Biosynthesis of Ag Nanoparticles Using Caralluma acutangula Extract and Its Catalytic Functionality towards Degradation of Hazardous Dye Pollutants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of CAExtract
2.3. GC-MS Analysis of CAExtract
2.4. Preparation of CA-AgNPs
2.5. Characterization of CA-AgNPs
2.6. Catalytic Degradation of MB and CR Dyes
3. Results and Discussion
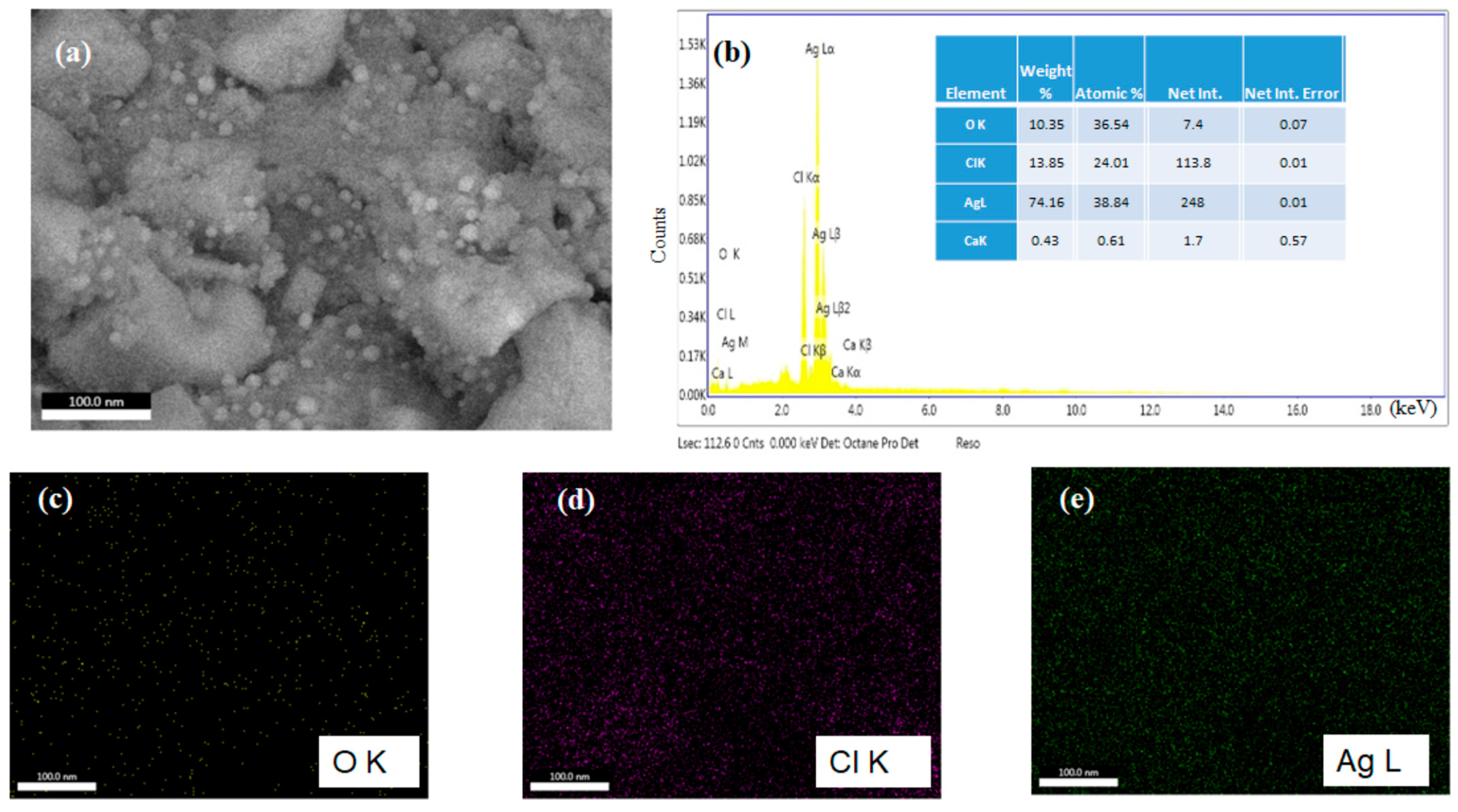
3.1. Phytochemical Analysis by GC-MS and CA-AgNPs Charecterization
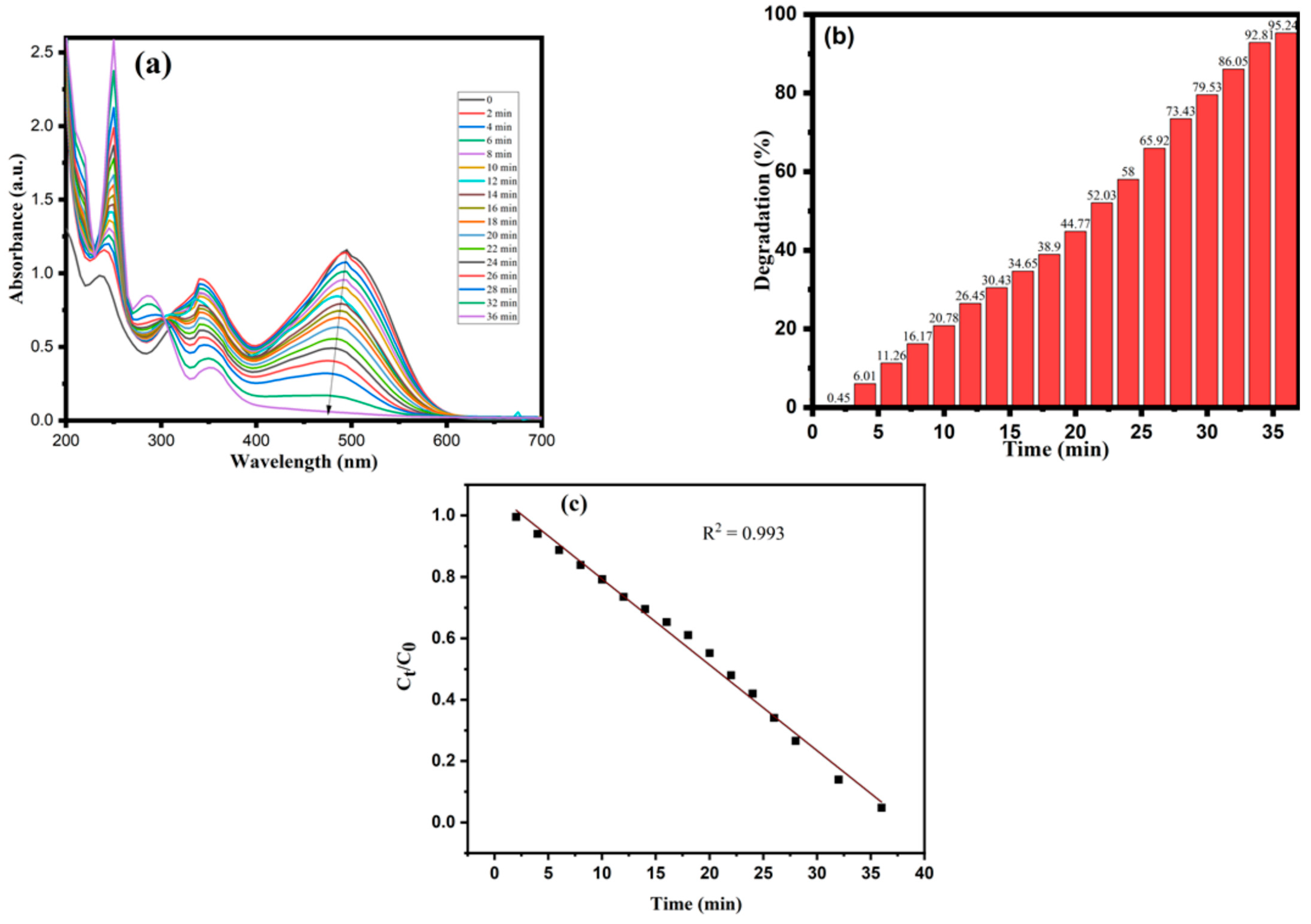
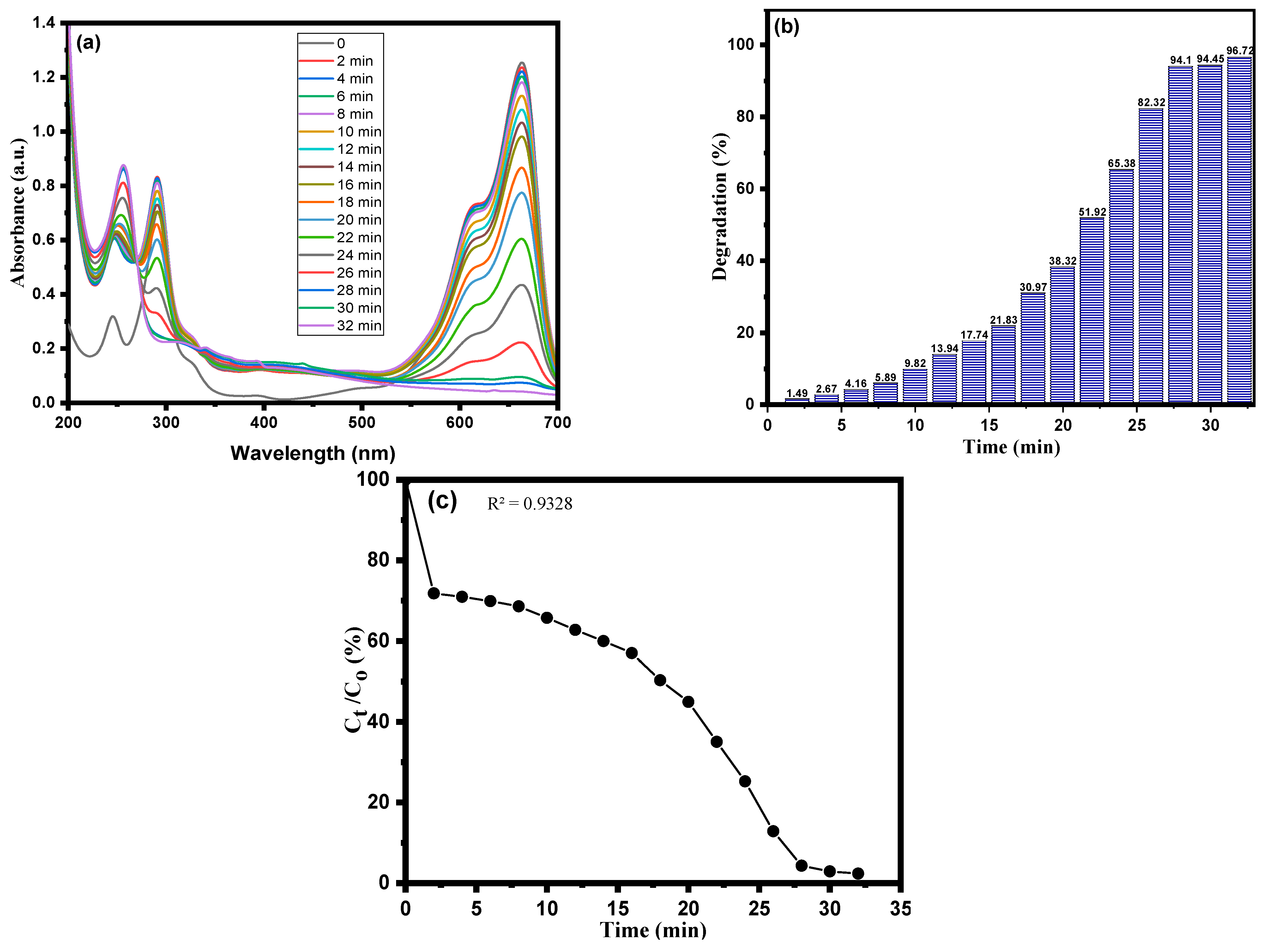
3.2. Dye Degradation by CA-AgNPs
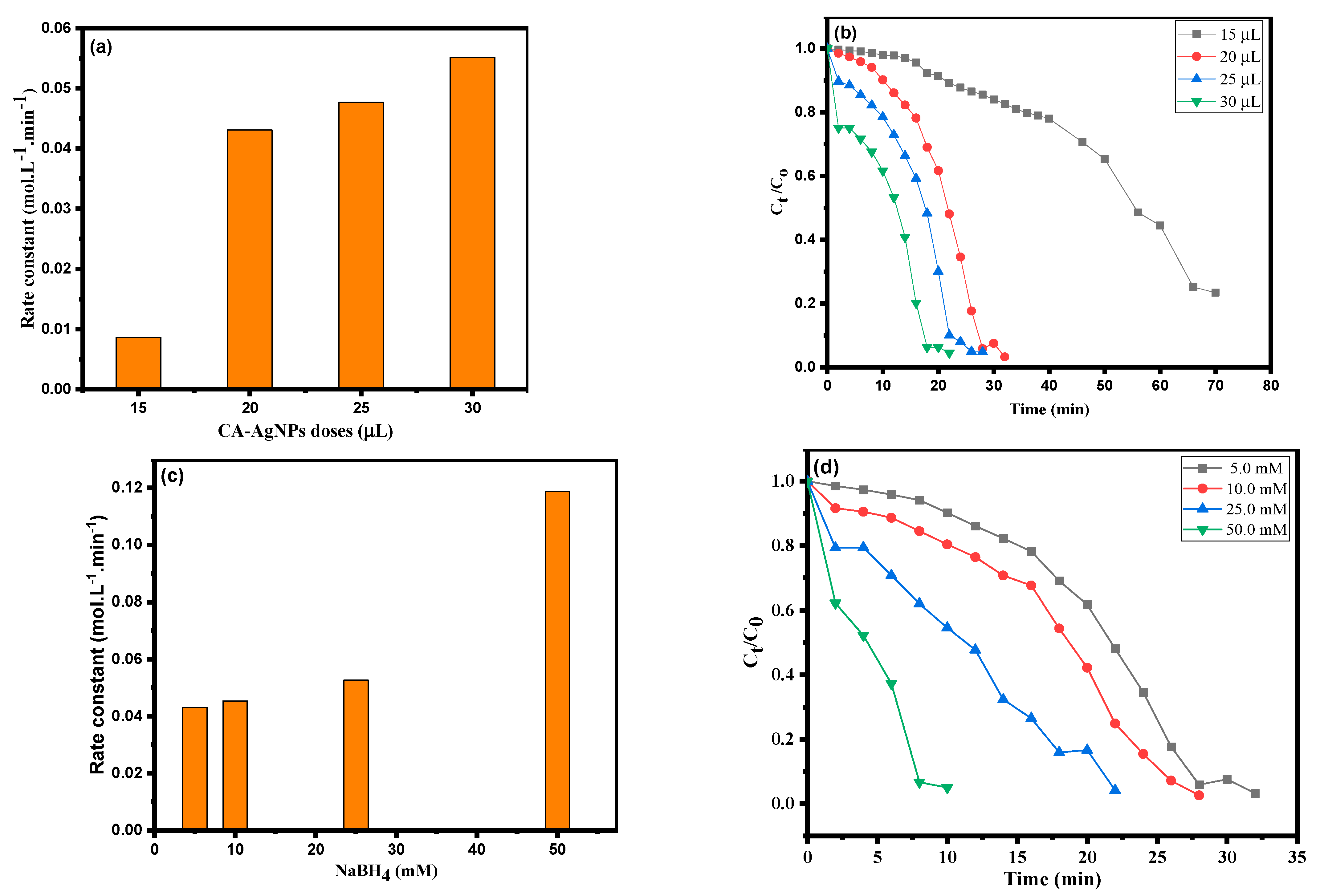
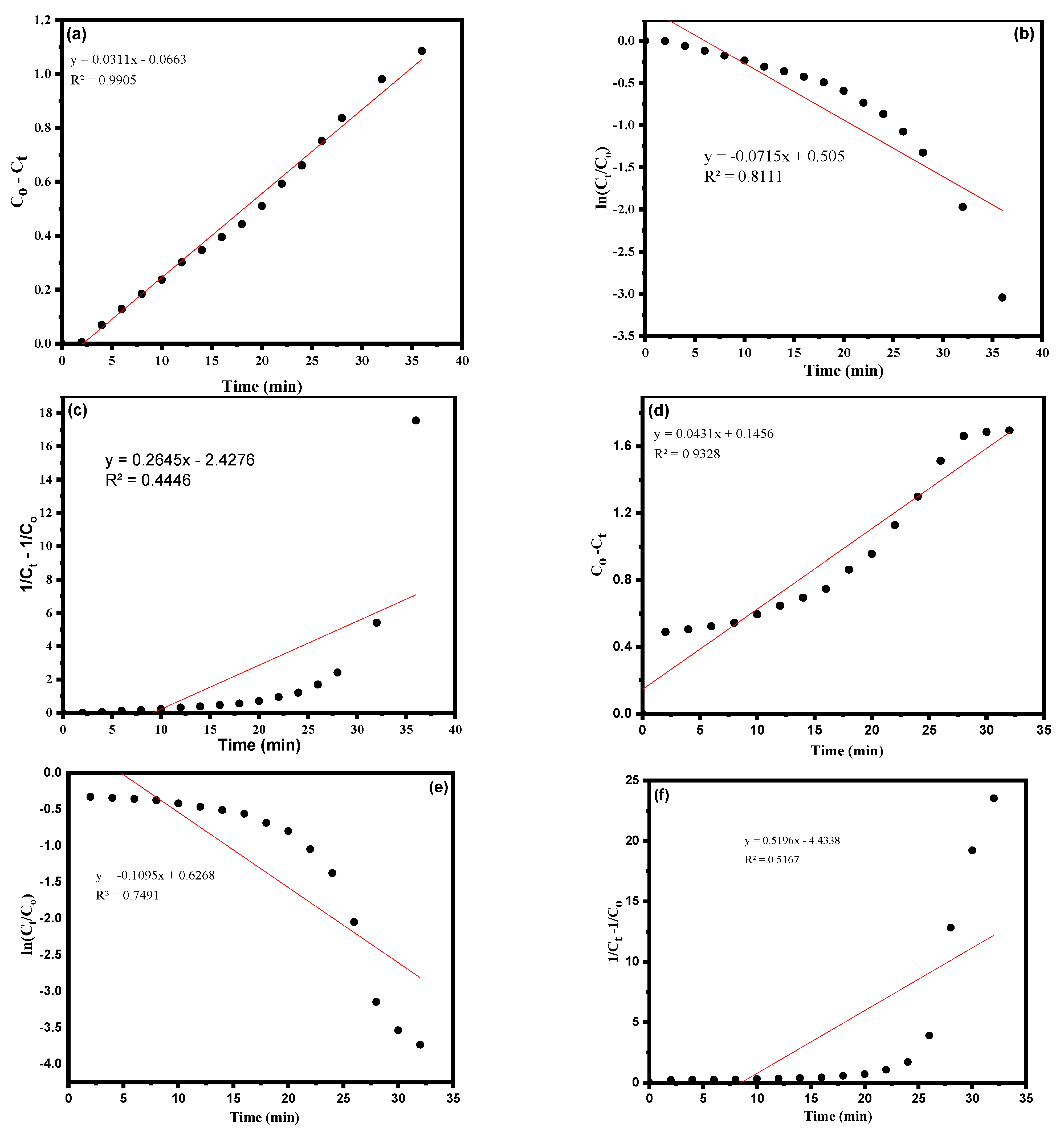
3.3. Kinetic Study Using CA-AgNPs Catalyst
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ali, F.; Khan, S.B.; Kamal, T.; Alamry, K.A.; Asiri, A.M. Chitosan-titanium oxide fibers supported zero-valent nanoparticles: Highly efficient and easily retrievable catalyst for the removal of organic pollutants. Sci. Rep. 2018, 8, 6260. [Google Scholar] [CrossRef] [Green Version]
- Tsunoyama, H.; Sakurai, H.; Ichikuni, N.; Negishi, Y.; Tsukuda, T. Colloidal gold nanoparticles as catalyst for carbon-carbon bond formation: Application to aerobic homocoupling of phenylboronic acid in water. Langmuir 2004, 20, 11293–11296. [Google Scholar] [CrossRef]
- Galletto, P.; Brevet, P.F.; Girault, H.H.; Antoine, R.; Broyer, M. Enhancement of the second harmonic response by adsorbates on gold colloids: The effect of aggregation. J. Phys. Chem. B 1999, 103, 8706–8710. [Google Scholar] [CrossRef]
- Maier, S.A.; Brongersma, M.L.; Kik, P.G.; Meltzer, S.; Requicha, A.A.G.; Atwater, H.A. Plasmonics a route to nanoscale optical devices. Adv. Mater. 2001, 13, 1501–1505. [Google Scholar] [CrossRef]
- Han, M.; Gao, X.; Nie, S.J.Z. Quantum-dot-tagged microbeads for multiplexed optical coding of biomolecules. Nat. Biotechnol. 2001, 19, 631–635. [Google Scholar] [CrossRef]
- West, J.L.; Halas, N.J. Engineered nanomaterials for biophotonics applications: Improving sensing, imaging, and therapeutics. Annu. Rev. Biomed. Eng. 2003, 5, 285–292. [Google Scholar] [CrossRef] [Green Version]
