Abstract
In the present study, the interaction between organophosphorus pesticides and cholinesterase enzymes was investigated by quantum chemical cluster model and hard-soft acid-base (HSAB) approaches. The computational results of the equilibrium structure and reaction enthalpy were used to decipher the mechanism of organophosphorus pesticides coumaphos, dicrotophos, phorate, and terbufos, which interacted with the molecular cluster models of acetylcholinesterase (AChE) and butyrylcholinesterase (BChE) enzymes. In addition, the HOMO-LUMO energy gap and the HSAB descriptors prove that AChE has outstanding electron acceptability, which is suitable as a biosensing material. In terms of the calculated electronic spectrum, because the energy level of the ground state and the excited state are changed after adding pesticides with enzymes, a significant red shift phenomenon will occur.
1. Introduction
Organophosphorus (OP) pesticides have adverse effects on neurodevelopment, including primordial reflex abnormalities, mental and motor development delays, attention deficit/hyperactivity disorder (ADHD) and autism spectrum disorder (ASD) symptoms, etc. [1]. They are not only used in agricultural production, but also in parks, homes, and public places, which cause people to be exposed to a harmful environment. The OP compounds have great affinity after binding with acetylcholinesterase (AChE) and are considered to be irreversible inhibitors [2]. Due to the acute toxicity, bioaccumulation, environmental persistence, and long-term side effects of OP compounds, rapid detection is becoming more and more important [3]. A biosensor is a detector that uses biomolecules as identifying components. Proteins and enzymes help to quantitatively detect pesticide pollutants in the environment and/or degrade them, which makes it ideal for toxicological measurement to determine health and safety implications [4]. It provides advantages such as high sensitivity, simple operation, and fast response speed [5,6].
AChE can be used as one of the neurotoxicity biomarkers of various environmental organic pollutants [7]. Biomarkers can not only evaluate their potential ecological effects in indicator organisms, but also be used in environmental toxicology research and pollution monitoring. AChE is often used as a biosensor component for the detection of OP compounds, which has the advantages of instant response, simple operation, low cost, and high sensitivity [8]. Molecular docking and kinetics studies have been used to investigate the binding modes of several pesticides with mAChE and hAChE. The calculated global minimum energy (Eglob_min) and binding free energy (ΔGbinding) are not only related to the acute toxicity (LD50) of pesticides to mice, but can also generate a model to predict the LD50 of pesticides to humans [9].
In addition, butyrylcholinesterase (BChE) is a potential detoxification enzyme, which can hydrolyze cocaine and other toxic esters or eliminate neurotoxic OP compounds [10]. The sequence homology between AChE and BChE is as high as 53%, but they are different in terms of substrate specificity and sensitivity to various inhibitors [11]. In previous studies, molecular simulation results showed that all oximes in the active sites of AChE and BChE have similar binding modes [12].
The quantum chemical cluster method relies on the use of relatively precise quantum chemical methods and is an effective method for studying enzymatic reactions. The active site finite model can predict the bond strength and spectral properties of the enzyme active site, and is a technique aimed at studying the reaction mechanism of the enzyme active site [13,14,15,16,17].
In this study, the structural basis of human AChE and BChE with four OP pesticides, including coumaphos (Cou), dicrotophos (Dic), phorate (Pho), or terbufos (Ter) was provided, and their interaction patterns were compared. The purpose is to calculate which enzyme–pesticide combination has the best binding efficiency and sensitivity, so as to design biosensors suitable for the detection of pesticides in the environment and food.
2. Computational Details
Using PubChem, 3D structures of all the investigated pesticides—i.e., coumaphos (Cou), dicrotophos (Dic), phorate (Pho), and terbufos (Ter)—are obtained. Cou, Dic, Pho, and Ter are selected since their statuses are registered for use and their hazard rankings are high in the US EPA. Among them, Cou, Pho, and Ter are organothiophosphates, and Dic is a dialkyl phosphate. These organophosphate insecticides act as EC 3.1.1.7 (acetylcholinesterase)/EC 3.1.1.8 (cholinesterase) inhibitors. The GHS classification and labelling of the above pesticides is an acute toxicity and environmental hazard with a danger signal. Cou is also an irritant.
The X-ray crystal structures of human acetylcholinesterase (EC 3.1.1.7) (PDB: 4PQE) and human butyryl cholinesterase (EC 3.1.1.8) (PDB: 1P0M) are retrieved from RCSB Protein Data Bank (www.rcsb.org, accessed on 13 June 2021) as starting geometry. All the ligands and the non-bonding water molecules present in the crystallographic structure of each enzyme were removed prior to docking.
In this work, the AutoDock Vina Extended SAMSON Extension is used to dock the organophosphorus pesticides with the AChE and BChE enzymes [18]. The AutoDock Vina Extended SAMSON Extension packages the protein–ligand docking program AutoDock Vina, and provides flexible side chains, rotatable bonds, and types of locked bonds. The search domain based on the receptor, a ligand, or a binding site is easily set up. For the present docking procedure, the setup search domain is based on the whole size of the AChE and BChE enzymes. The center of the grid is set so that the grid box covers the entire enzyme. Default values are used for other parameters. Among the first 200 docking poses, the best candidate is selected according to the standard scoring function.
The quantum chemical calculations are performed at the PM7 level using MOPAC 2016 software [19]. All amino acids in the cluster model of AChE and BChE enzymes are truncated at the α-carbon, and hydrogen atoms are manually added. The protonation state of residues comes from the experimental evidence. During geometry optimization, the truncated α-carbon remains fixed in its input position. The molecular mechanics correction for the amidic bonds (CONH) is applied. The geometries of all systems are optimized using tight optimization criteria with a gradient threshold of 4.2 × 10−4 kJ mol−1 Å−1. The solvent effect is implicitly treated by the conductor-like screening model (COSMO), which uses a dielectric constant of 78.4 for water [20]. MOPAC 2016 is available from http://openmopac.net, accessed on 14 June 2021. The results of MOPAC 2016 PM7(COSMO) with optimized geometry and Mulliken charge are illustrated by MoCalc 2012 software to study the role of electrostatic interaction and hydrogen bonding in the strength of the binding reaction. In addition, Gabedit software is used to read AUX files calculated by MOPAC 2016 to visualize HOMO and LUMO [21]. ORCA software is used to obtain the singlet and triplet excited state energies of the studied system through INDO/S-CIS calculations [22,23].
3. Results and Discussion
3.1. Computational Results Using Molecular Docking and Quantum Chemical Cluster Model Methods
Molecular docking is an efficient tool to simulate the binding pattern and provide interaction information and energy between protein receptors and small molecules [24]. However, many scoring functions used to evaluate docking poses try to express binding energy in some way, but are unable to due to the complexity of ligand–protein binding-related phenomena and their inability to incorporate quantum effects, such as charge transfer between protein and ligand and the wide differences of atomic charges between the different structures of ligands and proteins [25]. These functions generally reduce the level of correlation between experimental information related to biological activity (such as Ki or IC50) [26]. Previous studies have mentioned that using the PM6 method to calculate the partial charge between the ligand and the protein has the two major advantages of accurately processing the docking of metalloproteins and producing more accurate docking results. The PM6 method calculations subjoin the energy scoring function of AutoDock 4, including the evaluation of different secondary interactions, dispersion/repulsion, hydrogen bonding, static electricity, and desolvation, which improves the accuracy of docking calculations. Then, a large number of validation sets have shown that the semi-empirical method can not only calculate partial charge accurately, but can aslo reproduce the bond energies of the experimental homodimer and heterodimer hydrogen [27].
