EXtra-Xwiz: A Tool to Streamline Serial Femtosecond Crystallography Workflows at European XFEL
Abstract
:1. Introduction
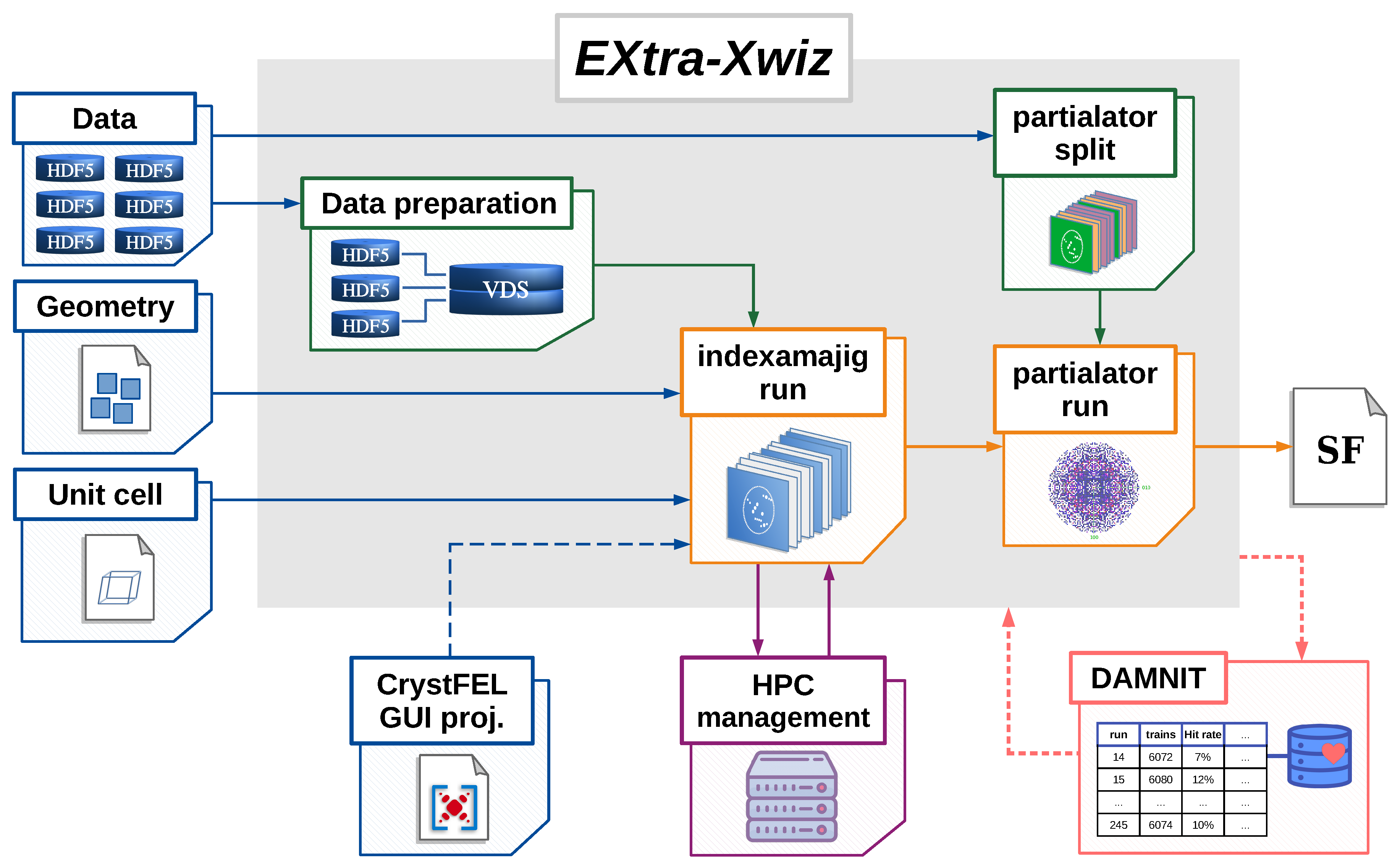
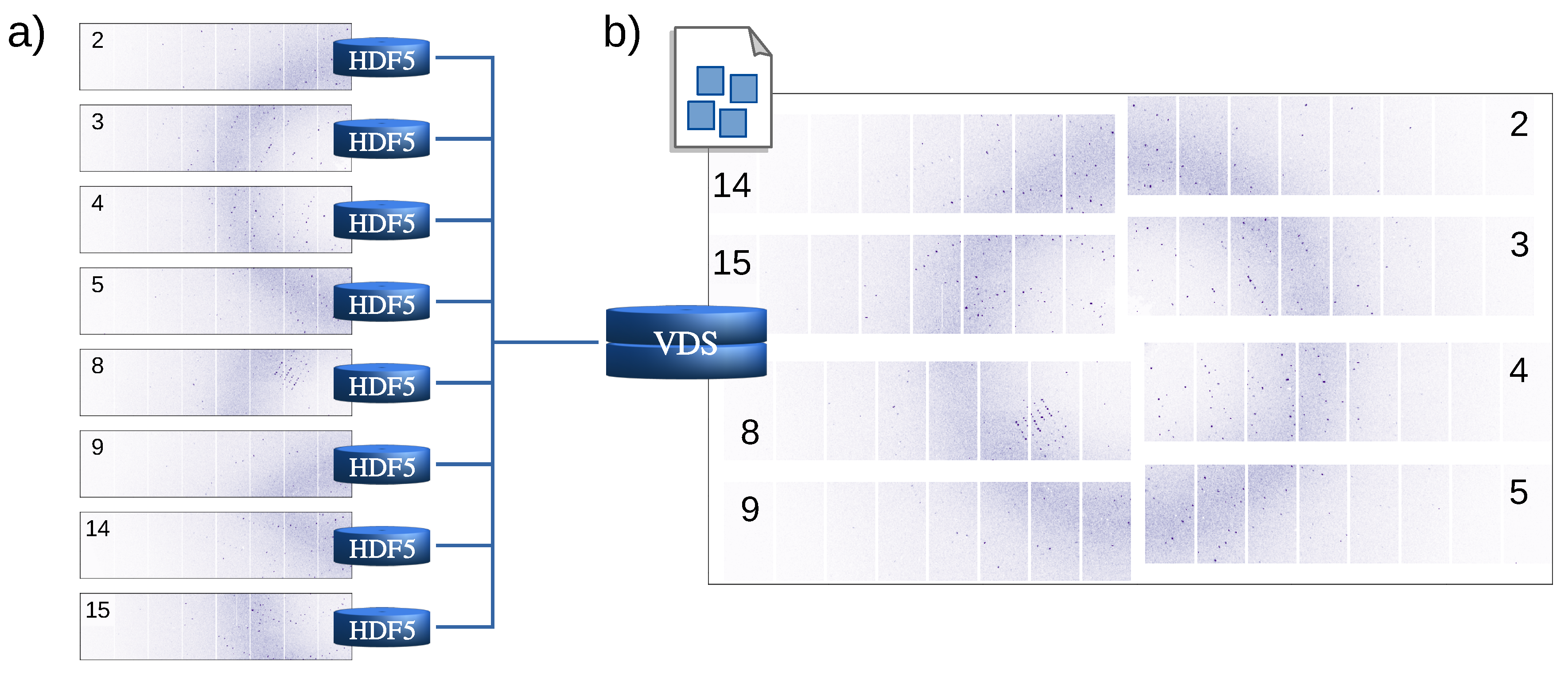
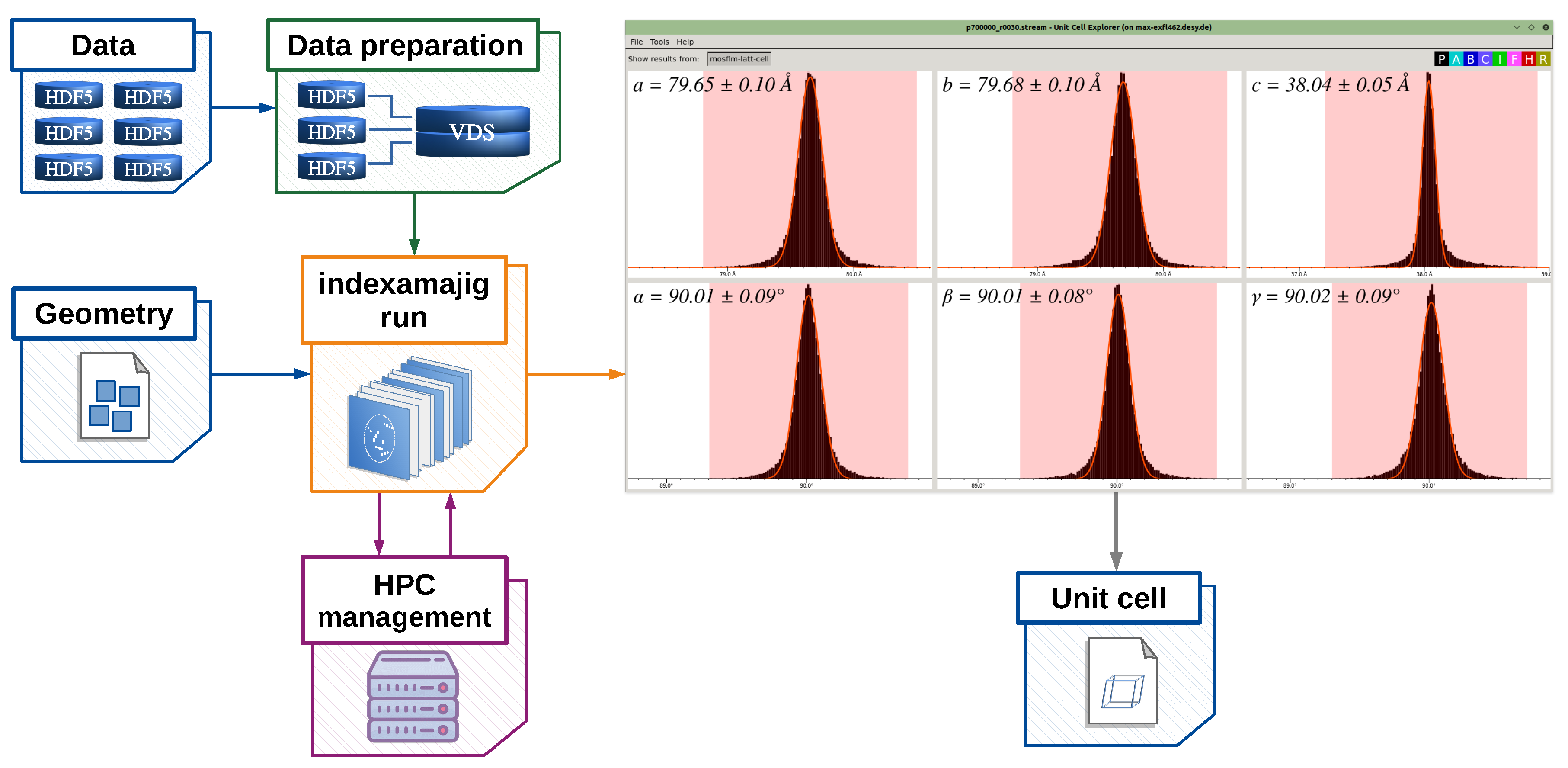
2. Design and Implementation of the EXtra-Xwiz Pipeline
- indexamajig—main command-line program for indexing and integrating diffraction patterns;
- cell_explorer—a tool for displaying and determining the unit cell parameters of the crystalline sample;
- partialator—used for scaling, merging, and postrefining reflections data;
- check_hkl—used for calculating FOMs based on the full set of merged reflections, such as completeness, average signal strength, and redundancy;
- compare_hkl—used for calculating the FOMs based on the merged reflections split into two sets, such as R factors and correlation coefficients.
