Crystal Structure of Candida antarctica Lipase B with a Putative Pro-Peptide Region
Abstract
1. Introduction
2. Materials and Methods
2.1. CalB Production
2.2. CalB Crystallization
2.3. Data Collection, Processing, Structure Solution, and Refinement of CalB Crystal
3. Results and Discussion
3.1. Overall Structure of CalB
3.2. Comparative Analysis of Conformational Changes in the Lid Domain
3.3. Comparison with the Pro-Peptide Region of Other Lipases
4. Conclusions
5. Patents
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CalB | Lipase B from Candida antarctica |
| DLS | Diamond Light Source |
| E. coli | Escherichia coli |
| IMAC | Immobilized metal affinity chromatography |
| PDB | Protein Data Bank |
| RcL | Rhizopus chinensis lipase |
| RmL | Lipase from Rhizomucor miehei |
| RMSD | Root mean square deviation |
| RoL | Lipase from Rhizopus oryzae |
| SEC | Size exclusion chromatography |
| SHyL | Staphylococcus hyicus lipase |
| TEV | Tobacco Etch Virus nuclear-inclusion-a endopeptidase |
References
- Uppenberg, J.; Hansen, M.T.; Patkar, S.; Jones, T.A. The sequence, crystal structure determination and refinement of two crystal forms of lipase B from Candida antarctica. Structure 1994, 2, 293–308. [Google Scholar] [CrossRef]
- Uppenberg, J.; Oehrner, N.; Norin, M.; Hult, K.; Kleywegt, G.J.; Patkar, S.; Waagen, V.; Anthonsen, T.; Jones, T.A. Crystallographic and molecular-modeling studies of lipase B from Candida antarctica reveal a stereospecificity pocket for secondary alcohols. Biochemistry 1995, 34, 16838–16851. [Google Scholar] [CrossRef]
- Anderson, E.M.; Larsson, K.M.; Kirk, O. One biocatalyst–many applications: The use of Candida antarctica B-lipase in organic synthesis. Biocatal Biotransfor. 1998, 16, 181–204. [Google Scholar] [CrossRef]
- Qian, Z.; Horton, J.R.; Cheng, X.; Lutz, S. Structural redesign of lipase B from Candida antarctica by circular permutation and incremental truncation. J. Mol. Biol. 2009, 393, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; An, J.; Yang, G.; Wu, G.; Zhang, Y.; Cui, L.; Feng, Y. Enhanced enzyme kinetic stability by increasing rigidity within the active site. J. Biol. Chem. 2014, 289, 7994–8006. [Google Scholar] [CrossRef]
- Stauch, B.; Fisher, S.J.; Cianci, M. Open and closed states of Candida antarctica lipase B: Protonation and the mechanism of interfacial activation. J. Lipid Res. 2015, 56, 2348–2358. [Google Scholar] [CrossRef] [PubMed]