In addition, the realistic model needs to be as consistent as possible with the conditions in vivo, which are chemically sensitive. For example, the model needs to have solvated substances and ideally temperature conditions where biochemical processes occur. Therefore, quantum mechanics calculation methods can be used to simulate common chemical and biochemical processes, such as minimum energy geometry, non-covalent interactions, and chemical reactions [28]. When using the PM7 method and solvent in the COSMO model to calculate protein–ligand energy, not only is the global energy minimum close to the position of the natural (crystalline) ligand (INN = 1), the ligand is also a minimum value (IN = 1) or almost the lowest energy (IN = 2–5) [29]. The comparison of the reference data and the results obtained through the semiempirical PM7 method implemented in MOPAC 2012 can be widely verified, such as the formation of heat (ΔHf), the geometry of small molecules and organic and inorganic solids, and the derivative properties (such as the energy of the intermolecular interaction) [28]. The literature also mentions that the enthalpy was derived from quantum mechanics and combined with some additional calculated thermodynamic terms to generate free energy data that are highly correlated with experimental data [21,22,23,24]. Although the protein–ligand affinity is affected by the enthalpy and entropy terms, the ligand is mainly driven by enthalpy, which is more selective for this target, because a larger enthalpy value may be related to the interaction between specific molecules [25,26]. Hence, compounds with the most favorable binding enthalpy can be found by ligand screening strategies with more selective reagents. For example, the molecular docking of 52 compounds (series 1) that selectively inhibit Shp2, combined with the semiempirical PM7 method, compared with Shp1, it can be shown that Shp2 has a more favorable interaction enthalpy (the confidence level is 99%) [30]. In summary, although molecular docking can calculate energy, the energetics calculation based on the quantum mechanics method through protein–ligand binding can improve its accuracy. In order to clarify the molecular interaction mechanism of enzymes and pesticides, this study explored the molecular docking and semiempirical PM7 methods. Figure 1 and Figure 2 are the molecular docking results of human AChE and BChE enzymes with organophosphorus pesticide coumaphos (Cou).
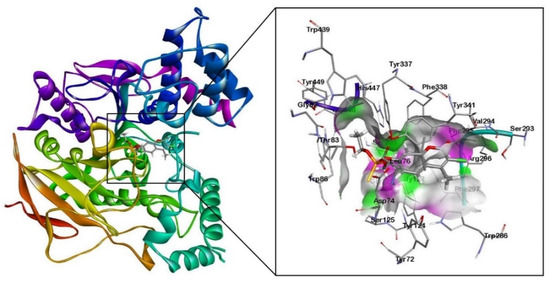
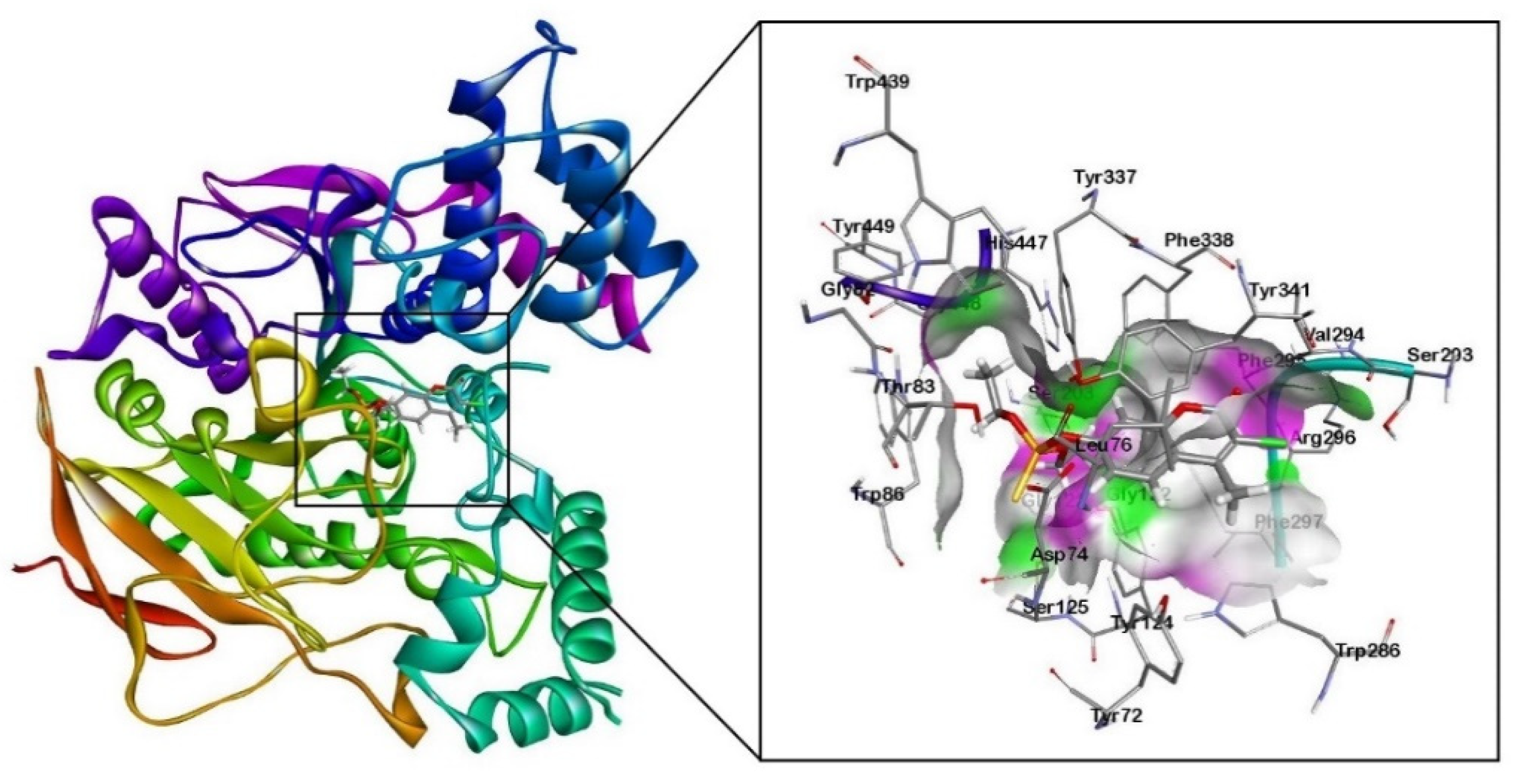
Figure 1.
Molecular docking results of human AChE enzyme (PDB: 4PQE) with organophosphorus pesticide coumaphos (Cou).
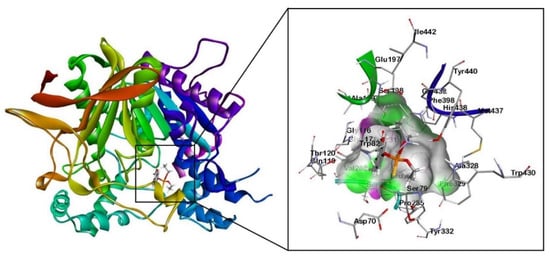
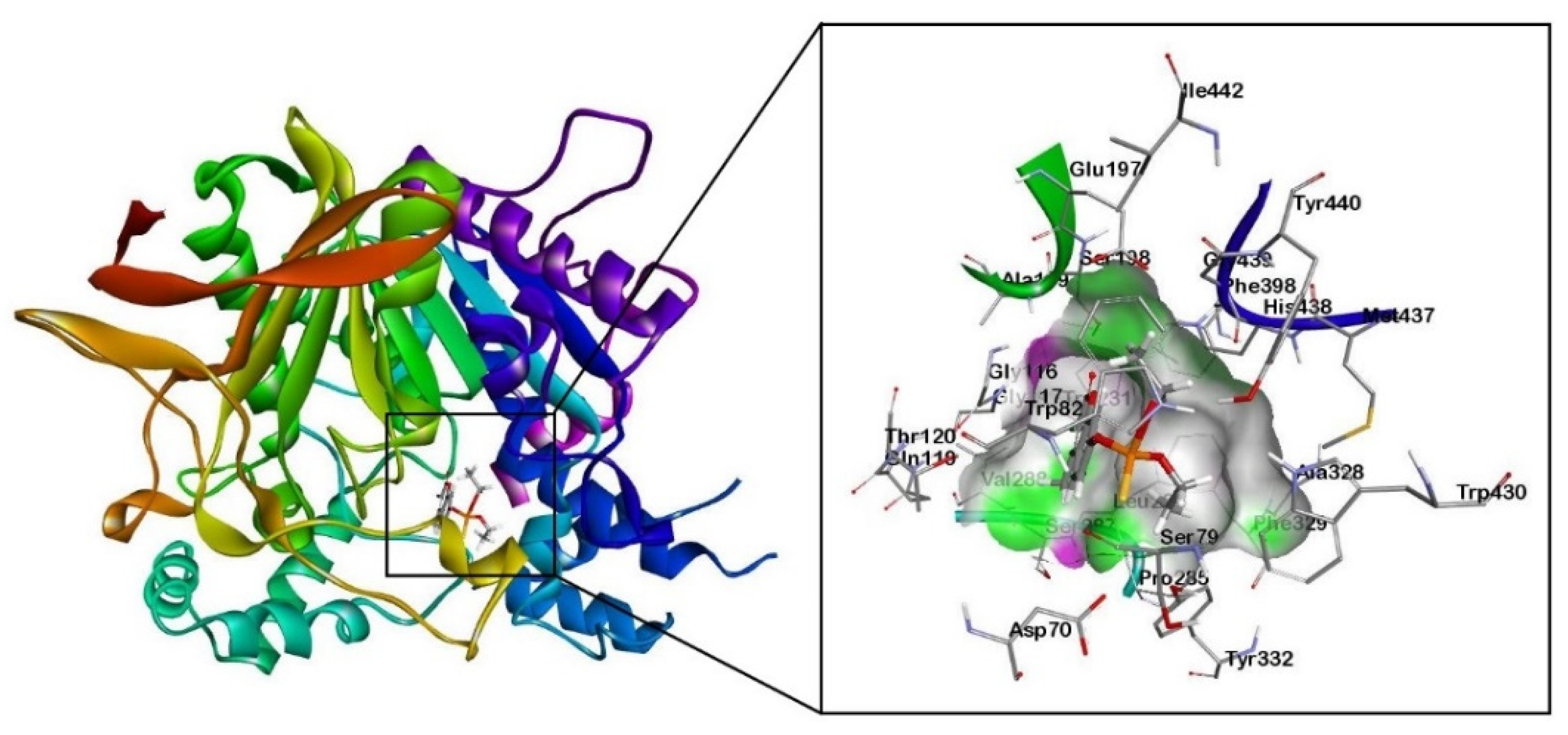
Figure 2.