Structure of the EXtra-Xwiz Pipeline
3. Data Processing with EXtra-Xwiz via Example
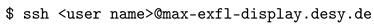
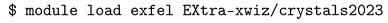
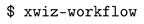
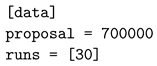
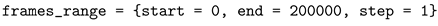








3.1. Automatic Scan over EXtra-Xwiz Configuration Parameters


3.2. Running EXtra-Xwiz Tutorial Using VISA
- Navigate to https://visa.xfel.eu, accessed on 16 October 2023 [65];
- Click “Create a new instance”;
- Click “Search for experiments”, and select the proposal “p700000-SFX on Hen egg-white lysozyme, AGIPD detector”;
- Click on the “EXtra-Xwiz_Crystals2023” environment;
- Choose the virtual hardware;
- Create the instance.


4. Discussion and Outlook
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SFX | Serial Femtosecond Crystallography |
| XFEL | X-Ray-Free Electron Laser |
| EuXFEL | European XFEL facility |
| EXDF | EuXFEL Data Format |
| AGIPD | Adaptive-Gain Integrating Pixel Detector |
| LPD | Large Pixel Detector |
| HPC | High-Performance Computing |
| HDF5 | Hierarchical Data Format v.5 |
| CBF | Crystallographic Binary Format |
| CXIDB | Coherent X-Ray Imaging Data Bank |
| GUI | Graphical User Interface |
| VDS | Virtual Data Set File |
| FOM | Figure Of Merit |
| SNR | Signal-To-Noise Ratio |
| HEWL | Hen Egg-White Lysozyme |
| VISA | Virtual Infrastructure for Scientific Analysis |
References
- Smyth, M.S.; Martin, J.H.J. X-ray crystallography. Mol. Pathol. 2000, 53, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y. A Glimpse of Structural Biology through X-ray Crystallography. Cell 2014, 159, 995–1014. [Google Scholar] [CrossRef] [PubMed]
- Maveyraud, L.; Mourey, L. Protein X-ray Crystallography and Drug Discovery. Molecules 2020, 25, 1030. [Google Scholar] [CrossRef] [PubMed]
- The Protein Data Bank: Statistics. Available online: https://www.rcsb.org/stats (accessed on 15 August 2023).
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Als-Nielsen, J.; McMorrow, D. Elements of Modern X-ray Physics, 2nd ed.; Wiley: Hoboken, NJ, USA, 2011. [Google Scholar]
- Perutz, M.F.; Rossmann, M.G.; Cullis, A.F.; Muirhead, H.; Will, G.; North, A.C.T. Structure of Hæmoglobin: A Three-Dimensional Fourier Synthesis at 5.5-Å. Resolution, Obtained by X-ray Analysis. Nature 1960, 185, 416–422. [Google Scholar] [CrossRef]
- Wimberly, B.T.; Brodersen, D.E.; Clemons, W.M.; Morgan-Warren, R.J.; Carter, A.P.; Vonrhein, C.; Hartsch, T.; Ramakrishnan, V. Structure of the 30S ribosomal subunit. Nature 2000, 407, 327–339. [Google Scholar] [CrossRef]
- Garman, E.F. Radiation damage in macromolecular crystallography: What is it and why should we care? Acta Crystallogr. Sect. D 2010, 66, 339–351. [Google Scholar] [CrossRef]
- Garman, E.F.; Owen, R.L. Cryocooling and radiation damage in macromolecular crystallography. Acta Crystallogr. Sect. D 2006, 62, 32–47. [Google Scholar] [CrossRef]
- Standfuss, J.; Spence, J. Serial crystallography at synchrotrons and X-ray lasers. IUCrJ 2017, 4, 100–101. [Google Scholar] [CrossRef]
- Neutze, R.; Wouts, R.; van der Spoel, D.; Weckert, E.; Hajdu, J. Potential for biomolecular imaging with femtosecond X-ray pulses. Nature 2000, 406, 752–757. [Google Scholar] [CrossRef]
- Chapman, H.N. X-ray Free-Electron Lasers for the Structure and Dynamics of Macromolecules. Annu. Rev. Biochem. 2019, 88, 35–58. [Google Scholar] [CrossRef] [PubMed]
