Abstract
Protopanaxadiol-type ginsenosides, the major bioactive components of Panax ginseng, exhibit diverse pharmacological activities, but suffer from low oral bioavailability due to poor water solubility and membrane permeability. Enzymatic deglycosylation has emerged as an effective strategy to enhance their therapeutic potential; however, most glucosidases lack sufficient thermostability for industrial applications. A β-glucosidase from the thermophilic bacterium Caldicellulosiruptor bescii (CbBGL) has demonstrated efficient conversion of major ginsenosides into compound K at elevated temperatures. In this study, the high-resolution crystal structure of CbBGL was determined at 1.9 Å. Structural analysis revealed that CbBGL adopts a classical (α/β)8 TIM barrel fold and functions as a homodimer. Comparative studies with other glucosidases highlighted structural features contributing to its thermostability, including moderate B-factor distribution and a limited hydrogen bond network. Docking analyses revealed a narrow, inverted conical substrate-binding cleft, which imposes specific binding orientations and underlies the enzyme’s stepwise deglycosylation mechanism. These insights provide a structural basis for CbBGL’s thermal resilience and substrate specificity, offering a valuable platform for the rational engineering of glucosidases in ginsenoside bioconversion processes.
1. Introduction
Ginsenosides, the glycosidic constituents of Panax ginseng saponins, have been widely recognized as the primary bioactive components responsible for the pharmacological effects of ginseng [1,2,3,4]. Among these, protopanaxadiol-type ginsenosides (PPD-type ginsenosides), including Rb1, Rb2, Rc, and Rd, account for over 80% of the total ginsenoside content [5,6,7] (Figure 1). These compounds generally possess one or two sugar moieties attached at the C3 and C20 positions of the protopanaxadiol backbone [2,6]. Numerous studies have reported that PPD-type ginsenosides exhibit a wide range of biological activities, such as cardioprotective, anticancer, antidiabetic, antioxidant, and neuroprotective effects [8,9,10,11].
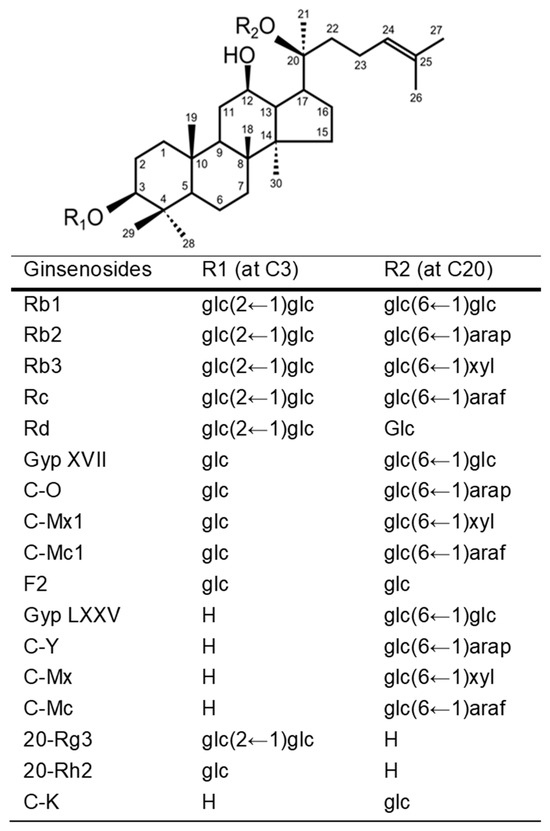
Figure 1.
Chemical structures of protopanaxadiol (PPD)-type ginsenosides. PPD-type ginsenosides contain glycosides at C3 and C20. C-, compound; Gyp, gypenoside; glc, β-D-glucopyranosyl; arap, α-L-arabinopyranosyl; xyl, β-D-xylopyranosyl; araf, α-L-arabinofuranosyl.
Despite their diverse bioactivities, ginsenosides exhibit low oral bioavailability and limited cellular uptake, as demonstrated in various pharmacokinetic studies [7,12,13]. These limitations are primarily attributed to their poor aqueous solubility and low membrane permeability, caused by the hydrophobic dammarane-type backbone and bulky glycosidic chains [11,13].
To enhance the pharmacological effectiveness of ginsenosides, multiple biotransformation strategies have been investigated. Among these, enzymatic deglycosylation has emerged as a particularly promising approach due to its high selectivity and efficiency [6,12,14,15,16,17,18,19,20,21,22]. However, the poor solubility of ginsenosides often requires elevated reaction temperatures to ensure sufficient substrate dissolution which, in turn, can result in enzyme denaturation and loss of catalytic activity [7,16,23,24]. These challenges have limited the practical application of enzymatic ginsenoside conversion in industrial settings [6,16,25].
To overcome these barriers, enzymes with inherent thermostability have been explored as potential biocatalysts for ginsenoside deglycosylation. Such thermostable enzymes retain catalytic activity and structural integrity under high-temperature conditions and also provide a suitable platform for protein engineering aimed at improving catalytic efficiency and substrate specificity. Nevertheless, glucosidases with ginsenoside-converting activity derived from thermophilic microorganisms remain scarce [16,22,23], and their structural and biochemical properties are not well characterized. This lack of information presents a significant limitation in both the mechanistic understanding and technological advancement of enzyme-based ginsenoside transformation.
In one study, a β-glucosidase isolated from the thermophilic bacterium Caldicellulosiruptor bescii DSM 6725 (CbBGL) demonstrated efficient conversion of ginsenosides in ginseng extract into compound K under optimal conditions of pH 5.5 and 75 °C [22] (Figure S3). This suggests that the enzyme is capable of cleaving sugar moieties at both the C3 and C20 positions (Figure S3). Moreover, CbBGL exhibits broad substrate specificity toward various sugars and ultimately produces compound K with most sugar units removed, underscoring its potential as a robust enzymatic platform for ginsenoside biotransformation [22].
In this study, the high-resolution (1.9 Å) crystal structure of CbBGL was determined, and structure-based analyses were conducted to identify the structural features that contribute to its substrate specificity and thermostability. These results provide fundamental insights into the molecular mechanisms underlying thermostable ginsenoside-deglycosylating enzymes and offer a basis for structure-guided enzyme engineering for industrial applications.