Molecular docking results of human BChE enzyme (PDB: 1P0M) with organophosphorus pesticide coumaphos (Cou).
3.1.1. The Interaction between Organophosphorus Pesticides and AChE
The molecular docking results revealed that the alkyl groups of the Cou pesticide formed π-alkyl interactions with the aromatic rings of tyrosine residues (Tyr124, Tyr337) and tryptophan residues (Trp86, Trp286). A π-πT interaction between the aromatic ring of the Cou pesticide and the aromatic ring of the tyrosine residue (Tyr124) was formed. The aromatic ring of the Cou pesticide formed π-π stacking interactions with the aromatic ring of tryptophan residues (Trp286) and tyrosine residues (Tyr341).
Using the quantum chemical cluster model method, Figure 3a shows hydrogen bonding interactions of two oxygen atoms of phosphorothioate in the Cou pesticide with the hydroxyl group of tyrosine (Tyr337). The exocyclic oxygen atom of chromone in the pesticides formed hydrogen bonding with the hydrogen atom on the amino group of phenylalanine (Phe295) and the amino group of arginine (Arg296).
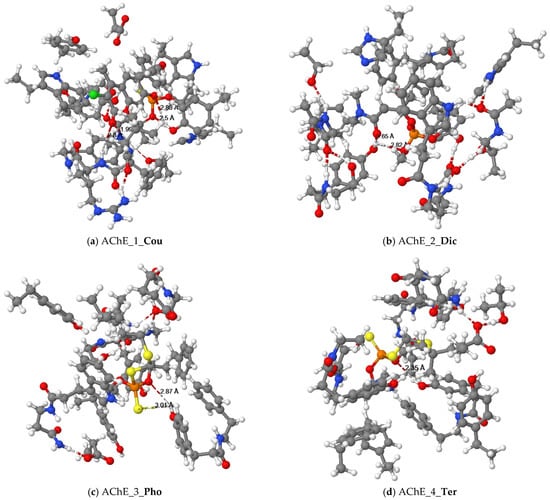
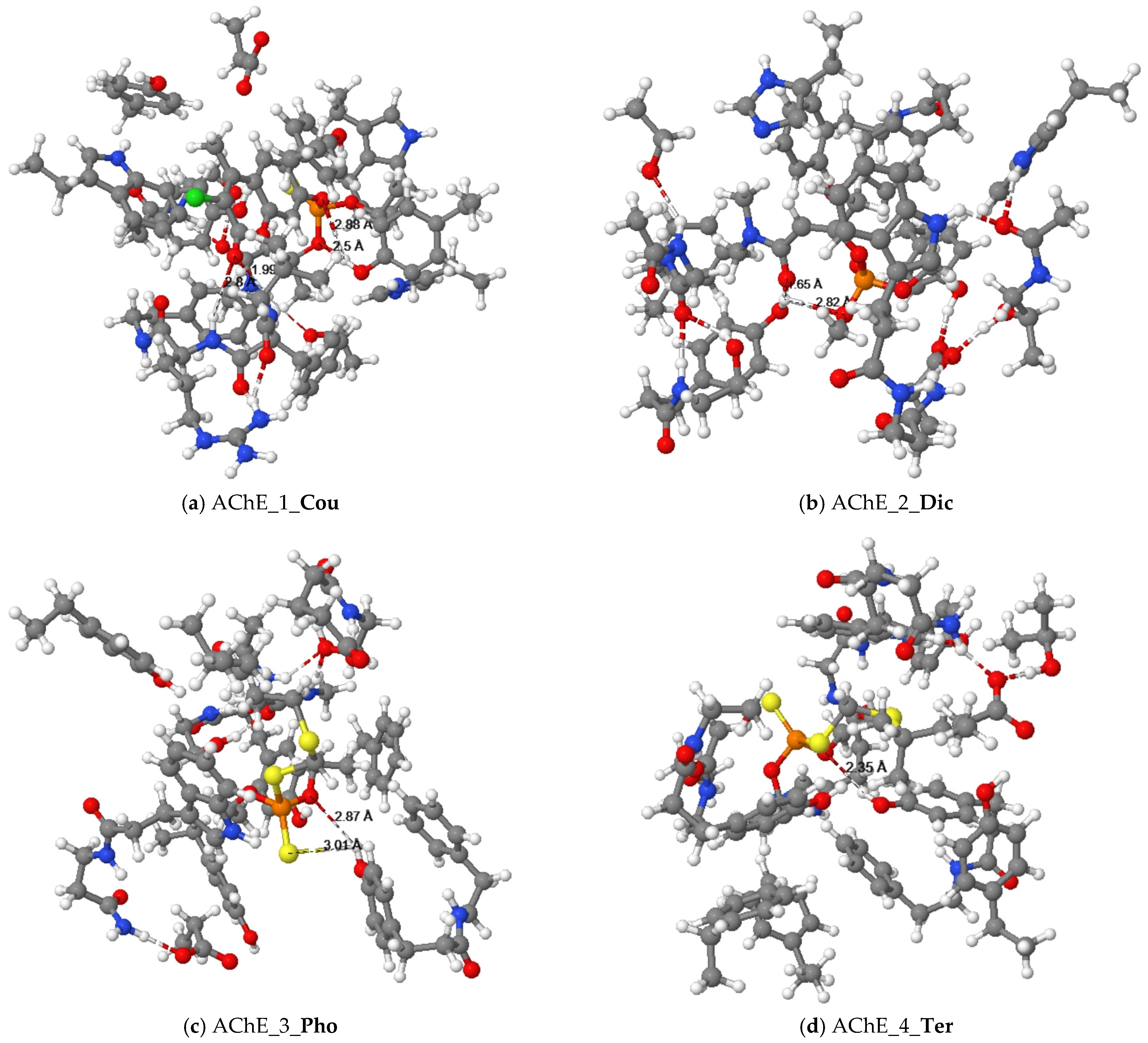
Figure 3.
Equilibrium structures of organophosphorus pesticides ((a) Cou, (b) Dic, (c) Pho and (d) Ter) binded with cluster models of the human AChE enzyme.