- Chapman, H.N.; Fromme, P.; Barty, A.; White, T.A.; Kirian, R.A.; Aquila, A.; Hunter, M.S.; Schulz, J.; DePonte, D.P.; Weierstall, U.; et al. Femtosecond X-ray protein nanocrystallography. Nature 2011, 470, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Fromme, P.; Spence, J.C. Femtosecond nanocrystallography using X-ray lasers for membrane protein structure determination. Curr. Opin. Struct. Biol. 2011, 21, 509–516. [Google Scholar] [CrossRef] [PubMed]
- Schlichting, I. Serial femtosecond crystallography: The first five years. IUCrJ 2015, 2, 246–255. [Google Scholar] [CrossRef]
- Barends, T.R.M.; Stauch, B.; Cherezov, V.; Schlichting, I. Serial femtosecond crystallography. Nat. Rev. Methods Prim. 2022, 2, 59. [Google Scholar] [CrossRef]
- Wiedorn, M.O.; Oberthür, D.; Bean, R.; Schubert, R.; Werner, N.; Abbey, B.; Aepfelbacher, M.; Adriano, L.; Allahgholi, A.; Al-Qudami, N.; et al. Megahertz serial crystallography. Nat. Commun. 2018, 9, 4025. [Google Scholar] [CrossRef]
- De Wijn, R.; Melo, D.V.M.; Koua, F.H.M.; Mancuso, A.P. Potential of Time-Resolved Serial Femtosecond Crystallography Usin High Repetition Rate XFEL Sources. Appl. Sci. 2022, 12, 2551. [Google Scholar] [CrossRef]
- Pandey, S.; Poudyal, I.; Malla, T.N. Pump-Probe Time-Resolved Serial Femtosecond Crystallography at X-ray Free Electron Lasers. Crystals 2020, 10, 628. [Google Scholar] [CrossRef]
- Aquila, A.; Hunter, M.S.; Doak, R.B.; Kirian, R.A.; Fromme, P.; White, T.A.; Andreasson, J.; Arnlund, D.; Bajt, S.; Barends, T.R.M.; et al. Time-resolved protein nanocrystallography using an X-ray free-electron laser. Opt. Express 2012, 20, 2706. [Google Scholar] [CrossRef]
- Kupitz, C.; Basu, S.; Grotjohann, I.; Fromme, R.; Zatsepin, N.A.; Rendek, K.N.; Hunter, M.S.; Shoeman, R.L.; White, T.A.; Wang, D.; et al. Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser. Nature 2014, 513, 261–265. [Google Scholar] [CrossRef]
- Orville, A.M. Recent results in time resolved serial femtosecond crystallography at XFELs. Curr. Opin. Struct. Biol. 2020, 65, 193–208. [Google Scholar] [CrossRef] [PubMed]
- Powell, H.R. X-ray data processing. Biosci. Rep. 2017, 37, BSR20170227. [Google Scholar] [CrossRef]
- Kabsch, W. XDS. Acta Cryst. D 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Battye, T.G.G.; Kontogiannis, L.; Johnson, O.; Powell, H.R.; Leslie, A.G.W. iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. Sect. D 2011, 67, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Winter, G.; Waterman, D.G.; Parkhurst, J.M.; Brewster, A.S.; Gildea, R.J.; Gerstel, M.; Fuentes-Montero, L.; Vollmar, M.; Michels-Clark, T.; Young, I.D.; et al. DIALS: Implementation and evaluation of a new integration package. Acta Cryst. 2018, D74, 85–97. [Google Scholar] [CrossRef]
- Brewster, A.S.; Waterman, D.G.; Parkhurst, J.M.; Gildea, R.J.; Young, I.D.; O’Riordan, L.J.; Yano, J.; Winter, G.; Evans, G.; Sauter, N.K. Improving signal strength in serial crystallography with DIALS Geom. Refinement. Acta Crystallogr. Sect. D 2018, 74, 877–894. [Google Scholar] [CrossRef]
- White, T.A.; Kirian, R.A.; Martin, A.V.; Aquila, A.; Nass, K.; Barty, A.; Chapman, H.N. CrystFEL: A software suite for snapshot serial crystallography. J. Appl. Cryst. 2012, 45, 335–341. [Google Scholar] [CrossRef]
- White, T. Processing serial crystallography data with CrystFEL: A step-by-step guide. Acta Cryst. 2019, D75, 219–233. [Google Scholar] [CrossRef]
- Lamzin, V.S.; Perrakis, A. Current state of automated crystallographic data analysis. Nat. Struct. Biol. 2000, 7, 978–981. [Google Scholar] [CrossRef]