2. Materials and Methods
2.1. Cloning, Protein Expression, and Purification
The gene encoding β-glucosidase from Caldicellulosiruptor bescii DSM 6725 (CbBGL, GenBank No. ACM59590.1) was synthesized (IDT Inc., Coralville, IA, USA) and cloned into the pET-28a expression vector (Novagen, Madison, WI, USA) using the NdeI and XhoI restriction sites. The sequence of the cloned gene was verified by DNA sequencing (Macrogen, Seoul, Republic of Korea). The verified plasmid was transformed into Escherichia coli BL21(DE3) cells. The transformed cells were cultured in LB medium at 37 °C until the optical density at 600 nm (OD600) reached 0.5. Protein expression was then induced by adding 1 μM IPTG, followed by further incubation at 37 °C for 4 h. After induction, the cultured cells were harvested by centrifugation at 7000 rpm for 30 min at 4 °C. The cell pellet was resuspended in 15 mL of lysis buffer containing 20 mM Tris-HCl (pH 7.4) and 400 mM NaCl. The cells were lysed using sonication, and the lysate was centrifuged again at 17,000 rpm for 1 h at 4 °C to collect supernatant. To selectively denature E. coli proteins while retaining CbBGL activity, the supernatant was heat-treated at 60 °C for 30 min. Following heat treatment, the sample was centrifuged again at 17,000 rpm for 1 h at 4 °C, and the resulting supernatant was collected as the crude CbBGL fraction. Further purification was carried out using Ni-NTA affinity chromatography, exploiting the N-terminal 6×His-tag of CbBGL. The final purification step involved size exclusion chromatography, during which the buffer was exchanged to 5 mM Tris-HCl (pH 7.4) containing 50 mM NaCl. The purity of the eluted protein solution was assessed using 15% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and protein concentration was determined using the Bradford assay. The purified protein was concentrated to ~30 mg/mL for crystallization. The purified protein was concentrated to 50 mg/mL, aliquoted and stored at −70 °C until crystallization.
2.2. Crystallization
Crystallization screening was performed using the sitting drop vapor diffusion method at 20 °C using a commercial crystallization kit, including the Crystal Screen 1, 2, MembFac and Natrix (Hampton Research, Aliso Viejo, CA, USA), and Wizard Classic 1, 2, 3, and 4 (Rigaku, The Woodlands, TX, USA). The protein solution (1 µL) was mixed with the crystallization screen solution (1 µL) and equilibrated with a reservoir solution (50 µL). Four different microcrystals were obtained from the following conditions: (i) 0.1 M lithium sulfate monohydrate, 0.1 M ADA (pH 6.5), 12% (w/v) polyethylene glycol 4000 and 2% (v/v) 2-Propanol; (ii) 0.1 M of Tris-HCl (pH 8.5) and 8% (w/v) polyethylene glycol 8000; (iii) 0.2 M lithium sulfate monohydrate, 0.1 M Tris-HCl (pH 8.5) and 30% (w/v) polyethylene glycol 4000; (iv) 0.2 M sodium acetate trihydrate, 0.1 M sodium cacodylate trihydrate (pH 6.5), and 30% (w/v) polyethylene glycol 8000. Suitable crystals for X-ray diffraction were obtained from condition (i) within 4 weeks.
2.3. X-Ray Diffraction Data Collection
X-ray diffraction data were collected at beamline 5C at Pohang Light Source II (PLS-II, Pohang, Republic of Korea) [26]. The CbBGL crystal was cryoprotected using a reservoir solution supplemented with 30% (v/v) glycerol for 5 s. The crystal was mounted on a goniometer under a 100 K nitrogen gas stream. Diffraction data were recorded on a Eiger2 S 9M detector (DECTRIS, Baden, Switzerland). Diffraction data were indexed, integrated, and scaled using HKL3000 [27]. X-ray diffraction images were visualized using ADXV (https://www.scripps.edu/tainer/arvai/adxv.html, accessed on 18 August 2023).
2.4. Structure Determination
The phase problem was solved by molecular replacement using Phaser in PHENIX [28]. The crystal structure of synthetic ancestral glucosidase (PDB code: 6Z1H) [29] was used as the search model. Coot was used for manual model building based on the electron density map [30]. Structure refinement was conducted with phenix.refine in PHENIX [28].
2.5. Bioinformatics and Structure Analysis
Amino acid sequence alignment was performed using Clustal Omega [31]. Structure-based multiple sequence alignment was visualized with ESPript 3.0 [32]. The packing densities of the protein cores were obtained by ProteinVolume 1.3 [33]. Hydrophobic clusters inside of the proteins were identified and visualized by ProteinTools [34]. The lengths of the mobile loops in protein structures were identified using STRIDE web server [35]. Molecular dockings of the two kind of protopanaxadiol (PPD); Rg3 and gypenoside XVII (GypLXXV), were performed by AutoDock vina (smina) [36]. Models of the substrates were provided by SMILES strings from PubChem (https://pubchem.ncbi.nlm.nih.gov, accessed on 12 December 2024). Protein structures were visualized using PyMOL (https://pymol.org, accessed on 1 March 2025).
3. Results
3.1. Structure Determination of CbBGL
The molecular mass of CbBGL was approximately 100 kDa, as determined by size exclusion chromatography, which closely matches the theoretical dimer mass of 106 kDa, indicating that CbBGL exists as a dimer in solution (Figure 2A). Initial crystallization trials yielded crystals under the following four conditions: (i) 0.1 M lithium sulfate monohydrate, 0.1 M ADA (pH 6.5), 12% (w/v) polyethylene glycol 4000 and 2% (v/v) 2-Propanol; (ii) 0.1 M of Tris-HCl (pH 8.5) and 8% (w/v) polyethylene glycol 8000; (iii) 0.2 M lithium sulfate monohydrate, 0.1 M Tris-HCl (pH 8.5) and 30% (w/v) polyethylene glycol 4000; (iv) 0.2 M sodium acetate trihydrate, 0.1 M sodium cacodylate trihydrate (pH 6.5) and 30% (w/v) polyethylene glycol 8000 (Figure 2B). After reproduction of these crystals, X-ray diffraction experiment proceeded and the crystal obtained from condition (i) diffracted up to a resolution of ~1.9 Å (Figure 2C). So, the high-resolution data collection was proceeded with this crystal. Although the completeness of the high-resolution shell was only 35%, the data cutoff was set at 1.9 Å, as the I/σ, Rmeas, and CC1/2 values for the high resolution shell were 5.0, 28.7%, and 0.946, respectively, all within generally accepted thresholds [37] (Table 1). The CbBGL crystal belonged to the space group P212121, with unit cell dimensions of a = 74.60 Å, b = 96.87 Å, and c = 143.01 Å (Table 1). The structure of CbBGL was determined at 1.9 Å resolution, with Rwork and Rfree values of 15.37% and 18.07%, respectively (Table 1).