- Park, A.; Kim, S.; Park, J.; Joe, S.; Min, B.; Oh, J.; Song, J.; Park, S.; Park, S.; Lee, H. Structural and experimental evidence for the enantiomeric recognition toward a bulky sec-alcohol by Candida antarctica lipase B. ACS Catal. 2016, 6, 7458–7465. [Google Scholar] [CrossRef]
- Cen, Y.; Singh, W.; Arkin, M.; Moody, T.S.; Huang, M.; Zhou, J.; Wu, Q.; Reetz, M.T. Artificial cysteine-lipases with high activity and altered catalytic mechanism created by laboratory evolution. Nat. Commun. 2019, 10, 3198. [Google Scholar] [CrossRef]
- Silvestrini, L.; Cianci, M. Principles of lipid–enzyme interactions in the limbus region of the catalytic site of Candida antarctica Lipase B. Int. J. Biol. Macromol. 2020, 158, 358–363. [Google Scholar] [CrossRef]
- Ollis, D.L.; Cheah, E.; Cygler, M.; Dijkstra, B.; Frolow, F.; Franken, S.M.; Harel, M.; Remington, S.J.; Silman, I.; Schrag, J. The alpha/beta hydrolase fold. Protein Eng. 1992, 5, 197–211. [Google Scholar] [CrossRef]
- Xu, J.; Cen, Y.; Singh, W.; Fan, J.; Wu, L.; Lin, X.; Zhou, J.; Huang, M.; Reetz, M.T.; Wu, Q. Stereodivergent protein engineering of a lipase to access all possible stereoisomers of chiral esters with two stereocenters. J. Am. Chem. 2019, 141, 7934–7945. [Google Scholar] [CrossRef]
- Strzelczyk, P.; Bujacz, G.D.; Kiełbasiński, P.; Błaszczyk, J. Crystal and molecular structure of hexagonal form of lipase B from Candida antarctica. Acta Biochim. Pol. 2016, 63, 103–109. [Google Scholar] [CrossRef]
- Høegh, I.; Patkar, S.; Halkier, T.; Hansen, M.T. Two lipases from Candida antarctica: Cloning and expression in Aspergillus oryzae. Canad. J. Bot. 1995, 73, 869–875. [Google Scholar] [CrossRef]
- Vadhana, A.K.P.; Samuel, P.; Berin, R.M.; Krishna, J.; Kamatchi, K.; Meenakshisundaram, S. Improved secretion of Candida antarctica lipase B with its native signal peptide in Pichia pastoris. Enzyme Microb. Technol. 2013, 52, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Ikemura, H.; Takagi, H.; Inouye, M. Requirement of pro-sequence for the production of active subtilisin E in Escherichia coli. J. Biol. Chem. 1987, 262, 7859–7864. [Google Scholar] [CrossRef]
- Eder, J.; Fersht, A.R. Pro-sequence-assisted protein folding. Mol. Microbiol. 1995, 16, 609–614. [Google Scholar] [CrossRef]
- Wang, Z.; Lv, P.; Luo, W.; Yuan, Z.; He, D. Expression in Pichia pastoris and characterization of Rhizomucor miehei lipases containing a new propeptide region. J. Gen. Appl. Microbiol. 2016, 62, 25–30. [Google Scholar] [CrossRef]