The docking results between Dic and AChE showed that the alkyl groups of pesticides formed π-alkyl interactions with the aromatic rings of tyrosine residues (Tyr124, Tyr337), tryptophan residues (Trp86, Trp439), amphetamine (Phe297), and histidine (His447). The quantum chemical cluster model is illustrated in Figure 3b. The oxygen atoms of the phosphate and the oxygen atoms of the amide in the Dic pesticide formed interactions with the hydroxyl group of tyrosine (Tyr124).
The alkyl groups of the Pho pesticide formed π-alkyl interactions with the aromatic rings of tyrosine residues (Tyr124), tryptophan residues (Trp86), and phenylalanine (Phe297). The sulfur atom of the ethylthio group in the Pho pesticide formed a π-sulfur interaction with the aromatic ring of the tryptophan residue (Trp86). Figure 3c illustrates that the quantum chemical results of the oxygen and sulfur atoms of the dithiophosphate group in the Pho pesticide interacted with the hydroxyl group of tyrosine (Tyr337).
The results of molecular docking showed that the alkyl groups of the Ter pesticide formed π-alkyl interactions with the aromatic rings of tyrosine residues (Tyr337), tryptophan residues (Trp86), phenylalanine (Phe295, Phe297, Phe338), and histidine (His447). After quantum chemical cluster model calculations, Figure 3d shows that the oxygen atom of the dithiophosphate group in the Ter pesticide and the hydroxyl group of tyrosine (Tyr337) formed a hydrogen bond interaction.
3.1.2. The Interaction between Organophosphorus Pesticides and BChE
The docking results showed that the alkyl groups of the Cou pesticide formed π-alkyl interactions with the aromatic rings of tyrosine residues (Tyr332), tryptophan residues (Trp82), and phenylalanine (Phe329). The aromatic ring of the Cou pesticide and the aromatic ring of Phe329 formed a π-πT interaction. The alkyl group of the pesticide interacts with the carbon atom of alanine (Ala328) to form an alkyl group interaction. Using the quantum chemical cluster model approach, Figure 4a shows that the sulfur atom of the phosphorothioate group in the Cou pesticide and the hydroxyl group of tyrosine (Tyr332) form a hydrogen bond. The exocyclic oxygen atoms of chromone in the Cou pesticide form hydrogen bonds with the hydroxyl group of aspartic acid (Asp70) and the hydrogen atoms on the amine group of tryptophan (Trp82), respectively.
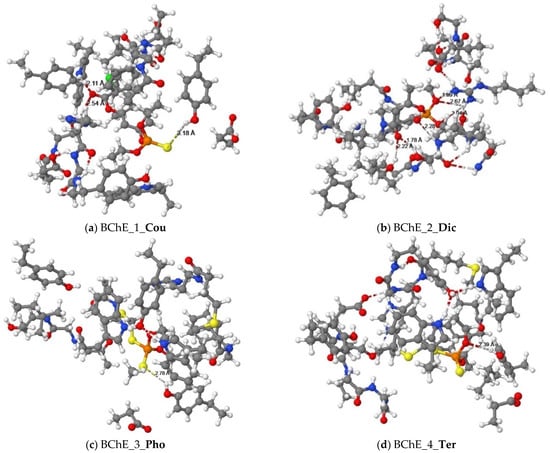
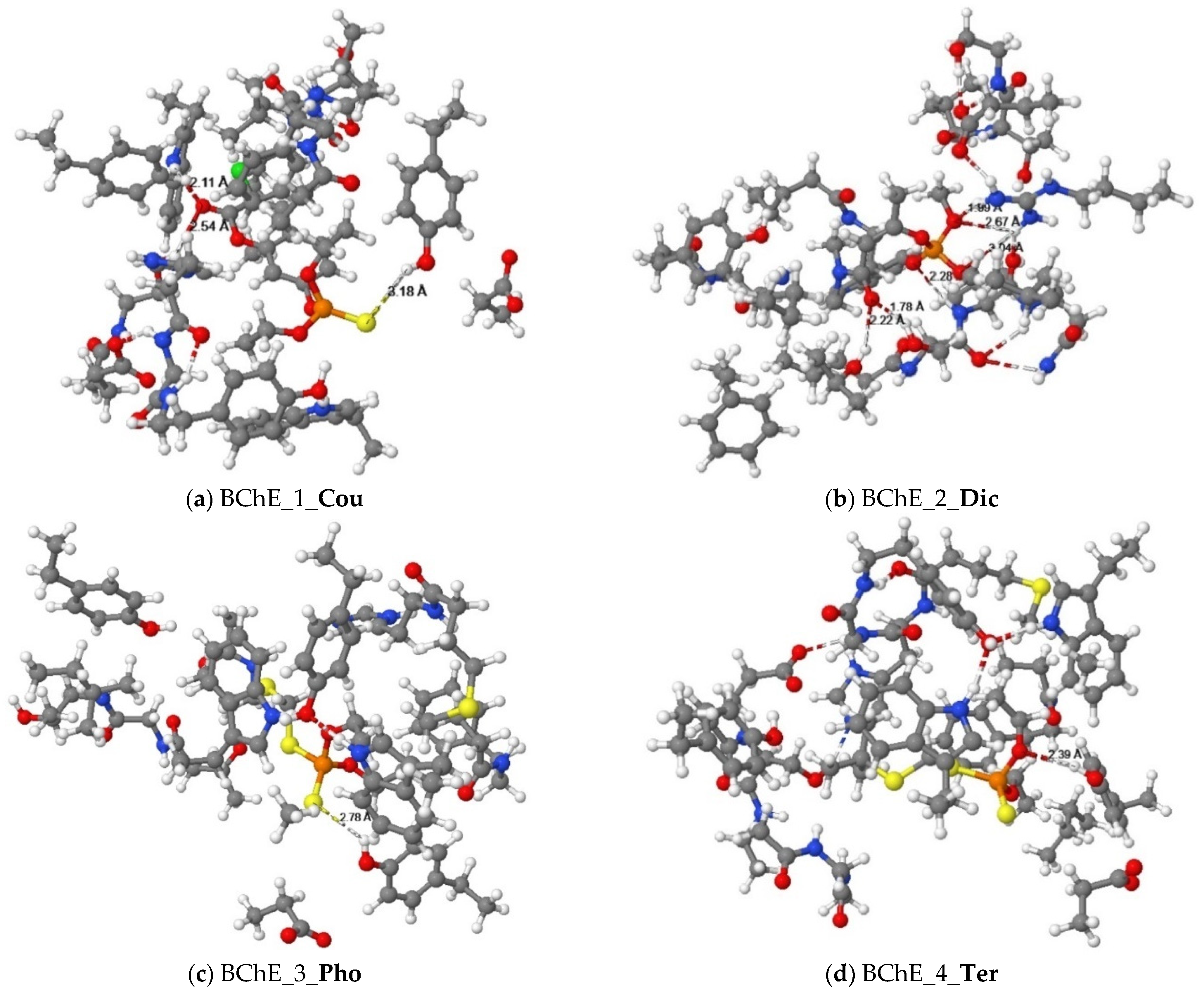
Figure 4.
Equilibrium structures of organophosphorus pesticides ((a) Cou, (b) Dic, (c) Pho, and (d) Ter) binded with cluster models of the human BChE enzyme.
The alkyl groups of the Dic pesticide interact with the carbon atoms of proline (Pro359), valine (Val280), and leucine (Leu286) to form alkyl group interactions. Using the quantum chemical method (Figure 4b), two oxygen atoms of phosphate ester in the Dic pesticide and the hydrogen atoms on the guanidine group of arginine (Arg242) formed three hydrogen bond interactions. Another oxygen atom of phosphate ester in the pesticides formed a hydrogen bond interaction with the hydrogen atom on the amine group of Valine (Val288). The oxygen atoms of amide in the pesticides formed hydrogen bonds with the hydroxyl group of serine (Ser287) and the hydroxyl group of propanol.
The alkyl groups of the Pho pesticide formed π-alkyl interactions with the aromatic rings of tyrosine residues (Tyr332, Tyr440), tryptophan residues (Trp82, Trp430), and amphetamine (Phe329). The alkyl group of the pesticide forms alkyl group interactions with the carbon atoms of alanine (Ala328), methionine (Met437), and leucine (Leu125). Using the quantum chemical method, Figure 4c shows that the sulfur atom of the dithiophosphate group in the Pho pesticide form a hydrogen bond interaction with the hydroxyl group of tyrosine (Tyr332).