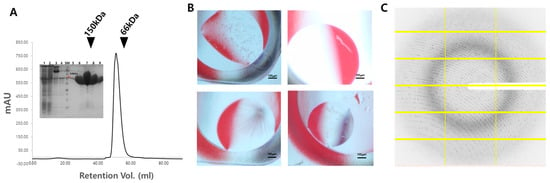
Figure 2.
Purification, crystallization, and diffraction of CbBGL. (A) Gel filtration profile of CbBGL indicating dimeric state. (Insert) Purity of CbBGL on SDS-PAGE after His-Affinity chromatography. (B) Photo of CbBGL crystals from condition (i) (top left), (ii) (top right), (iii) (bottom left), (iv) (bottom right). (C) Diffraction pattern of CbBGL from condition (i) in data collection.

Table 1.
Data collection and refinement statistics for CbBGL.
3.2. Crystal Structure of CbBGL
The crystal structure of CbBGL, determined by X-ray crystallography, revealed that the enzyme exists as a homodimer, consistent with its solution-state molecular weight as assessed by size exclusion chromatography (Figure 2A and Figure 3A). Each monomer adopts a canonical α8β8 TIM barrel fold, a structural hallmark of glycoside hydrolase family 1 (GH1) enzymes [38] (Figure 3A). For convenience, the secondary structure elements forming the core α8β8 TIM barrel are labeled numerically, while the additional secondary structures are denoted alphabetically (Figure 3B).
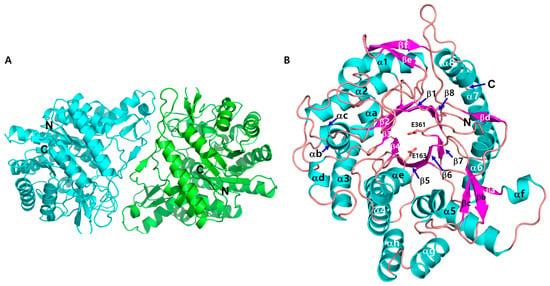
Figure 3.
Crystal structure of CbBGL. (A) Homo-dimeric form of CbBGL found in the asymmetric unit of crystal. (B) Overall structure of monomeric CbBGL. Annotation of each secondary structures were assigned on the structure. Catalytic residues are presented in stick model (E163 and E361). N and C terminal were assigned on the structure.
A comparison of the two monomers in the structure revealed negligible differences, with a root-mean-square deviation (RMSD) of 0.5 Å for the Cα atoms. The two monomers form a dimer by facing each other, with their respective axes parallel to the core β-barrel tilted at an angle of 112 degrees to each other (Figure 3A and Figure S1). The dimer interface is primarily formed along the region extending from βc through βd to α7. Notably, βd forms an antiparallel β-sheet with the βd of the opposing monomer via backbone-to-backbone hydrogen bonds. As a result, along the outer rim of the TIM barrel, one half is closed, forming the dimer interface, while the other half remains open and exposed to the solvent (Figure S1).
Structural homology searches using the resolved crystal structures, in conjunction with sequence alignment and structural superimposition with five other GH1 glucosidases, identified two highly conserved glutamate residues, Glu163 and Glu361, as putative catalytic residues (Figure 3B and Figure 4). Mapping these catalytic residues (Glu 163, 361) within the three-dimensional structure of CbBGL enabled the identification of the putative active site (Figure 3B). The catalytic acid/base Glu163 is located at the end of β4 and the catalytic nucleophile Glu361 is located at the end of β7 (Figure 3B).
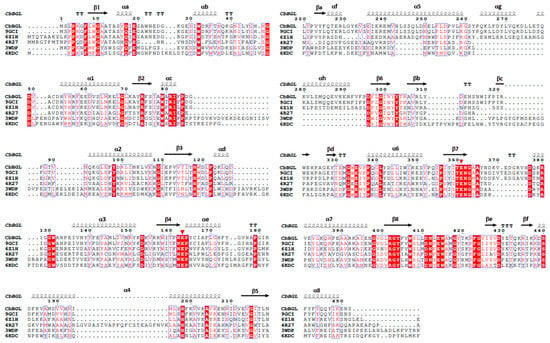
Figure 4.
Multiple sequence alignment of β-glycosidases. Each of the glucosidase denoted with their PDB ID except CbBGL. 9GCI: CsBGL, 6Z1H: anGL, 4R27: BGL167, 3WDP: PfBGL, 6KDC: FpBGL. Residues of the catalysis (active center) are represented in red triangles. Representations of the secondary structure stand for the CbBGL only. The α1 to α8 helices and β1 to β8 strands of a conventional TIM (β/α)8 barrel are represented.
3.3. Comparative Structural Investigation of CbBGL Thermostability
To investigate the structural determinants of CbBGL thermostability, comparative analyses were conducted using structurally related β-glucosidases from glycoside hydrolase family 1 (GH1) with varying thermal stabilities. As mesothermophilic references, an ancestral glucosidase (anGL, PDB ID: 6Z1H) and a β-glucosidase from Caldicellulosiruptor saccharolyticus (CsBGL, PDB ID: 9GCI) were selected, sharing 53.5% and 93.4% sequence identity with CbBGL, respectively. Both have an optimum activity temperature of 65 °C [29,39].
Two crystal structures have been reported for anGL: with and without bound heme [29]. As heme binding has not been observed in other GH1 structures, the heme-bound form was excluded from this study. For hyperthermophilic references, β-glucosidases from Fervidobacterium pennivorans (FpBGL, PDB ID: 6KDC) and Pyrococcus furiosus (PfBGL, PDB ID: 3WDP) were chosen. Despite sharing only 27.3% and 26.3% of sequence identity with CbBGL, respectively, both enzymes can deglycosylate ginsenosides, and FpBGL, like CbBGL, produces Compound K [40,41]. Their reported melting temperatures (Tm) are 98.8 °C for FpBGL and 109.5 °C for PfBGL [41,42]. The optimum temperature of CbBGL is 80 °C [22], placing it between the mesothermophilic and hyperthermophilic references in terms of thermostability.