- Nicewarner-Pena, S.R.; Freeman, R.G.; Reiss, B.D.; He, L.; Peña, D.J.; Walton, I.D.; Cromer, R.; Keating, C.D.; Natan, M.J. Submicrometer metallic barcodes. Science 2001, 294, 137–141. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, X.; Li, Y.; Liu, H.; Wang, Y.; Chang, Q.; Jiao, W.; Song, Y. Saturable absorption and reverse saturable absorption in platinum nanoparticles. Opt. Commun. 2005, 251, 429–433. [Google Scholar] [CrossRef]
- Yanez-Sedeno, P.; Pingarron, J.M. Gold nanoparticle-based electrochemical biosensors. Anal. Bioanal. Chem. 2005, 382, 884–886. [Google Scholar] [CrossRef]
- Dang, T.M.; Le, T.T.; Fribourg-Blanc, E.; Dang, M.C. The influence of solvents and surfactants on the preparation of copper nanoparticles by a chemical reduction method. Adv. Nat. Sci. Nanosci. Nanotechnol. 2011, 2, 025004. [Google Scholar] [CrossRef]
- Zhu, H.; Zhang, C.; Yin, Y. Rapid synthesis of copper nanoparticles by sodium hypophosphite reduction in ethylene glycol under microwave irradiation. J. Cryst. J. Growth 2004, 270, 722–728. [Google Scholar] [CrossRef]
- Thakkar, K.N.; Mhatre, S.S.; Parikh, R.Y. Biological synthesis of metallic nanoparticles. Nanomed. Nanotechnol. Biol. Med. 2010, 6, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, K.B.; Sakthivel, N. Biological synthesis of metal nanoparticles by microbes. Adv. Colloid Interface Sci. 2010, 156, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Halim, E.S.; El-Rafie, M.H.; Al-Deyab, S.S. Polyacrylamide/guar gum graft copolymer for preparation of silver nanoparticles. Carbohydr. Polym. 2011, 85, 692–697. [Google Scholar] [CrossRef]
- Jasbi, A.R. Chemistry and biological activity of secondary metabolites in Euphorbia from Iran. Phytochemistry 2006, 67, 1977–1984. [Google Scholar] [CrossRef]
- Dubey, S.; Lahtinen, M.; Sillanpa, M. Tansy fruit mediated greener synthesis of silver and gold nanoparticles. Process Biochem. 2010, 45, 1065–1071. [Google Scholar] [CrossRef]
- Zhan, G.; Huang, J.; Du, M.; Abdul-Rauf, I.; Li, Q.; Ma, Y. Green synthesis of Au–Pd bimetallic nanoparticles: Single-step bioreduction method with plant extract. Mat. Lett. 2011, 65, 2989–2991. [Google Scholar] [CrossRef]