- Beer, H.D.; Wohlfahrt, G.; Schmid, R.D.; McCarthy, J.E. The folding and activity of the extracellular lipase of Rhizopus oryzae are modulated by a prosequence. Biochem. J. 1996, 319, 351–359. [Google Scholar] [CrossRef]
- Rosenstein, R.; Götz, F. Staphylococcal lipases: Biochemical and molecular characterization. Biochimie 2000, 82, 1005–1014. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.W.; Wang, L.L.; Xu, Y. Rhizopus chinensis lipase: Gene cloning, expression in Pichia pastoris and properties. J. Mol. Catal. B Enzym. 2009, 57, 304–311. [Google Scholar] [CrossRef]
- Van Tassel, L.; Moilanen, A.; Ruddock, L.W. Efficient production of wild-type lipase B from Candida antarctica in the cytoplasm of Escherichia coli. Protein Expr. Purif. 2020, 165, 105498. [Google Scholar] [CrossRef] [PubMed]
- Sohail, A.A.; Gaikwad, M.; Khadka, P.; Saaranen, M.J.; Ruddock, L.W. Production of extracellular matrix proteins in the cytoplasm of E. coli: Making giants in tiny factories. Int. J. Mol. Sci. 2020, 21, 688. [Google Scholar] [CrossRef] [PubMed]
- Hatahet, F.; Nguyen, V.D.; Salo, K.E.; Ruddock, L.W. Disruption of reducing pathways is not essential for efficient disulfide bond formation in the cytoplasm of E. coli. Microb. Cell Factories 2010, 9, 67. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix. refine. Acta Cryst. D 2012, 68, 352–367. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Cryst. D 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Cryst. D 2010, 66, 12–21. [Google Scholar] [CrossRef]
- McNicholas, S.; Potterton, E.; Wilson, K.S.; Noble, M.E.M. Presenting your structures: The CCP4mg molecular-graphics software. Acta Cryst. D 2011, 67, 386–394. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Cryst. D 2004, 60, 2256–2268. [Google Scholar] [CrossRef]
- Krissinel, E. Stock-based detection of protein oligomeric states in jsPISA. Nucleic Acids Res. 2015, 43, W314–W319. [Google Scholar] [CrossRef] [PubMed]
- Liebschner, D.; Afonine, P.V.; Moriarty, N.W.; Poon, B.K.; Sobolev, O.V.; Terwilliger, T.C.; Adams, P.D. Polder maps: Improving OMIT maps by excluding bulk solvent. Biol. Crystallogr. 2017, 73, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Moroz, O.V.; Blagova, E.; Reiser, V.; Saikia, R.; Dalal, S.; Jørgensen, C.I.; Bhatia, V.K.; Baunsgaard, L.; Andersen, B.; Svendsen, A.; et al. Novel inhibitory function of the Rhizomucor miehei lipase propeptide and three-dimensional structures of its complexes with the enzyme. ACS Omega 2019, 4, 9964–9975. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Yu, X.W.; Xu, Y.; Guo, R.T.; Swapna, G.V.T.; Szyperski, T.; Hunt, J.F.; Montelione, G.T. Structural basis by which the N-terminal polypeptide segment of Rhizopus chinensis lipase regulates its substrate binding affinity. Biochemistry 2019, 58, 3943–3954. [Google Scholar] [CrossRef] [PubMed]

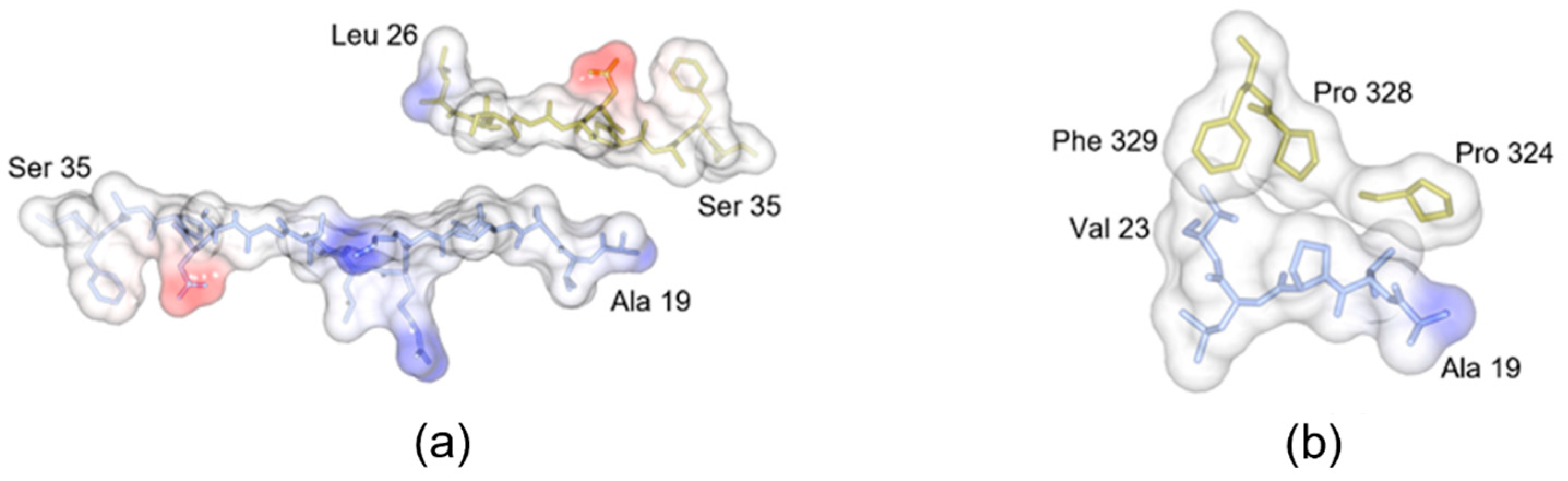
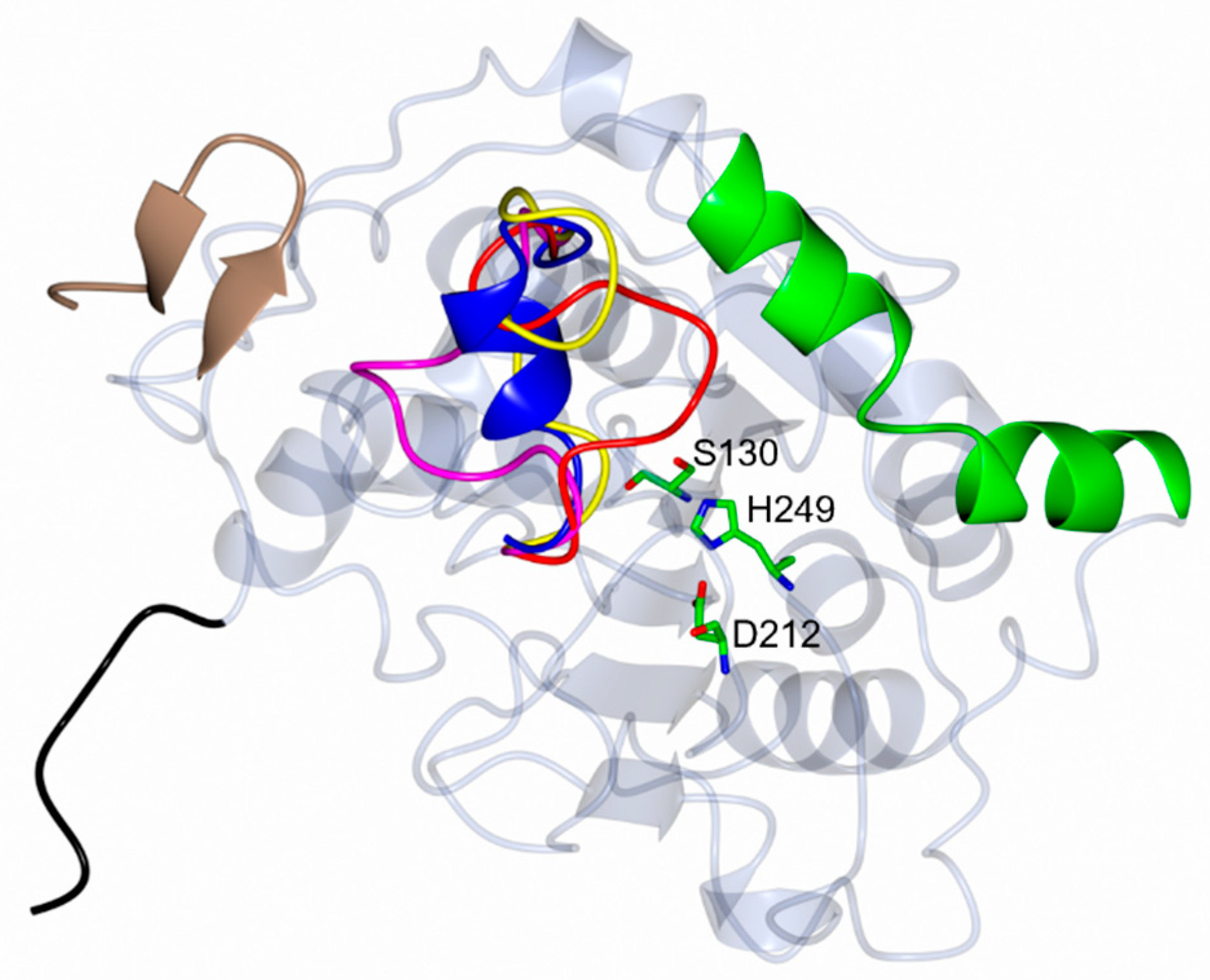
| Source Organism | Candida antarctica (Yeast) |
|---|---|
| DNA source | Codon optimized by GenScript |
| Cloning vector | Modified pET23-based with a pTac promoter |
| Expression vector | pAS88 and pMJS205 1 |
| Expression host | Escherichia coli BL21 (DE3) |
| Method | Sitting-Drop Vapor Diffusion |
|---|---|
| Plate type | 96-well TTP Lab Tech triple sitting-drop |
| Temperature | 295.15 K |
| Protein concentration | 10 mg/mL |
| Protein storage solution | 20 mM phosphate buffer, pH 7.4; 150 mM NaCl |
| Crystallization solution | 200 M potassium sodium tartrate tetrahydrate 100 M bis-tris propane, pH 7.5; 25% w/v PEG 3350 |
| Crystallization drop volume | Crystal drop volume is 400 nL, where 200 nL is protein, 160 nL of crystallization solution, and 40 nL is crystal seed |
| Data Collection Parameters | |
| Diffraction source | I04-DLS-UK |
| Wavelength (Å) | 0.9795 |
| Detector | Eiger2 XE 16 M |
| Temperature (K) | 100 |
| Data-processing software | XDS/Aimless |
| Data-processing statistics | |
| Unit cell parameters (Å,°) | a = 47.34, b = 80.98, c = 73.99 α = 90.00, β = 98.30, γ = 90.00 |
| Space group | P21 |
| Resolution range (Å) | 54.37–1.45 (1.45–1.41) 1 |
| Molecules per asymmetric unit | 2 |
| Number of observations (total) | 503,254 (27,177) 1 |
| Number of observations (unique) | 104,810 (7629) 1 |
| Redundancy | 4.4 (3.56) 1 |
| Completeness (%) | 99.86 (86.60) 1 |
| Rmerge (%) | 4.7 (41.4) 1 |
| I/σ (I) | 14.5 (1.3) 1 |
| CC (1/2) | 0.999 (0.905) 1 |
| Refinement statistics | |
| Resolution (Å) | 46.85–1.45 (1.51–1.45) 1 |
| Rwork (%) | 13.59 (19.45) 1 |
| Rfree (%) | 16.79 (24.78) 1 |
| Number of unique reflections | 97,613 |
| Number of non-hydrogen atoms | 4700 |
| Average B-factors (Å2) | 23.12 |
| r.m.s.d. bond length (Å) | 0.005 |
| r.m.s.d. bond angle (°) | 0.81 |
| Ramachandran plot | |
| Favored (%) | 97.17 |
| Allowed (%) | 2.83 |
| Outliers (%) | 0.00 |