To identify structural features associated with thermostability, the following parameters were evaluated for each enzyme: packing density of the protein core, internal hydrophobic clusters, number of mobile loops, intramolecular salt bridges, and hydrogen bond networks (Table 2). Among these, the number of salt bridges and hydrogen bond networks showed a clear positive correlation with thermostability, despite a slight decrease observed in PfBGL, the most thermostable enzyme. (Table 2).

Table 2.
Structural factors that contribute to the thermostability of β-glucosidases.
The length of mobile loops is generally inversely correlated with protein thermostability [43]. Among the five structures analyzed, only CbBGL and PfBGL contained relatively long loops (six and seven amino acids, respectively), while the remaining loops were all five residues or fewer. However, the number of mobile loops did not correlate as clearly with thermostability, likely due to an unresolved region in the anGL structure. This missing segment, spanning from the middle of helix αh to the end of strand βa in CbBGL (Figure 3B), is believed to be stabilized by heme binding [29]. Therefore, anGL may possess more and longer mobile loops than observed in the structure.
In contrast, no clear trend was observed in the packing density of the protein core or in the extent of hydrophobic clusters, although both features may still contribute to structural stability [44]. The packing density remained consistently high (~0.73) across all enzymes, exceeding the reported average value of ~0.55 [45]. While the overall extent of hydrophobic clusters did not correlate with thermostability, major clusters were consistently located between the inner β-barrel and surrounding α-helices of the TIM barrel, suggesting a stabilizing role in the protein core (Figure S2).
To further assess local flexibility and infer thermal stability, atomic displacement parameter (ADP), commonly referred to as the B-factor in crystallography, were mapped onto the crystal structures of CbBGL, anGL, CsBGL, FpBGL, and PfBGL (Figure 5). Atomic displacement parameters or B-factors, which indicate atomic displacement, are commonly used to assess structural rigidity, with lower values suggesting reduced thermal motion and greater stability [46]. All five enzymes showed low ADPs in the central α8β8 TIM barrel, indicating that this conserved core contributes to intrinsic stability (Figure 5). In contrast, peripheral helices and loop regions exhibited higher ADPs, reflecting increased flexibility and reduced thermal resistance (Figure 5).
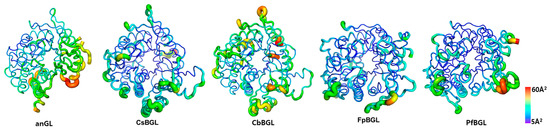
Figure 5.
ADP representations of the overall structures of anGL, CsBGL, CbBGL, FpBGL, and PfBGL.ADPs (B-factors) of Cα derived from the diffraction data of each crystal structure are visualized using both color and tube radius. The ADP values increase from blue with a narrow radius to red with a wide radius. Scale bars indicate the value of ADPs.
For a more detailed comparison, the structures with mapped ADPs were superimposed. Three regions in CbBGL were found to have relatively high ADP values, unlike the corresponding regions in the other enzymes. These include the C-terminal segments of helices α2 and α3, the C-terminal segments of helices α6 and α7, and the region spanning helices αg and αh.
In particular, the C-terminal regions of helices α2 and α3 showed significantly elevated ADP values in CbBGL, whereas the corresponding regions in the other enzymes displayed minimal variation (Figure 6A). These segments are directly connected to β2 and β3 of the central TIM barrel, suggesting that increased flexibility here may compromise overall structural stability.
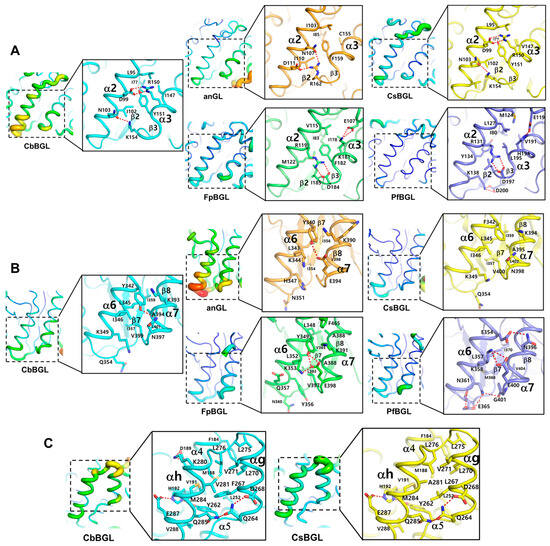
Figure 6.
ADP representations and close-up views of the corresponding regions in anGL, CsBGL, CbBGL, FpBGL, and PfBGL. (A) C-terminal regions of α2 and α3. (B) C-terminal regions of α6 and α7. (C) Region spanning αg and αh. Residues involved in interactions are shown as stick models. Hydrogen bonds and salt bridges are indicated by red dashed lines. ADP values are color-coded and radius-scaled, increasing from blue with a narrow radius to red with a wide radius.
At the molecular level, no significant differences were observed in the hydrophobic clusters among the five enzymes (Figure 6A). However, clear differences emerged in polar interactions. CbBGL forms a salt bridge-associated bifurcated hydrogen bond (Asp99–Arg150) and an additional hydrogen bond between Asn103 and Lys154. CsBGL shows the same interactions. In contrast, anGL features a distinct hydrogen bond network centered on Arg162, involving Asn107 and Asp111.
The two hyperthermophilic enzymes, FpBGL and PfBGL, exhibit more extensive stabilizing interactions. FpBGL forms two salt bridge-mediated bifurcated hydrogen bonds (Arg119–Asp184 and Lys181–Glu107), while PfBGL shows a similar pattern with two bifurcated salt bridges (Arg131–Asp197 and Lys138–Asp200) (Figure 6A). Additionally, PfBGL forms an extra hydrogen bond between His194 and Glu119, which may further enhance its structural stability.
In the C-terminal regions of helices α6 and α7, ADP values decreased with increasing enzyme thermostability (Figure 6B). These helices are connected to strand β7, which contains the catalytic residues and helps form the sugar-binding pocket. Instability in this region may affect both structural integrity and enzymatic function at high temperatures.