The docking result showed that the alkyl group of the Ter pesticide formed π-alkyl interactions with the aromatic rings of tyrosine residues (Tyr332), tryptophan residues (Trp82, Trp430), and phenylalanine (Phe329). The alkyl group of the pesticide interacts with the carbon atoms of alanine (Ala328) and methionine (Met437) to form alkyl group interactions. The hydrogen atom on the alkyl group of the pesticide formed a π-Sigma interaction with the aromatic ring of the tryptophan residue (Trp82). For the Ter pesticide, the sulfur atom of the dithiophosphate group formed a π-sulfur interaction with the aromatic ring of the histidine residue (His438). Using the quantum chemical method, the result is shown in Figure 4d. The oxygen atom of the dithiophosphate group in pesticide and the hydroxyl group of tyrosine (Tyr332) formed a hydrogen bond interaction.
Since “hydrophobicity” is a common phenomenon in aqueous solutions, it is reasonable to assume that AChE uses potentially favorable hydrophobic interactions in the binding step to recognize substrates and other ligands. In addition, the main contribution of binding energy is due to hydrophobic alkyl substituents [31]. According to Figure 1 and Figure 2, the main binding force between enzymes and pesticides is hydrophobic interaction, including π-π stacking, π-alkyl, π-πT, alkyl, and π-Sigma interactions. There is a myriad of aromatic residues in the AChE canyon. In order to bind the respective fragments of the inhibitor as well as the substrate of the alkyl groups, aryl groups, and cation, the residues provide an active site for Tyr337, Phe338, Tyr133, and Trp84, which are suitable for binding hydrophobic groups of the substrate. The concept is the same as the hydrophobic binding zone. However, only the removal of the hydrophobic residue from position 133 can eliminate the hydrolysis of the neutral substrate, and the replacement of Tyr337 and Phe338 with alanine will moderately reduce the reaction rate [31]. It can be seen that the reaction rate of BChE will be slower than that of AChE, and it will also reduce the sensing sensitivity. The hydrophobic region contains an acyl binding site and a choline binding site, which are important for the hydrophobic region, mainly composed of tryptophan (Trp), tyrosine (Tyr), and phenylalanine (Phe) in human AChE. For BChE, the choline binding site Tyr337 of AChE was replaced by Ala328, while the acyl binding sites Phe295 and Phe297 were replaced by Leu286 and Val288, respectively. The substitution of aromatic residues in BChE’s ABS and CBS allows it to bind larger substrates and inhibitors than AChE. In addition, ABS and CBS are mainly responsible for the specificity of these enzymes, so they are the main targets of research when synthesizing inhibitors (such as imidazole or pyridine derivatives) [32]. Among them, ABS is an important active site in promoting binding affinity and complex stability. Another important active site in promoting binding affinity and complex stability is the oxygen anion pore (OAH) site, which helps stabilize the interaction between the intermediate state and the transition state. Moreover, most hydrogen bonds are formed at the oxygen anion pore (OAH) site. Hydrogen bonds play an important role in the enzymatic catalysis of macromolecules, which lead to the maximum reaction enthalpy [33] and can stabilize the bond of pesticides with enzymes (AChE/BChE). Previous documents mentioned that at the lowest electron withdrawing group connected to OP, a hydrogen bond is formed between the hydrogen atom of AChE and the oxygen atom of OP [34]. Regardless of whether the pesticide is bound with AChE or BChE, the oxygen and sulfur atoms in the pesticide as the donor of the hydrogen bond interact with the hydrogen atoms on the amino acid residues (mostly hydroxyl group on the benzene ring) as the hydrogen bond acceptor. The key amino acid residues that cause pesticides to interact with acetylcholinesterase are Tyr337, Phe295, Arg296, and Tyr124, and each pesticide forms hydrogen bonds with hydrophilic tyrosine. However, in the hydrogen bond interaction between pesticides and BChE, the key amino acid residues are Tyr332, Asp70, Trp86, Arg242, Val288, and Ser287. Except for Dic, they all form hydrogen bonds with hydrophilic tyrosine. Figure 3 and Figure 4 show that the hydrogen bond length between pesticides with AChE is shorter than that of BChE, except for Pho. Therefore, the enthalpy of reaction between pesticides with AChE is greater than that of BChE (Table 1). Nevertheless, the hydrogen bond length between Ter and BChE is slightly shorter than that of AChE, but the enthalpy of the reaction between Ter and BChE is greater than the enthalpy of reaction between Ter and AChE because of its greater electron affinity. The greater the binding affinity between protein–ligand complexes, the greater the reaction enthalpy, and the more stable [35]. These hydrophobic interactions and hydrogen bond interactions confirm the stability of the binding structure of pesticides and enzymes, because molecular docking provides the best docking posture. As mentioned in previous studies, the model analysis of AChE binding OP and carbamate shows that the stability is due to the covalent attachment of the phosphate and carbamate moieties to the active site serine, mainly through the oxygen anion pores, the hydrogen-bonding interaction of the acyl group, and the hydrophobic interaction of the acyl binding site [36,37].

Table 1.
Reaction enthalpy (∆Hf in kcal/mol) of organophosphorus pesticides (Cou, Dic, Pho, and Ter) binded with cluster models of enzymes (AChE and BChE).
3.2. Hard and Soft Acid and Base (HSAB)
The potential of forming adducts between toxic electrophiles and bionucleophiles can be described by Pearson’s theory of hard-soft acid-base (HSAB) [38]. Thus, this study uses HSAB to discuss the molecular reactivity between OP pesticides and AChE/BChE enzymes. Reactivity descriptors include the ionization energy (I), the electron affinity (A), and the energy gap (GAP) between the energy of the lowest unoccupied molecular orbital (LUMO) and the energy of the highest occupied molecular orbital (HOMO), electronegativity (χ), chemical hardness (η), chemical potential (μ), chemical softness (S), and electrophilicity index (ω) (Table 2 and Table 3).

Table 2.
The ionization potential (I = -HOMO), electron affinity (A = -LUMO), and energy gap (GAP = LUMO-HOMO) in the present study (in eV).

Table 3.
The HSAB reactivity descriptors of organophosphorus pesticides (Cou, Dic, Pho and Ter) in the present study.
It can be seen from the results in Table 2 that after the enzyme is bonded with the OP pesticide, the ionization energy becomes smaller, and the ionization energy change of AChE is greater than that of BChE. After the addition of OP compounds, the greater the energy gap change, the greater the chance of becoming the sensor material choice. The energy gap change of AChE is larger than that of BChE in this study, so AChE is more suitable as a biosensor material than BChE. Among them, AChE_1_Cou has the largest change. Coupled with the hydrophobic and hydrogen bond interactions between Cou and AChE, this leads to a large reaction enthalpy and a stable structure of AChE_1_Cou. The greater the reaction enthalpy between the pesticide and the enzyme, the greater the binding affinity and the tighter the hydrogen bond interaction [33].
Since OP pesticides are electrophilic and have highly electronegative atoms such as oxygen and chlorine atoms, the most electronegative Cou can easily accept electrons from the enzyme (Table 3). In previous studies, the more electron-withdrawing groups in OP compounds, the lower the LD50 value, indicating the importance of electron-withdrawing groups in OP compounds. Having an electron-withdrawing group can generate a partial positive charge on the phosphorus atom, which, when bound to AChE, forms an irreversible and/or stable complex with strong affinity [34].
The order of reaction enthalpy between the OP pesticide and AChE was Cou > Pho > Dic > Ter. The larger the chemical hardness value, the smaller the reaction enthalpy, but the order of Dic and Ter is the opposite, because Dic has more hydrogen bonds and a shorter bond length. In addition, the order of reaction enthalpy between pesticides and BChE was Cou > Ter > Dic > Pho. Among them, Pho has fewer hydrogen bonds, longer bond length, and the smallest binding affinity, so the resulting reaction enthalpy is the smallest.