- Huang, X.; Wu, H.; Pu, S.; Zhang, W.; Liao, X.; Shi, B. One-step room-temperature synthesis of Au@Pd core–shell nanoparticles with tunable structure using plant tannin as reductant and stabilizer. Green Chem. 2011, 13, 950–957. [Google Scholar] [CrossRef]
- Raveendran, P.; Fu, J.; Wallen, S.L. A simple and “green” method for the synthesis of Au, Ag, and Au–Ag alloy nanoparticles. Green Chem. 2006, 8, 34–38. [Google Scholar] [CrossRef]
- Nam, J.; Won, N.; Jin, H.; Chung, H.; Kim, S. pH-induced aggregation of gold nanoparticles for photothermal cancer therapy. J. Am. Chem Soc. 2009, 131, 13639–13645. [Google Scholar] [CrossRef]
- Narayanan, K.B.; Sakthivel, N. Synthesis and characterization of nano-gold composite using Cylindrocladium floridanum and its heterogeneous catalysis in the degradation of 4-nitrophenol. J. Hazard. Mater. 2011, 189, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, X.; Ai, N.; Hao, J.; Chen, Q.; Strauf, S.; Shi, Y. Silver nanoparticle doped TiO2 nanofiber dye sensitized solar cells. Chem. Phys. Lett. 2011, 514, 141–145. [Google Scholar] [CrossRef]
- Fayaz, A.M.; Girilal, M.; Mahdy, S.A.; Somsundar, S.S.; Venkatesan, R.; Kalaichelvan, P.T. Vancomycin bound biogenic gold nanoparticles: A different perspective for development of anti VRSA agents. Process Biochem 2011, 46, 636–641. [Google Scholar] [CrossRef]
- Raj, S.; Singh, H.; Trivedi, R.; Soni, V. Biogenic synthesis of AgNPs employing Terminalia arjuna leaf extract and its efficacy towards catalytic degradation of organic dyes. Sci. Rep. 2020, 10, 9616. [Google Scholar] [CrossRef]
- Atarod, M.; Nasrollahzadeh, M.; Sajadi, S.M. Euphorbia heterophylla leaf extract mediated green synthesis of Ag/TiO2 nanocomposite and investigation of its excellent catalytic activity for reduction of variety of dyes in water. J. Colloid Interface Sci. 2015, 462, 272–279. [Google Scholar] [CrossRef]