Further analysis revealed that anGL, CsBGL, CbBGL, and FpBGL share a similar hydrophobic cluster architecture (Figure 6B). However, notable differences were found at the residue corresponding to Lys349 in CbBGL. Lys349 contributes to a hydrophobic cluster through its aliphatic side chain atoms (Cβ–Cδ), which are buried and engage in van der Waals contacts with nearby nonpolar residues, rather than through its charged amino group. This residue is substituted with His347 in anGL and Tyr356 in FpBGL. Given the differences in size and hydrophobicity, the His substitution in anGL may weaken hydrophobic interactions, reducing stability, while the Tyr substitution in FpBGL may enhance hydrophobic packing, contributing to increased thermostability.
Regarding polar interactions in this region, CbBGL forms a single hydrogen bond between Tyr342 and Asn397, and anGL shows a comparable bond between Tyr340 and Glu394. In CsBGL, Tyr342 is replaced by Phe, eliminating the hydrogen bond due to the absence of a hydroxyl group. In contrast, FpBGL forms a more complex hydrogen bond network involving Glu398, Lys353, and Tyr349 (Figure 6B).
PfBGL, distinct from the others, exhibits multiple polar interactions in this region. Tyr342 in CbBGL, engaged in both hydrogen bonding and hydrophobic interactions, is replaced by Glu354 in PfBGL, which forms a hydrogen bond with Asn396. Additionally, PfBGL forms a salt bridge-associated bifurcated hydrogen bond between Lys358 and Glu400, and an extended hydrogen bond network involving Asn361, Glu365, Gly401, and Asn396. The residue corresponding to Lys349 in CbBGL is substituted with Asn361 in PfBGL, further enhancing the polar interaction network (Figure 6B).
Previous studies have shown that salt bridges and hydrogen bond networks enhance protein stability not simply through the sum of individual interactions, but through synergistic effects that provide greater stabilization than the additive energy of each bond alone [47,48]. These observations support the notion that PfBGL’s superior thermal resistance arises not only from the number of stabilizing interactions, but also from their cooperative nature.
The region spanning helices αg and αh is found only in CbBGL and CsBGL. In anGL, this region is unresolved in the crystal structure and is reported to be stabilized by heme binding [29]. In FpBGL and PfBGL, it is absent due to a direct connection between helix α5 and strand β6. To further explore this unique feature, a comparison was made between CbBGL and CsBGL (Figure 6C). In both enzymes, helices αg and αh display relatively high ADP values, indicating increased flexibility. However, interaction analysis suggests greater stability in CbBGL, which forms an additional salt bridge-associated bifurcated hydrogen bond (Lys280–Asp189), and exhibits a more extensive hydrophobic interface (Phe267–Val281 in CbBGL vs. Leu267–Ala281 in CsBGL) (Figure 6C).
3.4. Structural Characterization of the Substrate Binding Cleft of CbBGL
To gain insight into the substrate-binding mode of CbBGL, a comparative analysis of its substrate-binding cleft was performed with BGL167 and FpBGL, two GH1 family β-glucosidases known to act on ginsenosides [6,41] (Figure 7). Surface representations revealed a single solvent-accessible cavity in CbBGL (Figure 7B). When oriented with the cavity opening facing upward, a well-defined cleft was visible, containing the catalytic residues and the sugar-binding pocket at its base. This region is inferred to serve as the substrate-binding cleft, facilitating substrate recognition and catalysis (Figure 7B).
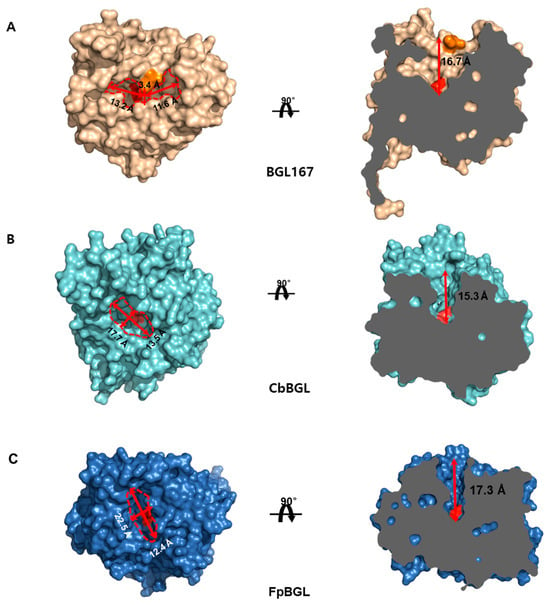
Figure 7.
Comparison of substrate binding clefts among ginsenoside glucosidases. Surface representations of BGL167 (A), CbBGL (B), and FpBGL (C). Catalytic residues were colored in red on the surfaces. Central protrusion region in the entrance of substrate binding cleft of BGL167 is colored orange.
Notably, the inner surface of this cleft is lined with hydrophobic residues, including Trp and Met, which are likely involved in stabilizing interactions with the protopanaxadiol (PPD) backbone of the ginsenoside substrate. These hydrophobic contacts are presumed to aid in substrate positioning and binding specificity.
Structurally, the substrate-binding cleft entrance differs among the three enzymes. In BGL167, the entrance is a narrow, elongated oval divided by a central protrusion (Figure 7A) into two segments measuring 13.2 Å and 11.6 Å, respectively (Figure 7A). In contrast, CbBGL features a more circular opening, measuring 17.7 Å in length and 13.5 Å in width (Figure 7B). FpBGL shows a cleft similar to that of CbBGL, though slightly narrower and more elongated, with dimensions of 22.5 Å by 12.5 Å (Figure 7C). The depth from the entrance to the base of the active site is comparable across the three enzymes, 16.7 Å in BGL167, 17.3 Å in FpBGL, and 15.3 Å in CbBGL. The interior shape of the cleft also varies significantly. BGL167 displays a broad, inverted cloverleaf-shaped cavity (Figure 7A), while CbBGL and FpBGL have clefts that narrow toward the base, forming an inverted conical shape (Figure 7B,C).
To further investigate substrate binding during catalysis, molecular dockings were performed using two ginsenoside substrates, Rg3 and GypLXXV, each bearing two sugar moieties attached to either the C3 (Rg3) or C20 (GypLXXV) position of the PPD backbone (Figure 8). Both substrate models were found to fit into the substrate-binding cleft of CbBGL in a nearly vertical orientation relative to the longitudinal axis, likely due to the narrowing geometry of the substrate-binding cleft. In contrast, the inverted cloverleaf-shaped substrate-binding cleft of BGL167 has been reported to allow for more variable binding orientations and conformations of ginsenosides [6].