- Winter, G.; McAuley, K.E. Automated data collection for macromolecular crystallography. Methods 2011, 55, 81–93. [Google Scholar] [CrossRef]
- Perrakis, A.; Morris, R.; Lamzin, V.S. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 1999, 6, 458–463. [Google Scholar] [CrossRef] [PubMed]
- Alharbi, E.; Bond, P.S.; Calinescu, R.; Cowtan, K. Comparison of automated crystallographic model-building pipelines. Acta Crystallogr. Sect. D Struct. Biol. 2019, 75, 1119–1128. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, W.C. The Revolution in Crystallography. Science 1970, 169, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Abola, E.; Kuhn, P.; Earnest, T.; Stevens, R.C. Automation of X-ray crystallography. Nat. Struct. Biol. 2000, 7, 973–977. [Google Scholar] [CrossRef]
- Sauter, N.K. XFEL diffraction: Developing processing methods to optimize data quality. J. Synchrotron Radiat. 2015, 22, 239–248. [Google Scholar] [CrossRef]
- Decking, W.; Abeghyan, S.; Abramian, P.; Abramsky, A.; Aguirre, A.; Albrecht, C.; Alou, P.; Altarelli, M.; Altmann, P.; Amyan, K.; et al. A MHz-repetition-rate hard X-ray free-electron laser driven by a superconducting linear accelerator. Nat. Photonics 2020, 14, 391–397. [Google Scholar] [CrossRef]
- Tschentscher, T. Investigating ultrafast structural dynamics using high repetition rate x-ray FEL radiation at European XFEL. Eur. Phys. J. Plus 2023, 138, 274. [Google Scholar] [CrossRef]
- Allahgholi, A.; Becker, J.; Bianco, L.; Bradford, R.; Delfs, A.; Dinapoli, R.; Goettlicher, P.; Gronewald, M.; Graafsma, H.; Greiffenberg, D.; et al. The adaptive gain integrating pixel detector. JINST 2016, 11, C02066. [Google Scholar] [CrossRef]
- Hart, M.; Angelsen, C.; Burge, S.; Coughlan, J.; Halsall, R.; Koch, A.; Kuster, M.; Nicholls, T.; Prydderch, M.; Seller, P.; et al. Development of the LPD, a high dynamic range pixel detector for the European XFEL. In Proceedings of the 2012 IEEE Nuclear Science Symposium and Medical Imaging Conference Record (NSS/MIC), Anaheim, CA, USA, 27 October–3 November 2012; pp. 534–537. [Google Scholar] [CrossRef]
- Mozzanica, A.; Andrä, M.; Barten, R.; Bergamaschi, A.; Chiriotti, S.; Brückner, M.; Dinapoli, R.; Fröjdh, E.; Greiffenberg, D.; Leonarski, F.; et al. The JUNGFRAU Detector for Applications at Synchrotron Light Sources and XFELs. Synchr. Rad. News 2018, 31, 16–20. [Google Scholar] [CrossRef]
- Turkot, O.; Dall’Antonia, F.; Bean, R.J.; E, J.; Fangohr, H.; Ferreira de Lima, D.E.; Kantamneni, S.; Kirkwood, H.; Koua, F.H.M.; Mancuso, A.P.; et al. Towards automated analysis of serial crystallography data at the European XFEL. In Proceedings of the X-ray Free-Electron Lasers: Advances in Source Development and Instrumentation VI, Prague, Czech Republic, 24–28 April 2023; Tschentscher, T., Patthey, L., Tiedtke, K., Zangrando, M., Eds.; International Society for Optics and Photonics, SPIE: Bellingham, WA, USA, 2023; Volume 12581, p. 125810M. [Google Scholar] [CrossRef]
- Karplus, P.A.; Diederichs, K. Linking crystallographic model and data quality. Science 2012, 336, 1030–1033. [Google Scholar] [CrossRef]
- White, T.A.; Mariani, V.; Brehm, W.; Yefanov, O.; Barty, A.; Beyerlein, K.R.; Chervinskii, F.; Galli, L.; Gati, C.; Nakane, T.; et al. Recent developments in CrystFEL. J. Appl. Crystallogr. 2016, 49, 680–689. [Google Scholar] [CrossRef] [PubMed]
- White, T. CrystFEL: Data Processing for FEL Crystallography. Available online: https://www.desy.de/~twhite/crystfel (accessed on 15 September 2023).
