Botër, M.; Amigues, B.; Peart, J.; Breuer, C.; Kadota, Y.; Casais, C.; Moore, G.; Kleanthous, C.; Ochsenbein, F.; Shirasu, K.; et al. Structural and Functional Analysis of SGT1 Reveals That Its Interaction with HSP90 Is Required for the Accumulation of Rx, an R Protein Involved in Plant Immunity. Plant Cell 2007, 19, 3791–3804. [Google Scholar] [CrossRef]
- Kadota, Y.; Shirasu, K. The HSP90 Complex of Plants. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2012, 1823, 689–697. [Google Scholar] [CrossRef]
- Zheng, N.; Schulman, B.A.; Song, L.; Miller, J.J.; Jeffrey, P.D.; Wang, P.; Chu, C.; Koepp, D.M.; Elledge, S.J.; Pagano, M.; et al. Structure of the Cul1–Rbx1–Skp1–F boxSkp2 SCF Ubiquitin Ligase Complex. Nature 2002, 416, 703–709. [Google Scholar] [CrossRef]
- Willhoft, O.; Kerr, R.; Patel, D.; Zhang, W.; Al-Jassar, C.; Daviter, T.; Millson, S.H.; Thalassinos, K.; Vaughan, C.K. The Crystal Structure of the Sgt1-Skp1 Complex: The Link between Hsp90 and Both SCF E3 Ubiquitin Ligases and Kinetochores. Sci. Rep. 2017, 7, 41626. [Google Scholar] [CrossRef] [PubMed]
- Nyarko, A.; Mosbahi, K.; Rowe, A.J.; Leech, A.; Boter, M.; Shirasu, K.; Kleanthous, C. TPR-Mediated Self-Association of Plant SGT1. Biochemistry 2007, 46, 11331–11341. [Google Scholar] [CrossRef]

| Beamline | RRSF BL18U1 |
|---|---|
| Wavelength | 0.9792 |
| Resolution range | 22.46–1.53 (1.59–1.53) a |
| Space group | P 3 |
| Unit cell | 93.493 93.493 32.941 90 90 120 |
| Total reflections | 478,266 |
| Unique reflections | 48,592 (4851) |
| Multiplicity | 18.9 (17.0) |
| Completeness (%) | 99.95 (100.00) |
| Mean I/sigma(I) | 48.41 (3.40) |
| Wilson B-factor | 20.8 |
| Rmerge b | 0.075 (0.780) |
| Rmeas | 0.091 (0.748) |
| Rp.i.m | 0.021 (0.180) |
| CC1/2 | 0.998 (0.879) |
| Reflections used in refinement | 48,589 (4851) |
| Reflections used for Rfree | 2001 (198) |
| Rwork c | 0.176 (0.209) |
| Rfree | 0.190 (0.213) |
| Number of non-hydrogen atoms | 2039 |
| Macromolecules | 1777 |
| Solvent | 262 |
| No. of protein residues | 231 |
| R.m.s. deviations bond lengths (Å) (bonds) | 0.006 |
| R.m.s. deviations bond angles (°) (angles) | 0.84 |
| Ramachandran plot (%) d | |
| Ramachandran favored | 97.36 |
| Ramachandran allowed | 2.64 |
| Ramachandran outliers | 0.00 |
| Rotamer outliers | 0.00 |
| Clashscore | 3.43 |
| Average B-factor | 28.1 |
| Macromolecules | 26.75 |
| Solvent | 37.57 |
| Number of TLS groups | 8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, Y.; Ji, L.; Qin, Y.; Yi, Y.; Qian, C.; Jiang, J.; Liu, T.; Liu, J.; Zhang, X. Crystal Structural Analysis of Oryza sativa SGT1-TPR Domain. Crystals 2025, 15, 543. https://doi.org/10.3390/cryst15060543
Chang Y, Ji L, Qin Y, Yi Y, Qian C, Jiang J, Liu T, Liu J, Zhang X. Crystal Structural Analysis of Oryza sativa SGT1-TPR Domain. Crystals. 2025; 15(6):543. https://doi.org/10.3390/cryst15060543
Chicago/Turabian StyleChang, Yongqi, Lifeng Ji, Yiling Qin, Yaqi Yi, Chen Qian, Jie Jiang, Tian Liu, Junfeng Liu, and Xin Zhang. 2025. "Crystal Structural Analysis of Oryza sativa SGT1-TPR Domain" Crystals 15, no. 6: 543. https://doi.org/10.3390/cryst15060543
APA StyleChang, Y., Ji, L., Qin, Y., Yi, Y., Qian, C., Jiang, J., Liu, T., Liu, J., & Zhang, X. (2025). Crystal Structural Analysis of Oryza sativa SGT1-TPR Domain. Crystals, 15(6), 543. https://doi.org/10.3390/cryst15060543