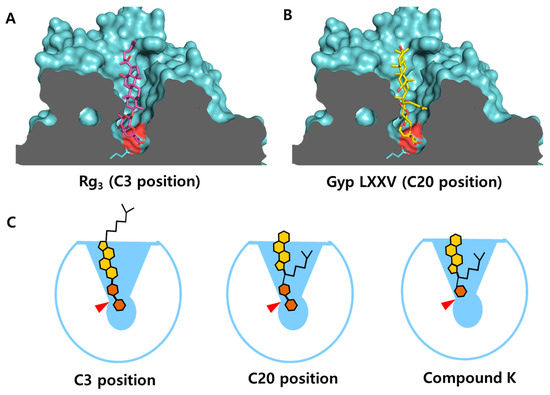
Figure 8.
Docking of the substrate model (ginsenoside) according to the positions of glycosylation. Representative docking result of Rg3 (A) and GypLXXV (B) of substrate models to the substrate-binding cleft of CbBGL. Catalytic residues were colored in red on the surfaces. (C) Schematic diagram of substrate binding to the substrate-binding cleft of CbBGL. Catalytic residues are denoted in red triangle.
Notably, when the sugar moiety at the C20 position was docked into the substrate-binding cleft (Figure 8B), the PPD backbone exhibited fewer interactions with the enzyme compared to docking at the C3 position (Figure 8A). This difference appears to result from steric hindrance between the acyl group at the C20 position of the PPD backbone and the narrow, lower region of CbBGL’s inverted conical substrate-binding cleft (Figure 8C). Such positional variation in enzyme–substrate interactions is expected to influence catalytic efficiency.
This structural observation is consistent with previous biochemical findings, in which CbBGL was shown to hydrolyze the sugar moiety at the C20 position of major ginsenosides first to generate Rd, followed by hydrolysis of the C3-linked sugar to produce F2 (Figure S3) [22]. The preferential removal of the C20 sugar can thus be explained by the relatively less stable interaction between the enzyme and substrate in this orientation, facilitating faster release of the intermediate product. Interestingly, a similar deglycosylation pattern has been reported for FpBGL, which shares a comparable substrate-binding cleft structure (Figure 7B,C) [41]. In that case as well, Rd is generated as an intermediate, further supporting this observation.
Furthermore, the steric hindrance between the acyl group at the C20 position and the lower cavity of the active site appears to influence other aspects of CbBGL’s catalytic behavior. In the reported ginsenoside bioconversion pathway of CbBGL and FpBGL, the enzymes were unable to remove the final sugar moiety at the C20 position, resulting in the accumulation of compound K as the final product [22,41]. These are likely due to the inability of the terminal sugar to access the sugar-binding pocket as a result of spatial interference from the acyl group.
4. Discussion
This study presents the high-resolution crystal structure (1.9 Å) of CbBGL, a thermostable β-glucosidase from Caldicellulosiruptor bescii, and provides an in-depth structural analysis to elucidate its thermostability and substrate recognition mechanisms. CbBGL adopts a canonical (α/β)8 TIM barrel architecture typical of GH1 enzymes and forms a homodimer in both the crystalline and solution states.
Comparative analyses with mesophilic (anGL and CsBGL) and hyperthermophilic (FpBGL and PfBGL) GH1 enzymes revealed several structural features associated with thermostability. While the central TIM barrel core exhibited uniformly low ADP values across all enzymes, peripheral regions, specifically the C-terminal ends of helices α2/α3 and α6/α7, were more flexible in CbBGL, suggesting these as potential targets for thermal optimization. Stabilizing interactions such as salt bridges and hydrogen bond networks were more abundant in hyperthermophilic enzymes. PfBGL, in particular, displayed an extensive network of salt bridges and bifurcated hydrogen bonds that likely contribute synergistically to its exceptional thermal resilience. In contrast, CbBGL harbored fewer polar interactions, but still maintained moderate thermostability, possibly due to conserved core packing and hydrophobic clustering. Interestingly, ADP analysis also highlighted the αg/αh region, unique to CbBGL and CsBGL, as flexible. However, this region in CbBGL is stabilized by an additional bifurcated salt bridge (Lys280–Asp189) and a more extensive hydrophobic interface than in CsBGL, potentially compensating for its flexibility.
Structural comparison of substrate-binding clefts between CbBGL, BGL167, and FpBGL revealed key differences in geometry. The inverted conical shape of CbBGL’s substrate-binding cleft, lined with hydrophobic residues, imposes a vertically oriented substrate binding mode. This contrasts with the cloverleaf-shaped cleft of BGL167, which allows for greater conformational flexibility.
Docking simulations further demonstrated that steric hindrance near the C20 position of the PPD backbone in CbBGL limits stable interaction, explaining the enzyme’s preference for initial cleavage at C20-linked sugars. This structural preference aligns with biochemical data showing sequential hydrolysis of C20 then C3 sugars to yield Rd and F2, respectively [22]. A similar stepwise mechanism is also reported for FpBGL [41], which shares a comparable cleft geometry, thereby reinforcing the proposed substrate-binding model. Furthermore, the inability of CbBGL to completely remove terminal sugars at C20, leading to Compound K accumulation, can be attributed to the spatial restriction caused by the acyl group at the C20 site, which impairs access to the sugar-binding pocket.
Together, these findings establish CbBGL as a thermostable and catalytically versatile β-glucosidase suitable for industrial ginsenoside biotransformation. Its dual activity at C3 and C20 positions and tolerance to elevated temperatures are advantageous for large-scale processes. Nevertheless, insights from comparison with PfBGL suggest potential avenues for protein engineering to further enhance its thermal resilience and broaden substrate scope.
These structural insights contribute to a growing body of knowledge on thermostable glycosidases and provide a rational framework for future efforts in enzyme design for pharmaceutical and nutraceutical applications involving ginsenoside modification.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cryst15050447/s1, Figure S1: Dimer Dimer structure of structure of CbBGL CbBGL; Figure S2: Hydrophobic clusters inside of the proteins; Figure S3: Biotransformation pathway for the conversion of PPD Biotransformation pathway for the conversion of PPD-type ginsenosides by CbBGL.