- European XFEL Data Analysis group. Code Repository of EXtra-Xwiz: Pipeline for SFX Data Analysis at European XFEL. Available online: https://github.com/European-XFEL/EXtra-Xwiz/tree/crystals2023 (accessed on 17 September 2023).
- Bernstein, H.J.; Hammersley, A.P. Specification of the Crystallographic Binary File (CBF/imgCIF). In International Tables for Crystallography Volume G: Definition and Exchange of Crystallographic Data; Springer: Dordrecht, The Netherlands, 2005; pp. 37–43. [Google Scholar] [CrossRef]
- The HDF Group. Hierarchical Data Format, Version 5. 1997–2023. Available online: https://www.hdfgroup.org/HDF5/ (accessed on 18 October 2023).
- Könnecke, M.; Akeroyd, F.A.; Bernstein, H.J.; Brewster, A.S.; Campbell, S.I.; Clausen, B.; Cottrell, S.; Hoffmann, J.U.; Jemian, P.R.; Männicke, D.; et al. The NeXus data format. J. Appl. Crystallogr. 2015, 48, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, H.J.; Förster, A.; Bhowmick, A.; Brewster, A.S.; Brockhauser, S.; Gelisio, L.; Hall, D.R.; Leonarski, F.; Mariani, V.; Santoni, G.; et al. Gold Standard for macromolecular crystallography diffraction data. IUCrJ 2020, 7, 784–792. [Google Scholar] [CrossRef] [PubMed]
- Maia, F.R.N.C. The Coherent X-ray Imaging Data Bank. Nat. Methods 2012, 9, 854–855. [Google Scholar] [CrossRef] [PubMed]
- European XFEL Data Analysis Group. EXtra-Data: Library for Accessing Data at European XFEL. Available online: https://extra-data.readthedocs.io (accessed on 15 September 2023).
- Yoo, A.B.; Jette, M.A.; Grondona, M. SLURM: Simple Linux Utility for Resource Management. In Proceedings of the Job Scheduling Strategies for Parallel Processing, Seattle, WA, USA, 24 June 2003; Springer: Berlin/Heidelberg, Germany, 2003; pp. 44–60. [Google Scholar]
- White, T.A. Post-refinement method for snapshot serial crystallography. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130330. [Google Scholar] [CrossRef]
- European XFEL Data Analysis Group. DAMNIT: Tool for Interactive Data and Metadata Inspection at European XFEL. Available online: https://rtd.xfel.eu/docs/damnit/en/latest/ (accessed on 7 May 2023).
- Götz, A.; Konrad, U.; Le Gall, E.; Ounsy, M.; Servan, S. VISA Sustainability Sheet. Zenodo. 2023. Available online: https://zenodo.org/records/7788840 (accessed on 18 October 2023).
- European XFEL Data Analysis group. EuXFEL Data Analysis User Documentation. Available online: https://rtd.xfel.eu/docs/data-analysis-user-documentation/en/latest/ (accessed on 15 September 2023).
- Preston-Werner, T. TOML: Tom’s Obvious Minimal Language. Available online: https://toml.io/en/ (accessed on 15 September 2023).
- European XFEL Data Analysis group. Documentation on EXtra-Xwiz: Pipeline for SFX Data Analysis at European XFEL. Available online: https://rtd.xfel.eu/docs/data-analysis-user-documentation/en/latest/software/extra-xwiz/ (accessed on 15 September 2023).