- Awad, M.A.; Hendi, A.A.; Ortashi, K.M.; Alzahrani, B.; Soliman, D.; Alanazi, A.; Alenazi, W.; Taha, R.M.; Ramadan, R.; El-Tohamy, M.; et al. Biogenic synthesis of silver nanoparticles using Trigonella foenum-graecum seed extract: Characterization, photocatalytic and antibacterial activities. Sens. Actuators A Phys. 2021, 323, 112670. [Google Scholar] [CrossRef]
- Radini, I.A.; Hasan, N.; Khan, M.A.M.Z. Biosynthesis of iron nanoparticles using Trigonella foenum-graecum seed extract for photo catalytic methyl orange dye degradation and antibacterial applications. J. Photochem. Photobiol. B Biol. 2018, 183, 154–163. [Google Scholar] [CrossRef]
- Reddy, N.K.; Hoskote, B.R.; Kumar, A.K.; Mandal, B.K. Bio fabricated silver nanoparticles as green catalyst in the degradation of different textile dyes. J. Environ. Chem. Eng. 2016, 4, 56–64. [Google Scholar]
- Al-Zaban, M.I.; Mohamad, A.M.; Al-Harbi, M.A. Catalytic Degradation of Methylene Blue Using Silver Nanoparticles Synthesized by Honey. Saudi J. Biol. Sci. 2021, 28, 2007–2013. [Google Scholar] [CrossRef] [PubMed]
- Edison, T.N.J.I.; Sethuraman, M.G. Instant green synthesis of silver nanoparticles using Terminalia chebula fruit extract and evaluation of their catalytic activity on reduction of methylene blue. Process Biochem. 2012, 47, 1359–5113. [Google Scholar] [CrossRef]
- Edison, T.N.J.I.; Atchudan, R.; Kamal, C.; Lee, Y.R. Caulerpa racemosa: A marine green alga for eco-friendly synthesis of silver nanoparticles and its catalytic degradation of methylene blue. Bioprocess Biosyst. Eng. 2016, 39, 1401–1408. [Google Scholar] [CrossRef] [PubMed]
- Rostami-Vartooni, A.; Nasrollahzadeh, M.; Alizadeh, M. Green synthesis of seashell supported silver nanoparticles using Bunium persicum seeds extract: Application of the particles for catalytic reduction of organic dyes. J. Colloid Interface Sci. 2016, 470, 268–275. [Google Scholar] [CrossRef]
- Roy, K.; Sarkar, C.K.; Ghosh, K. Photocatalytic activity of biogenic silver nanoparticles synthesized using yeast (Saccharomyces cerevisiae) extract. Appl. Nanosci. 2015, 15, 953–959. [Google Scholar] [CrossRef]