The electrophilic index has been shown to relate to the rate constants of adduct formation reactions and can be used to model chemical reactivity. Stronger and more electrophilic compounds are more likely to react with biological targets [39]. The electrophilic index was positively correlated with the change in energy gap (AChE/BChE) after the pesticide was bonded with the enzyme. Among them, Cou has the largest electrophilic index and easily reacts with enzymes (AChE/BChE). Therefore, the sensing sensitivity to Cou is excellent. It was previously mentioned that the charge distribution on the surface of rhBChE via the protein database can be used to assess the key adsorption sites that maintain enzyme activity and inhibitor sensitivity [40]. Experimental studies have shown that charged surfaces have a great influence on the detection capability of AChE-based biosensors. This indicates that the highly efficient biocatalytic reaction and fast direct electron transfer (DET) between the positively charged surface of TcAChE and the electrode surface provides a better microenvironment, which determines the analytical performance of AChE-based biosensors [41].
3.3. Frontier Molecular Orbital (FMO)
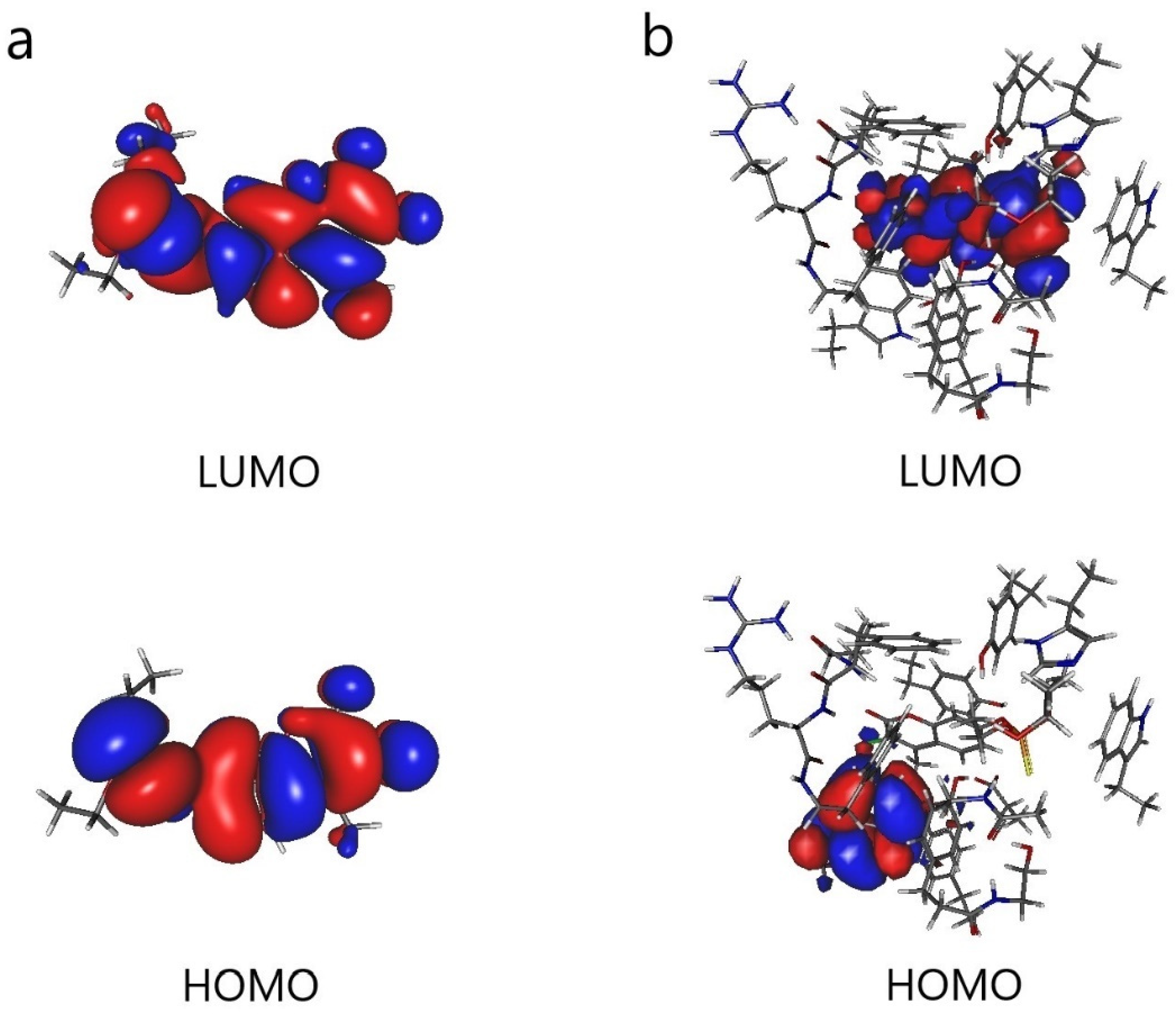
The results are shown in Figure 5. When Cou is bonded with AChE, the orbital lobes in the HOMO diagram are concentrated on tryptophan. It can be seen that the main electron donor in AChE is tryptophan. In addition, the orbital lobes in the LUMO diagram are concentrated on Cou, which is the main electron acceptor. As pointed out by the above HSAB analysis, OP pesticides are electrophiles and enzymes are nucleophiles. In addition, the smaller the energy gap, the easier it is for the transition of electrons from the HOMO of enzymes to the LUMO of OP pesticides, which lays the foundation for the development of biosensors.
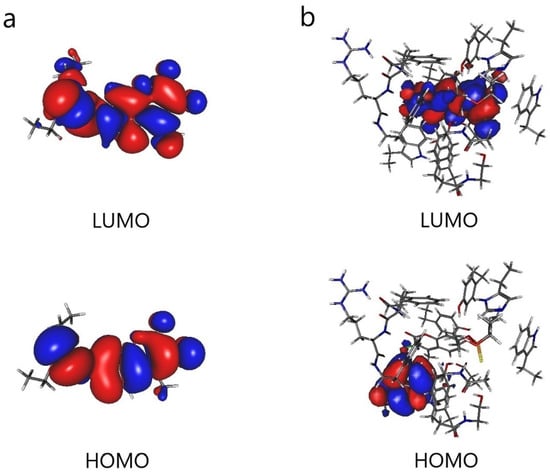
Figure 5.
Frontier molecular orbitals (HOMO and LUMO): (a) coumaphos (Cou) and (b) AChE cluster model with coumaphos (AChE_1_Cou).