- Yefanov, O.; Oberthür, D.; Bean, R.J.; Wiedorn, M.O.; Knoska, J.; Pena, G.; Awel, S.; Gumprecht, L.; Domaracky, M.; Sarrou, I.; et al. Evaluation of serial crystallographic structure determination within megahertz pulse trains. Struct. Dyn. 2019, 6, 064702. [Google Scholar] [CrossRef]
- European XFEL Data Analysis Group. EXtra-Geom: Library for Describing Physical Layout of Multi-Module Detectors at European XFEL. Available online: https://extra-geom.readthedocs.io (accessed on 15 May 2023).
- White, T. Symmetry Classification for Serial Crystallography Experiments. Available online: https://www.desy.de/~twhite/crystfel/twin-calculator.pdf (accessed on 15 September 2023).
- Götz, A.; Konrad, U.; Le Gall, E.; Ounsy, M.; Servan, S. Documentation on VISA: Virtual Infrastructure for Scientific Analysis. Available online: https://visa.readthedocs.io/en/latest/index.html (accessed on 15 September 2023).
- European XFEL Data Analysis Group. VISA Instance at European XFEL. Available online: https://visa.xfel.eu/ (accessed on 18 October 2023).
- Ferreira de Lima, D.E.; Davtyan, A.; Turkot, O.; Yefanov, O.; White, T.A.; Galchenkova, M.; Dall’Antonia, F.; Lane, T.J.; Fedrizzi, T.; Gelisio, L. Automatic online data analysis optimization: Application to serial femtosecond crystallography. Int. Union Crystallogr. 2014. in preparation. [Google Scholar]
- Yefanov, O.; Mariani, V.; Gati, C.; White, T.A.; Chapman, H.N.; Barty, A. Accurate determination of segmented X-ray detector geometry. Opt. Express 2015, 23, 28459. [Google Scholar] [CrossRef]
- European XFEL. Example Data at European XFEL. Available online: https://in.xfel.eu/metadata/doi/10.22003/XFEL.EU-DATA-700000-00 (accessed on 19 October 2023).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Turkot, O.; Dall’Antonia, F.; Bean, R.J.; E, J.; Fangohr, H.; Ferreira de Lima, D.E.; Kantamneni, S.; Kirkwood, H.J.; Koua, F.H.M.; Mancuso, A.P.; et al. EXtra-Xwiz: A Tool to Streamline Serial Femtosecond Crystallography Workflows at European XFEL. Crystals 2023, 13, 1533. https://doi.org/10.3390/cryst13111533
Turkot O, Dall’Antonia F, Bean RJ, E J, Fangohr H, Ferreira de Lima DE, Kantamneni S, Kirkwood HJ, Koua FHM, Mancuso AP, et al. EXtra-Xwiz: A Tool to Streamline Serial Femtosecond Crystallography Workflows at European XFEL. Crystals. 2023; 13(11):1533. https://doi.org/10.3390/cryst13111533
Chicago/Turabian StyleTurkot, Oleksii, Fabio Dall’Antonia, Richard J. Bean, Juncheng E, Hans Fangohr, Danilo E. Ferreira de Lima, Sravya Kantamneni, Henry J. Kirkwood, Faisal H. M. Koua, Adrian P. Mancuso, and et al. 2023. "EXtra-Xwiz: A Tool to Streamline Serial Femtosecond Crystallography Workflows at European XFEL" Crystals 13, no. 11: 1533. https://doi.org/10.3390/cryst13111533
APA StyleTurkot, O., Dall’Antonia, F., Bean, R. J., E, J., Fangohr, H., Ferreira de Lima, D. E., Kantamneni, S., Kirkwood, H. J., Koua, F. H. M., Mancuso, A. P., Melo, D. V. M., Round, A., Schuh, M., Sobolev, E., de Wijn, R., Wrigley, J. J., & Gelisio, L. (2023). EXtra-Xwiz: A Tool to Streamline Serial Femtosecond Crystallography Workflows at European XFEL. Crystals, 13(11), 1533. https://doi.org/10.3390/cryst13111533