- Saha, J.; Begum, A.; Mukherjee, A.; Kumar, S. A novel green synthesis of silver nanoparticles and their catalytic action in reduction of Methylene Blue dye. Sustain. Environ. Res. 2017, 27, 245–250. [Google Scholar] [CrossRef]
- Sharma, P.; Pant, S.; Rai, S.; Yadav, R.B.; Dave, V. Green Synthesis of Silver Nanoparticle Capped with Allium cepa and Their Catalytic Reduction of Textile Dyes: An Ecofriendly Approach. J. Polym. Environ. 2018, 26, 1795–1803. [Google Scholar] [CrossRef]
- Suvith, V.S.; Philip, D. Catalytic degradation of methylene blue using biosynthesized gold and silver nanoparticles. Mol. Biomol. Spectrosc. 2014, 118, 526–532. [Google Scholar] [CrossRef]
- Al-Yahya, M.; Al-Meshal, I.; Mossa, J.; Al-Badr, A.; Tariq, M. Saudi Plants: A Phytochemical and Biological Approach; King Saud University Press: Riyadh, Saudi Arabia, 1990; p. 523. [Google Scholar]
- Al-Faifi, Z.I.A.; Yahya, M.; Aly, M.S.; Al-Turki, T.A. In vitro Anticancer Activity of Caralluma acutangula (Decne.) N.E.Br. Extract. Int. J. Pharm. Sci. Rev. Res. 2016, 38, 59–63. [Google Scholar]
- Chaudhary, S.A.; Al-Wataniya, M.; Al-Watani, A.M. Flora of the Kingdom of Saudi Arabia; National Agriculture and Water Research Center: Riyadh, Saudi Arabia, 2001; p. 2. [Google Scholar]
- Halaweish, F.; Huntimer, E.; Khalil, A. Polyoxy pregnane glycosides from Caralluma retrospiciens. Phytochem. Anal. 2004, 15, 189–194. [Google Scholar] [CrossRef] [PubMed]
- DParé; N’do, J.Y.; Hilou, A.; Sombie, N.E.; Kpemissi, M.; Bogdan, S. Anti-Obesogenic Potential of Caralluma acutangula Bioactive Fractions. Sch. Int. J. Biochem. 2020, 3, 183–192. [Google Scholar]
- Liu, J.; Yong, H.; Yao, X.; Hu, H.; Yun, D.; Xiao, L. Recent advances in phenolic–protein conjugates: Synthesis, characterization, biological activities and potential applications. RSC Adv. 2019, 9, 35825–35840. [Google Scholar] [CrossRef] [Green Version]
- Karimova, N.V.; Luo, M.; Grassian, V.H.; Gerber, B. Absorption spectra of benzoic acid in water at different pH and in the presence of salts: Insights from the integration of experimental data and theoretical cluster models. Phys. Chem. Chem. Phys. 2020, 22, 5046–5056. [Google Scholar] [CrossRef] [PubMed]