3.4. Electronic Excitation Spectrum
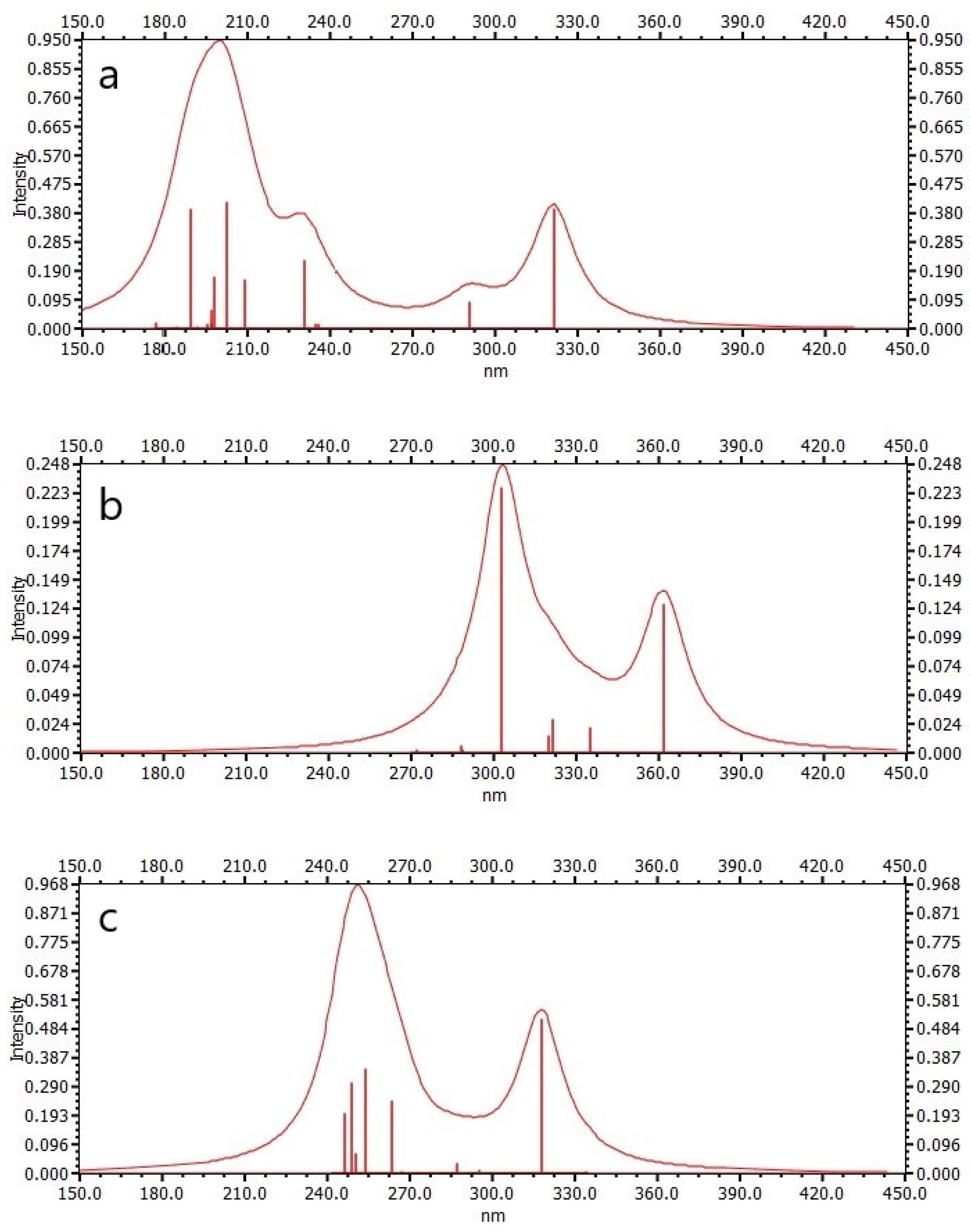
UV-Vis absorption spectroscopy is widely used to explore the structural changes of proteins and study the formation of protein–ligand complexes. The interaction of Cou, AChE_1_cou, and BChE_1_cou was studied by UV-VIS spectroscopy here. As shown in Figure 6, the two peak wavelengths of Cou are 200 nm and 320 nm, the two peak wavelengths of AChE_1_cou are 303 nm and 363 nm, and the two peak wavelengths of BChE_1_cou are 250 nm and 316 nm, respectively. It can be observed that after binding the enzyme (AChE/BChE) with Cou, the wavelengths of both peaks will be red-shifted to the long wavelengths. It can also be observed that AChE produces a hypochromic effect, while BChE produces a hyperchromic effect. This means that due to changes in protein conformation, the polarity and hydrophilicity of the microenvironment surrounding aromatic amino acid residues of endogenous enzymes have changed [24]. The wavelength change of AChE is larger than that of BchE, because there are more hydrophobic interactions and hydrogen bonding interactions between Cou and AchE, and the electron transfer ability is stronger, resulting in the maximum reaction enthalpy.
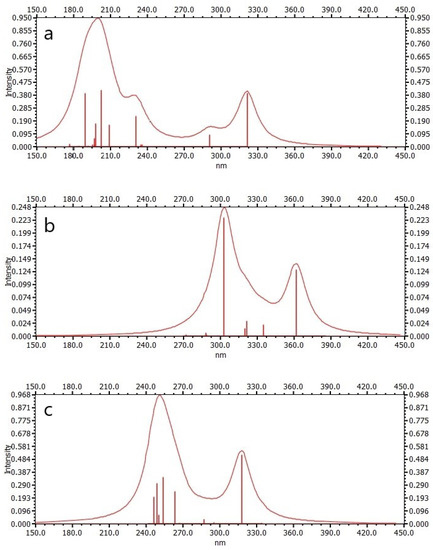
Figure 6.
Electronic excitation spectra: (a) coumaphos (Cou), (b) AchE cluster model with coumaphos (AchE_1_Cou), and (c) BChE cluster model with coumaphos (BChE_1_Cou).
4. Conclusions
The equilibrium structure, reaction enthalpy, molecular orbital, and electronic spectra of OP pesticides that interacted with molecular cluster models of AChE and BChE enzymes were calculated by the quantum chemical method. The oxygen and sulfur atoms in the OP pesticides act as the acceptor of the hydrogen bonds, which form stable hydrogen bonds with the hydrogen atoms in the amino acid residues. The binding tendency of AChE is greater than that of BChE due to the hydrogen bonding interaction. According to the HSAB principle, AChE has good electron transfer ability and a larger energy gap change. Furthermore, the red shift phenomenon of the calculated electronic spectrum can also prove that AChE is a suitable biosensing material for detecting the coumaphos (Cou) pesticide.
Author Contributions
Conceptualization, S.-C.C. and C.M.C.; methodology, S.-C.C. and C.M.C.; software, S.-C.C.; validation, C.M.C.; formal analysis, S.-C.C. and C.M.C.; investigation, S.-C.C.; resources, C.M.C.; data curation, S.-C.C.; writing—original draft preparation, S.-C.C.; writing—review and editing, C.M.C.; visualization, S.-C.C. and C.M.C.; supervision, C.M.C.; project administration, C.M.C.; funding acquisition, C.M.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Science Council of Taiwan, Republic of China and grant number [MOST 111-2321-B-005-004].
Data Availability Statement
Not applicable.
Acknowledgments
The authors thank the National Science Council of Taiwan, Republic of China, MOST 111-2321-B-005-004 for providing financial support. Computer time was provided by the National Center for High-Performance Computing.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hertz-Picciotto, I.; Sass, J.B.; Engel, S.; Bennett, D.H.; Bradman, A.; Eskenazi, B.; Lanphear, B.; Whyatt, R. Organophosphate exposures during pregnancy and child neurodevelopment: Recommendations for essential policy reforms. PLoS Med. 2018, 15, e1002671. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Sun, C.; Vijayaraghavan, R.; Zhou, F.; Zhang, X.; MacFarlane, D.R. Long lifetime photoluminescence in N, S co-doped carbon quantum dots from an ionic liquid and their applications in ultrasensitive detection of pesticides. Carbon 2016, 104, 33–39. [Google Scholar] [CrossRef]
- Jain, M.; Yadav, P.; Joshi, A.; Kodgire, P. Advances in detection of hazardous organophosphorus compounds using organophosphorus hydrolase based biosensors. Crit. Rev. Toxicol. 2019, 49, 387–410. [Google Scholar] [CrossRef] [PubMed]
- Pachapur, P.K.; Larios Martínez, A.D.; Pulicharla, R.; Pachapur, V.L.; Brar, S.K.; Galvez-Cloutier, R. Chapter 11—Advances in protein/enzyme-based biosensors for the detection of pesticide contaminants in the environment. In Tools, Techniques and Protocols for Monitoring Environmental Contaminants; Kaur Brar, S., Hegde, K., Pachapur, V.L., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 231–243. [Google Scholar]
- Kaur, J.; Singh, P.K. Enzyme-based optical biosensors for organophosphate class of pesticide detection. Phys. Chem. Chem. Phys. 2020, 22, 15105–15119. [Google Scholar] [CrossRef]
- Dennison, M.J.; Turner, A.P.F. Biosensors for environmental monitoring. Biotechnol. Adv. 1995, 13, 1–12. [Google Scholar] [CrossRef]
- Fu, H.; Xia, Y.; Chen, Y.; Xu, T.; Xu, L.; Guo, Z.; Xu, H.; Xie, H.Q.; Zhao, B. Acetylcholinesterase Is a Potential Biomarker for a Broad Spectrum of Organic Environmental Pollutants. Environ. Sci. Technol. 2018, 52, 8065–8074. [Google Scholar] [CrossRef]
- Reshma, R.; Gupta, B.; Sharma, R.; Ghosh, K.K. Facile and visual detection of acetylcholinesterase inhibitors by carbon quantum dots. New J. Chem. 2019, 43, 9924–9933. [Google Scholar] [CrossRef]
- Mladenović, M.; Arsić, B.B.; Stanković, N.; Mihović, N.; Ragno, R.; Regan, A.; Milićević, J.S.; Trtić-Petrović, T.M.; Micić, R. The Targeted Pesticides as Acetylcholinesterase Inhibitors: Comprehensive Cross-Organism Molecular Modelling Studies Performed to Anticipate the Pharmacology of Harmfulness to Humans In Vitro. Molecules 2018, 23, 2192. [Google Scholar] [CrossRef]
- Nicolet, Y.; Lockridge, O.; Masson, P.; Fontecilla-Camps, J.C.; Nachon, F. Crystal structure of human butyrylcholinesterase and of its complexes with substrate and products. J. Biol. Chem. 2003, 278, 41141–41147. [Google Scholar] [CrossRef]
- Sussman, J.L.; Silman, I. Acetylcholinesterase: Structure and use as a model for specific cation-protein interactions. Curr. Opin. Struct. Biol. 1992, 2, 721–729. [Google Scholar] [CrossRef]