Funding
This research received no external funding.
Data Availability Statement
Coordinates and structure factor amplitudes for the structure have been deposited in the PDB under the accession code 9UCT (https://www.rcsb.org/structure/(9UCT)).
Acknowledgments
I would like to thank the beamline staff at the 5C beamline at the Pohang.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Jia, L.; Zhao, Y. Current evaluation of the millennium phytomedicine–ginseng (I): Etymology, pharmacognosy, phytochemistry, market and regulations. Curr. Med. Chem. 2009, 16, 2475–2484. [Google Scholar] [CrossRef] [PubMed]
- Lü, J.M.; Yao, Q.; Chen, C. Ginseng compounds: An update on their molecular mechanisms and medical applications. Curr. Vasc. Pharmacol. 2009, 7, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 2012, 75, 311–335. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.R.; Leung, C.Y.; Ho, H.M.; Chai, S.; Yau, L.F.; Zhao, Z.Z.; Jiang, Z.H. Quantitative comparison of ginsenosides and polyacetylenes in wild and cultivated American ginseng. Chem. Biodivers. 2010, 7, 975–983. [Google Scholar] [CrossRef]
- Chen, W.; Balan, P.; Popovich, D.G. Analysis of Ginsenoside Content (Panax ginseng) from Different Regions. Molecules 2019, 24, 3491. [Google Scholar] [CrossRef]
- Park, S.J.; Choi, J.M.; Kyeong, H.H.; Kim, S.G.; Kim, H.S. Rational design of a β-glycosidase with high regiospecificity for triterpenoid tailoring. ChemBioChem 2015, 16, 854–860. [Google Scholar] [CrossRef]
- Zhu, H.; Zhang, R.; Huang, Z.; Zhou, J. Progress in the Conversion of Ginsenoside Rb1 into Minor Ginsenosides Using β-Glucosidases. Foods 2023, 12, 397. [Google Scholar] [CrossRef]
- Chen, R.J.; Chung, T.Y.; Li, F.Y.; Lin, N.H.; Tzen, J.T. Effect of sugar positions in ginsenosides and their inhibitory potency on Na+/K+-ATPase activity. Acta Pharmacol. Sin. 2009, 30, 61–69. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, X.; Qin, J.J.; Voruganti, S.; Nag, S.A.; Wang, M.H.; Wang, H.; Zhang, R. Natural product ginsenoside 25-OCH3-PPD inhibits breast cancer growth and metastasis through down-regulating MDM2. PLoS ONE 2012, 7, e41586. [Google Scholar] [CrossRef]
- Wu, Y.L.; Wan, Y.; Jin, X.J.; OuYang, B.Q.; Bai, T.; Zhao, Y.Q.; Nan, J.X. 25-OCH3-PPD induces the apoptosis of activated t-HSC/Cl-6 cells via c-FLIP-mediated NF-kappaB activation. Chem. Biol. Interact. 2011, 194, 106–112. [Google Scholar] [CrossRef]
- Xu, Q.F.; Fang, X.L.; Chen, D.F. Pharmacokinetics and bioavailability of ginsenoside Rb1 and Rg1 from Panax notoginseng in rats. J. Ethnopharmacol. 2003, 84, 187–192. [Google Scholar] [CrossRef]
- Cheng, L.Q.; Kim, M.K.; Lee, J.W.; Lee, Y.J.; Yang, D.C. Conversion of major ginsenoside Rb1 to ginsenoside F2 by Caulobacter leidyia. Biotechnol. Lett. 2006, 28, 1121–1127. [Google Scholar] [CrossRef]
- Weymouth-Wilson, A.C. The role of carbohydrates in biologically active natural products. Nat. Prod. Rep. 1997, 14, 99–110. [Google Scholar] [CrossRef]
- Chang, Y.H.; Ng, P.K. Effects of extrusion process variables on extractable ginsenosides in wheat-ginseng extrudates. J. Agric. Food Chem. 2009, 57, 2356–2362. [Google Scholar] [CrossRef] [PubMed]
- Han, B.H.; Park, M.H.; Han, Y.N.; Woo, L.K.; Sankawa, U.; Yahara, S.; Tanaka, O. Degradation of ginseng saponins under mild acidic conditions. Planta Med. 1982, 44, 146–149. [Google Scholar] [CrossRef] [PubMed]
- Li, W.N.; Fan, D.D. Biocatalytic strategies for the production of ginsenosides using glycosidase: Current state and perspectives. Appl. Microbiol. Biotechnol. 2020, 104, 3807–3823. [Google Scholar] [CrossRef]
- Li, W.; Jiang, Y.; Liu, Y.; Li, C.; Fan, D. Biocatalytic strategies in producing ginsenoside by glycosidase-a review. Sheng Wu Gong Cheng Xue Bao 2019, 35, 1590–1606. [Google Scholar] [PubMed]
- Park, C.S.; Yoo, M.H.; Noh, K.H.; Oh, D.K. Biotransformation of ginsenosides by hydrolyzing the sugar moieties of ginsenosides using microbial glycosidases. Appl. Microbiol. Biotechnol. 2010, 87, 9–19. [Google Scholar] [CrossRef]
- Cui, C.H.; Kim, J.K.; Kim, S.C.; Im, W.T. Characterization of a Ginsenoside-Transforming beta-glucosidase from Paenibacillus mucilaginosus and Its Application for Enhanced Production of Minor Ginsenoside F2. PLoS ONE 2014, 9, e85727. [Google Scholar]
- Kim, J.K.; Cui, C.H.; Liu, Q.; Yoon, M.H.; Kim, S.C.; Im, W.T. Mass production of the ginsenoside Rg3(S) through the combinative use of two glycoside hydrolases. Food Chem. 2013, 141, 1369–1377. [Google Scholar] [CrossRef]
- Kim, M.-J.; Upadhyaya, J.; Yoon, M.-S.; Ryu, N.S.; Song, Y.E.; Park, H.-W.; Kim, Y.-H.; Kim, M.-K. Highly regioselective biotransformation of ginsenoside Rb2 into compound Y and compound K by β-glycosidase purified from Armillaria mellea mycelia. J. Ginseng Res. 2018, 42, 504–511. [Google Scholar] [CrossRef]
- Shin, K.C.; Kim, T.H.; Choi, J.H.; Oh, D.K. Complete Biotransformation of Protopanaxadiol-Type Ginsenosides to 20-O-β-Glucopyranosyl-20(S)-protopanaxadiol Using a Novel and Thermostable β-Glucosidase. J. Agric. Food Chem. 2018, 66, 2822–2829. [Google Scholar] [CrossRef]