- Sastry, M.; Mayya, K.S.; Bandyopadhyay, K. pH Dependent changes in the optical properties of carboxylic acid derivatized silver colloidal particles. Colloids Surf. A Physicochem. Eng. Asp. 1997, 127, 221–228. [Google Scholar] [CrossRef]
- Sankar, R.; Karthik, A.; Prabu, A.; Karthik, S.; Shivashangari, S.K.; Ravikumar, S.V. Origanum vulgare mediated biosynthesis of silver nanoparticles for its antibacterial and anticancer activity. Colloids Surf. B Biointerfaces 2013, 108, 80–84. [Google Scholar] [CrossRef]
- Sireesha, M.; Venkata, N.R.; Suresh, B.K.; Sreenivasulu, M. Phytochemical library of Carulluma genus. Int. J. Res. Pharm. Sci. 2018, 9, 1201–1213. [Google Scholar]
- Gusrizal, G.; Santosa, S.J.; Kunarti, E.S.; Rusdiarso, B. Dual Function of p-Hydroxybenzoic Acid as Reducing and Capping Agent in Rapid and Simple Formation of Stable Silver Nanoparticles. Int. J. ChemTech Res. 2016, 9, 472–482. [Google Scholar]
- Gusrizal, G.; Santosa, S.J.; Kunarti, E.S.; Rusdiarso, B. Synthesis of Silver Nanoparticles by Reduction of Silver Ion with m-Hydroxybenzoic Acid. Asian J. Chem. 2017, 29, 1417–1422. [Google Scholar] [CrossRef]
- Susanthy, D.; Wahyuni, E.T.F.; Santosa, S. Synthesis of silver nanoparticles using o-hydroxybenzoic, p-hydroxybenzoic, and o,p-dihydroxybenzoic acids as reducing agents. Mater. Sci. Forum 2017, 901, 26–31. [Google Scholar] [CrossRef]
- Marimuthu, S.; Rahuman, A.A.; Rajakumar, G.; Santhoshkumar, T.; Kirthi, A.V.; Jayaseelan, C.; Bagavan, A.; Zahir, A.A.; Elango, G.; Kamaraj, C. Evaluation of green synthesized silver nanoparticles against parasites. Parasitol. Res. 2011, 108, 1541–1549. [Google Scholar] [CrossRef]
- Azira, W.M.K.W.; Kamyar, K.; Davoud, S.J.S.; Azizi, O.N.; Wahyuny, C.J.N.; Mohd, H.N. Biosynthesized silver nanoparticles by aqueous stem extract of entada spiralis and screening of their biomedical activity. Front. Chem. 2020, 8, 620. [Google Scholar]
- Lin-Vien, D.; Colthup, N.B.; Fateley, W.G.; Grasselli, J.G. Chapter 9—Compounds containing the carbonyl group. In The Handbook of Infrared and Raman Characteristic Frequencies of Organic Molecules; Lin-Vien, D., Colthup, N.B., Fateley, W.G., Grasselli, J.G., Eds.; Academic Press: Cambridge, MA, USA, 1991; pp. 117–154. [Google Scholar]