- Bušić, V.; Katalinić, M.; Šinko, G.; Kovarik, Z.; Gašo-Sokač, D. Pyridoxal oxime derivative potency to reactivate cholinesterases inhibited by organophosphorus compounds. Toxicol. Lett. 2016, 262, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Himo, F.; de Visser, S.P. Status report on the quantum chemical cluster approach for modeling enzyme reactions. Commun. Chem. 2022, 5, 29. [Google Scholar] [CrossRef]
- Prejanò, M.; Marino, T.; Russo, N. QM Cluster or QM/MM in Computational Enzymology: The Test Case of LigW-Decarboxylase. Front. Chem. 2018, 6, 249. [Google Scholar] [CrossRef] [PubMed]
- Himo, F. Recent Trends in Quantum Chemical Modeling of Enzymatic Reactions. J. Am. Chem. Soc. 2017, 139, 6780–6786. [Google Scholar] [CrossRef] [PubMed]
- Siegbahn, P.E.M.; Himo, F. The quantum chemical cluster approach for modeling enzyme reactions. WIREs Comput. Mol. Sci. 2011, 1, 323–336. [Google Scholar] [CrossRef]
- Siegbahn, P.E.M.; Himo, F. Recent developments of the quantum chemical cluster approach for modeling enzyme reactions. JBIC J. Biol. Inorg. Chem. 2009, 14, 643–651. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Stewart, J.J.P. Optimization of parameters for semiempirical methods VI: More modifications to the NDDO approximations and re-optimization of parameters. J. Mol. Model. 2013, 19, 1–32. [Google Scholar] [CrossRef]
- Klamt, A.; Schüürmann, G. COSMO: A new approach to dielectric screening in solvents with explicit expressions for the screening energy and its gradient. J. Chem. Soc. Perkin Trans. 1993, 2, 799–805. [Google Scholar] [CrossRef]
- Allouche, A.-R. Gabedit—A graphical user interface for computational chemistry softwares. J. Comput. Chem. 2011, 32, 174–182. [Google Scholar] [CrossRef]
- Neese, F. The ORCA program system. WIREs Comput. Mol. Sci. 2012, 2, 73–78. [Google Scholar] [CrossRef]
- Neese, F. Software update: The ORCA program system, version 4.0. WIREs Comput. Mol. Sci. 2018, 8, e1327. [Google Scholar] [CrossRef]
- Cao, Q.; Huang, Y.; Zhu, Q.F.; Song, M.; Xiong, S.; Manyande, A.; Du, H. The mechanism of chlorogenic acid inhibits lipid oxidation: An investigation using multi-spectroscopic methods and molecular docking. Food Chem. 2020, 333, 127528. [Google Scholar] [CrossRef] [PubMed]
- Verdonk, M.L.; Cole, J.C.; Hartshorn, M.J.; Murray, C.W.; Taylor, R.D. Improved Protein–Ligand Docking Using GOLD. Proteins Struct. Funct. Bioinform. 2003, 52, 609–623. [Google Scholar] [CrossRef] [PubMed]
- Plewczynski, D.; Lazniewski, M.; Augustyniak, R.; Ginalski, K. Can we trust docking results? Evaluation of seven commonly used programs on PDBbind database. J. Comput. Chem. 2011, 32, 742–755. [Google Scholar] [CrossRef]
- Bikadi, Z.; Hazai, E. Application of the PM6 semi-empirical method to modeling proteins enhances docking accuracy of AutoDock. J. Cheminform. 2009, 1, 15. [Google Scholar] [CrossRef]
- Martin, B.P.; Brandon, C.J.; Stewart, J.J.; Braun-Sand, S.B. Accuracy issues involved in modeling in vivo protein structures using PM7. Proteins 2015, 83, 1427–1435. [Google Scholar] [CrossRef]
- Sulimov, A.V.; Kutov, D.C.; Katkova, E.V.; Sulimov, V.B. Combined Docking with Classical Force Field and Quantum Chemical Semiempirical Method PM7. Adv. Bioinform. 2017, 2017, 7167691. [Google Scholar] [CrossRef]
- Rocha, S.; Sant’Anna, C.M.R. A procedure combining molecular docking and semiempirical method PM7 for identification of selective Shp2 inhibitors. Biopolymers 2019, 110, e23320. [Google Scholar] [CrossRef]
- Tõugu, V. Acetylcholinesterase: Mechanism of Catalysis and Inhibition. Curr. Med. Chem. Cent. Nerv. Syst. Agents 2001, 1, 155–170. [Google Scholar] [CrossRef]
- De Boer, D.; Nguyen, N.; Mao, J.; Moore, J.; Sorin, E.J. A Comprehensive Review of Cholinesterase Modeling and Simulation. Biomolecules 2021, 11, 580. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.-H.; Li, Y.; Wang, Y.-T.; Li, B.; Zhang, Y.-H. Investigation of interaction modes involved in alkaline phosphatase and organophosphorus pesticides via molecular simulations. Food Chem. 2018, 254, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Yasser, E.-N. Reactions of Acetylcholinesterase with Organophosphorus Insecticides. J. Pharmacol. Clin. Toxicol. 2018, 6, 1108. [Google Scholar]
- Naine, S.J.; Devi, C.S.; Mohanasrinivasan, V.; Doss, C.G.P.; Kumar, D.T. Binding and molecular dynamic studies of sesquiterpenes (2R-acetoxymethyl-1,3,3-trimethyl-4t-(3-methyl-2-buten-1-yl)-1t-cyclohexanol) derived from marine Streptomyces sp. VITJS8 as potential anticancer agent. Appl. Microbiol. Biotechnol. 2015, 100, 2869–2882. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Barron, M.G. A mechanism-based 3D-QSAR approach for classification and prediction of acetylcholinesterase inhibitory potency of organophosphate and carbamate analogs. J. Comput. Aided Mol. Des. 2016, 30, 347–363. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.Q.; Zhu, K.K.; Zhang, J.; Song, J.L.; Muehlmann, L.A.; Jiang, C.S.; Liu, C.L.; Zhang, H. Molecular-docking-guided design and synthesis of new IAA-tacrine hybrids as multifunctional AChE/BChE inhibitors. Bioorg. Chem. 2019, 83, 277–288. [Google Scholar] [CrossRef]
- LoPachin, R.M.; Geohagen, B.C.; Nordstroem, L.U. Mechanisms of soft and hard electrophile toxicities. Toxicology 2019, 418, 62–69. [Google Scholar] [CrossRef]
- Melnikov, F.; Geohagen, B.C.; Gavin, T.; LoPachin, R.M.; Anastas, P.T.; Coish, P.; Herr, D.W. Application of the hard and soft, acids and bases (HSAB) theory as a method to predict cumulative neurotoxicity. Neurotoxicology 2020, 79, 95–103. [Google Scholar] [CrossRef]
- Waiskopf, N.; Shweky, I.; Lieberman, I.; Banin, U.; Soreq, H. Quantum dot labeling of butyrylcholinesterase maintains substrate and inhibitor interactions and cell adherence features. ACS Chem. Neurosci. 2011, 2, 141–150. [Google Scholar] [CrossRef]
- Yang, S.; Liu, J.; Zheng, H.; Zhong, J.; Zhou, J. Simulated revelation of the adsorption behaviours of acetylcholinesterase on charged self-assembled monolayers. Nanoscale 2020, 12, 3701–3714. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).