- Noh, K.-H.; Son, J.-W.; Kim, H.-J.; Oh, D.-K. Ginsenoside Compound K Production from Ginseng Root Extract by a Thermostable β-Glycosidase from Sulfolobus solfataricus. Biosci. Biotechnol. Biochem. 2009, 73, 316–321. [Google Scholar] [CrossRef]
- Yang, L.; Zou, H.; Gao, Y.; Luo, J.; Xie, X.; Meng, W.; Zhou, H.; Tan, Z. Insights into gastrointestinal microbiota-generated ginsenoside metabolites and their bioactivities. Drug Metab. Rev. 2020, 52, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Sun, L.W.; Zhao, D.Q. Current Status and Problem-Solving Strategies for Ginseng Industry. Chin. J. Integr. Med. 2019, 25, 883–886. [Google Scholar] [CrossRef]
- Park, S.-Y.; Ha, S.-C.; Kim, Y.-G. The protein crystallography beamlines at the pohang light source II. Biodesign 2017, 5, 30–34. [Google Scholar]
- Minor, W.; Cymborowski, M.; Otwinowski, Z.; Chruszcz, M. HKL-3000: The integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr. Sect. D Biol. Crystallogr. 2006, 62 Pt 8, 859–866. [Google Scholar] [CrossRef] [PubMed]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkóczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.-W.; Jain, S.; McCoy, A.J. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Biol. Crystallogr. 2019, 75, 861–877. [Google Scholar] [CrossRef] [PubMed]
- Gamiz-Arco, G.; Gutierrez-Rus, L.I.; Risso, V.A.; Ibarra-Molero, B.; Hoshino, Y.; Petrovic, D.; Justicia, J.; Cuerva, J.M.; Romero-Rivera, A.; Seelig, B.; et al. Heme-binding enables allosteric modulation in an ancient TIM-barrel glycosidase. Nat. Commun. 2021, 12, 380. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018, 27, 135–145. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.R.; Makhatadze, G.I. ProteinVolume: Calculating molecular van der Waals and void volumes in proteins. BMC Bioinform. 2015, 16, 101. [Google Scholar] [CrossRef] [PubMed]
- Ferruz, N.; Schmidt, S.; Höcker, B. ProteinTools: A toolkit to analyze protein structures. Nucleic Acids Res. 2021, 49, W559–W566. [Google Scholar] [CrossRef] [PubMed]
- Heinig, M.; Frishman, D. STRIDE: A web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res. 2004, 32, W500–W502. [Google Scholar] [CrossRef]
- Koes, D.R.; Baumgartner, M.P.; Camacho, C.J. Lessons learned in empirical scoring with smina from the CSAR 2011 benchmarking exercise. J. Chem. Inf. Model. 2013, 53, 1893–1904. [Google Scholar] [CrossRef]
- Karplus, P.A.; Diederichs, K. Assessing and maximizing data quality in macromolecular crystallography. Curr. Opin. Struct. Biol. 2015, 34, 60–68. [Google Scholar] [CrossRef]
- Henrissat, B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 1991, 280 Pt 2, 309–316. [Google Scholar] [CrossRef]
- Sotiropoulou, A.I.; Hatzinikolaou, D.G.; Chrysina, E.D. Structural studies of β-glucosidase from the thermophilic bacterium Caldicellulosiruptor saccharolyticus. Biol. Crystallogr. 2024, 80, 733–743. [Google Scholar] [CrossRef]
- Oh, H.-J.; Shin, K.-C.; Oh, D.-K. Production of ginsenosides Rg1 and Rh1 by hydrolyzing the outer glycoside at the C-6 position in protopanaxatriol-type ginsenosides using β-glucosidase from Pyrococcus furiosus. Biotechnol. Lett. 2014, 36, 113–119. [Google Scholar] [CrossRef]
- Zhou, K.; Zhang, Y.; Xu, M.; Zhou, Y.; Sun, A.; Zhou, H.; Han, Y.; Zhao, D.; Yu, S. A GH1 β-glucosidase from the Fervidobacterium pennivorans DSM9078 showed extraordinary thermostability and distinctive ability in the efficient transformation of ginsenosides. Bioorg. Chem. 2025, 154, 108049. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, M.; Kataoka, M.; Mishima, Y.; Maeno, Y.; Ishikawa, K. Structural analysis of beta-glucosidase mutants derived from a hyperthermophilic tetrameric structure. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70 Pt 3, 877–888. [Google Scholar] [CrossRef]
- Linse, S.; Thulin, E.; Nilsson, H.; Stigler, J. Benefits and constrains of covalency: The role of loop length in protein stability and ligand binding. Sci. Rep. 2020, 10, 20108. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.; Sumreen, A.; Bibi, A.; Batool, K. In silico approach to elucidate factors associated with GH1 β-Glucosidase thermostability. J. Pure Appl. Microbiol. 2019, 13, 1953–1968. [Google Scholar] [CrossRef]
- Grigas, A.T.; Liu, Z.; Logan, J.A.; Shattuck, M.D.; O’Hern, C.S. Protein Folding as a Jamming Transition. PRX Life 2025, 3, 013018. [Google Scholar] [CrossRef]
- Sun, Z.; Liu, Q.; Qu, G.; Feng, Y.; Reetz, M.T. Utility of B-factors in Protein Science: Interpreting Rigidity, Flexibility, and Internal Motion and Engineering Thermostability. Chem. Rev. 2019, 119, 1626–1665. [Google Scholar] [CrossRef]
- Hung, C.-L.; Kuo, Y.-H.; Lee, S.W.; Chiang, Y.-W. Protein Stability Depends Critically on the Surface Hydrogen-Bonding Network: A Case Study of Bid Protein. J. Phys. Chem. B 2021, 125, 8373–8382. [Google Scholar] [CrossRef]
- Xu, D.; Tsai, C.J.; Nussinov, R. Hydrogen bonds and salt bridges across protein-protein interfaces. Protein Eng. 1997, 10, 999–1012. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).