- Ramirez, D.; Jaramillo, F. Facile one-pot synthesis of uniform silver nanoparticles and growth mechanism. Dyna 2016, 83, 165–170. [Google Scholar] [CrossRef]
- Aziz, A.; Khalid, M.; Akhtar, S.M.; Nadeem, M.; Gilani, Z.; Khan, M.; Rehman, J.; Ullah, Z.; Saleem, M. Structural, morphological and optical investigations of silver nanoparticles synthesized by sol-gel auto-combustion method. Dig. J. Nanomater. Biostructures 2018, 13, 679–683. [Google Scholar]
- Moldovan, B.; Sincari, V.; Perde-Schrepler, M.; David, L. Biosynthesis of silver nanoparticles using Ligustrum Ovalifolium fruits and their cytotoxic effects. Nanomaterials 2018, 8, 627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Q.; Cai, X.; Li, J.; Zheng, M.; Chen, Z.; Yu, C.P. Green synthesis of silver nanoparticles using tea leaf extract and evaluation of their stability and antibacterial activity. Coll. Surf. A Physicochem. Eng. Asp. 2014, 444, 226–231. [Google Scholar] [CrossRef]
- Yallappa, S.; Dhananjaya, J.M.L. Phytosynthesis of stable Au, Ag and Au-Ag alloy nanoparticles using J. sambac leaves extract, and their enhanced antimicrobial activity in presence of organic antimicrobials. Spectrochim. Acta. A Mol. Biomol. Spectrosc. 2015, 137, 236–243. [Google Scholar]
- David, L.; Moldovan, B. Green Synthesis of Biogenic Silver Nanoparticles for Efficient Catalytic Removal of Harmful Organic Dyes. Nanomaterials 2020, 10, 202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preetha, D.; Prachi, K.; Chirom, A.; Arun, R. Synthesis and characterization of silver nanoparticles using cannonball leaves and their cytotoxic activity against MCF-7 cell line. J. Nanotechnol. 2013, 2013, 598328. [Google Scholar]
- Roy, K.; Sarkar, C.K.; Ghosh, C.K. Plant-mediated synthesis of silver nanoparticles using parsley (Petroselinum crispum) leaf extract: Spectral analysis of the particles and antibacterial study. Appl. Nanosci. 2015, 5, 945–951. [Google Scholar] [CrossRef] [Green Version]