| PDB code | 9EVI |
| # | PDB | Chain | Lid Domain chain | Z-Score | RMSD | Reference |
|---|---|---|---|---|---|---|
| 1 | 1LBS | A–F | Helical A–F | 57.2–57.1 | 0.5 | [2] |
| 2 | 1LBT | A–B | Helical A–B | 57.5 | 0.6 | |
| 3 | 1TCA | A | Helical A | 57.2 | 0.6 | [1] |
| 4 | 1TCB | A–B | Helical A–B | 57.3–57.2 | 0.6 | |
| 5 | 1TCC | A–B | Helical A–B | 57.8–57.7 | 0.5–0.6 | |
| 6 | 3ICV | A | Unmodelled A | 46.2 | 1.0 | [4] |
| 7 | 3ICW | A | Unmodelled A | 45.9 | 1.0 | |
| 8 | 3W9B | A–D | Helical A–D | 57.1–56.3 | 0.6–0.7 | Unpublished |
| 9 | 4K6G | A–B | Unmodelled A, Open B | 58.3–58.1 | 0.4–0.8 | [5] |
| 10 | 4K6H | A–B | Open A, Closed B | 59.4–58.2 | 0.4–0.9 | |
| 11 | 4K6K | A–B | Unmodelled A–B | 57.6–57.1 | 0.4–0.5 | |
| 12 | 4K5Q | A | Open A | 57.9 | 0.9 | |
| 13 | 4ZV7 | A | Helical A | 57.9 | 0.6 | [12] |
| 14 | 5A6V | A–B | Helical A, Closed B | 57.4 | 0.6–0.7 | [6] |
| 15 | 5A71 | A–B | Helical A, Closed B | 57.9–57.7 | 0.6–0.7 | |
| 16 | 5GV5 | A–H | Helical A,B,F,G,H, Unmodelled D,E, Open C | 57.7–56.7 | 0.6–0.9 | [7] |
| 17 | 6ISP | A–D | Closed A–D | 57.3–57.2 | 0.7–1.0 | [8] |
| 18 | 6ISQ | A–B | Open A–B | 55.3–55.2 | 1.3 | |
| 19 | 6ISR | A–B | Open A–B | 53.6–53.6 | 1.2–1.3 | |
| 20 | 6J1P | A–B | Closed A, Open B | 59.5–57 | 0.4–0.9 | [11] |
| 21 | 6J1Q | A–B | Closed A, Open B | 59.3–57.6 | 0.4–0.8 | |
| 22 | 6J1R | A–B | Open A, Closed B | 58.9–58 | 0.5–0.9 | |
| 23 | 6J1S | A–B | Closed A, Open B | 59.2–57.3 | 0.4–0.9 | |
| 24 | 6J1T | A–B | Open A, Closed B | 58.2–57.8 | 0.7–0.9 | |
| 25 | 6TP8 | A–C | Helical A–C | 58.1 | 0.6 | [9] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sohail, A.A.; Recacha, R.; Ruddock, L.W. Crystal Structure of Candida antarctica Lipase B with a Putative Pro-Peptide Region. Crystals 2025, 15, 927. https://doi.org/10.3390/cryst15110927
Sohail AA, Recacha R, Ruddock LW. Crystal Structure of Candida antarctica Lipase B with a Putative Pro-Peptide Region. Crystals. 2025; 15(11):927. https://doi.org/10.3390/cryst15110927
Chicago/Turabian StyleSohail, Anil A., Rosario Recacha, and Lloyd W. Ruddock. 2025. "Crystal Structure of Candida antarctica Lipase B with a Putative Pro-Peptide Region" Crystals 15, no. 11: 927. https://doi.org/10.3390/cryst15110927
APA StyleSohail, A. A., Recacha, R., & Ruddock, L. W. (2025). Crystal Structure of Candida antarctica Lipase B with a Putative Pro-Peptide Region. Crystals, 15(11), 927. https://doi.org/10.3390/cryst15110927