- Karuppiah, M.; Rajmohan, R. Green synthesis of silver nanoparticles using Ixora coccinea leaves extract. Mater. Lett. 2013, 97, 141–143. [Google Scholar] [CrossRef]
- Singha, S.; Neog, K.; Kalita, P.P.; Talukdar, N.; Sarma, M.P. Biological synthesis of silver nanoparticles by Neptunia oleraceae. Int. J. Basic Appl. Biol. 2014, 2, 55–59. [Google Scholar]
- Manikandan, V.; Velmurugan, P.; Park, J.H.; Chang, W.S.; Park, Y.J.; Jayanthi, P.; Cho, M.; Oh, B.T. Green synthesis of silver oxide nanoparticles and its antibacterial activity against dental pathogens. 3 Biotech 2017, 7, 72. [Google Scholar] [CrossRef] [Green Version]
- Sami, A.; David, E.; Fréchette, M. Procedure for evaluating the crystallinity from X-ray diffraction scans of high and low density polyethylene/SiO2 composites. In Annual Report Conference on Electrical Insulation and Dielectic Phenomena; IEEE: Manhattan, NY, USA, 2010; pp. 1–4. [Google Scholar]
- Naseem, K.; Begum, R.; Al-Sehemi, A.G.; Farooqi, Z.H. Catalytic reduction of toxic dyes in the presence of silver nanoparticles impregnated core-shell composite microgels. J. Clean. Prod. 2019, 211, 855–864. [Google Scholar] [CrossRef]
- Sengan, M.; Veeramuthu, D.; Veerappan, A. Photosynthesis of silver nanoparticles using Durio zibethinus aqueous extract and its application in catalytic reduction of nitroaromatics, degradation of hazardous dyes and selective colorimetric sensing of mercury ions. Mater. Res. Bull. 2018, 100, 386–393. [Google Scholar] [CrossRef]
- Fairuzi, A.A.; Bonnia, N.N.; Akhir, R.M.; Abrani, M.A.; Akil, H.M. Degradation of methylene blue using silver nanoparticles synthesized from imperata cylindrica aqueous extract. IOP Conf. Ser Earth Environ. Sci. 2018, 105, 012018. [Google Scholar] [CrossRef]
- Laouini, S.E.; Laouini, X.A.; Soldatov, A.V.; Algarni, H.; Tedjani, M.L.; Ali, G.A.M.; Barhoum, A. Green Synthesized of Ag/Ag2O Nanoparticles Using Aqueous Leaves Extracts of Phoenix dactylifera L. and Their Azo Dye Photodegradation. Membranes 2021, 11, 468. [Google Scholar] [CrossRef] [PubMed]

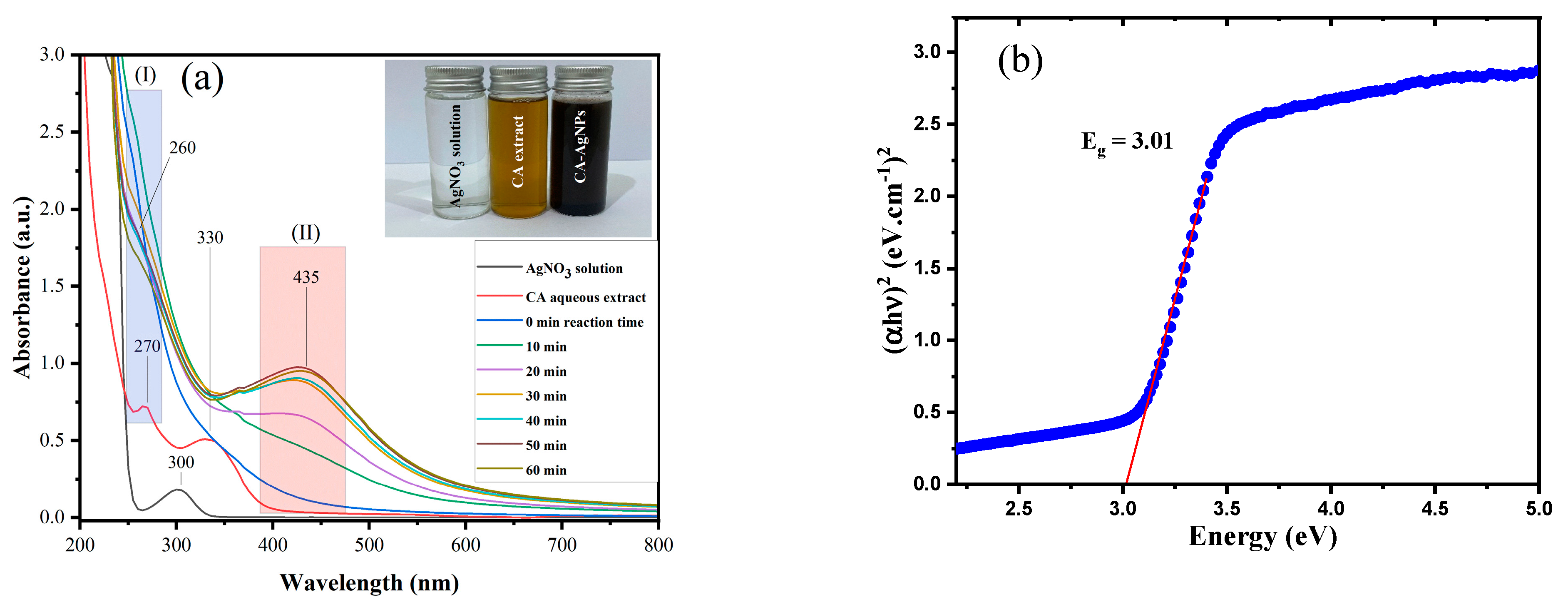
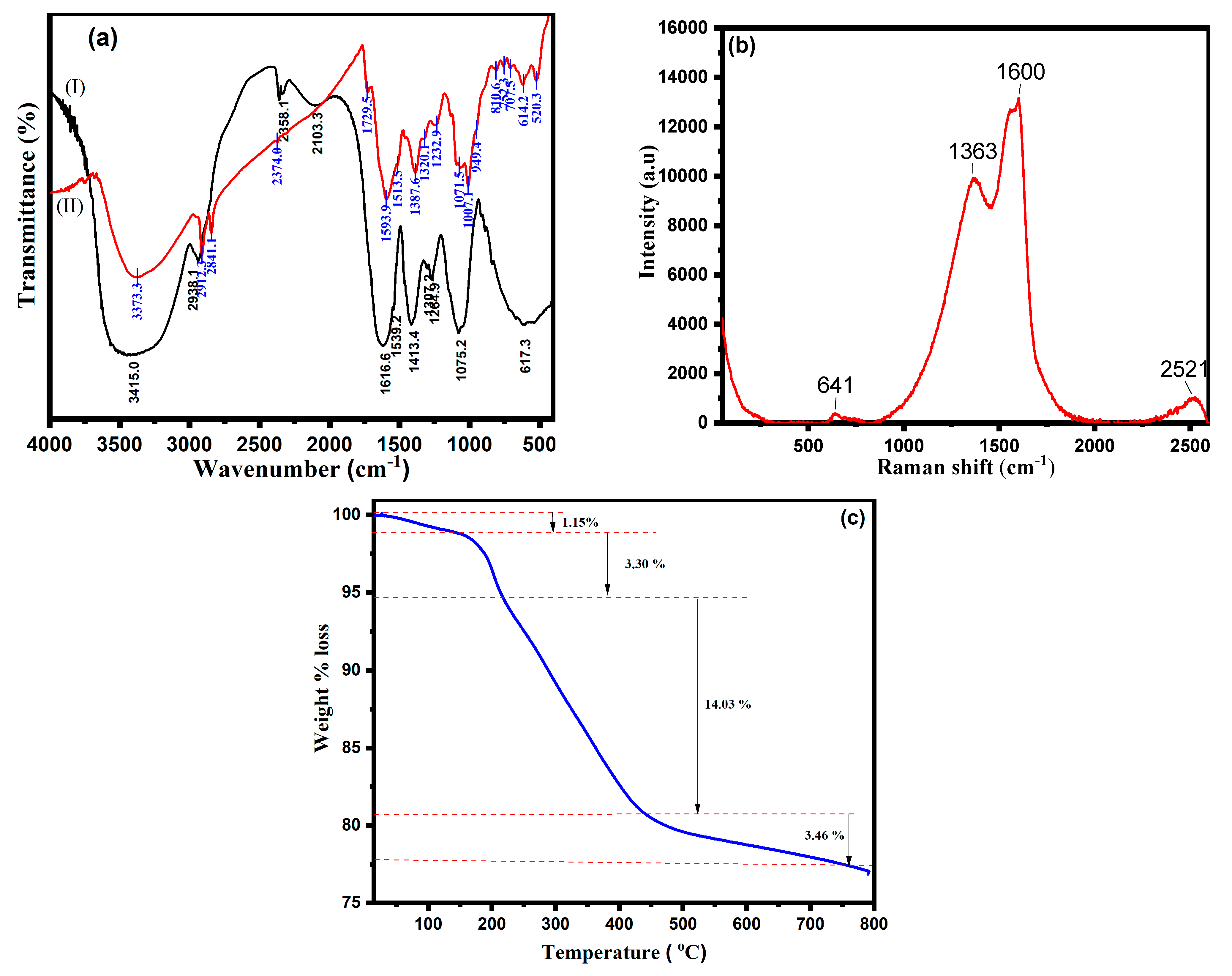
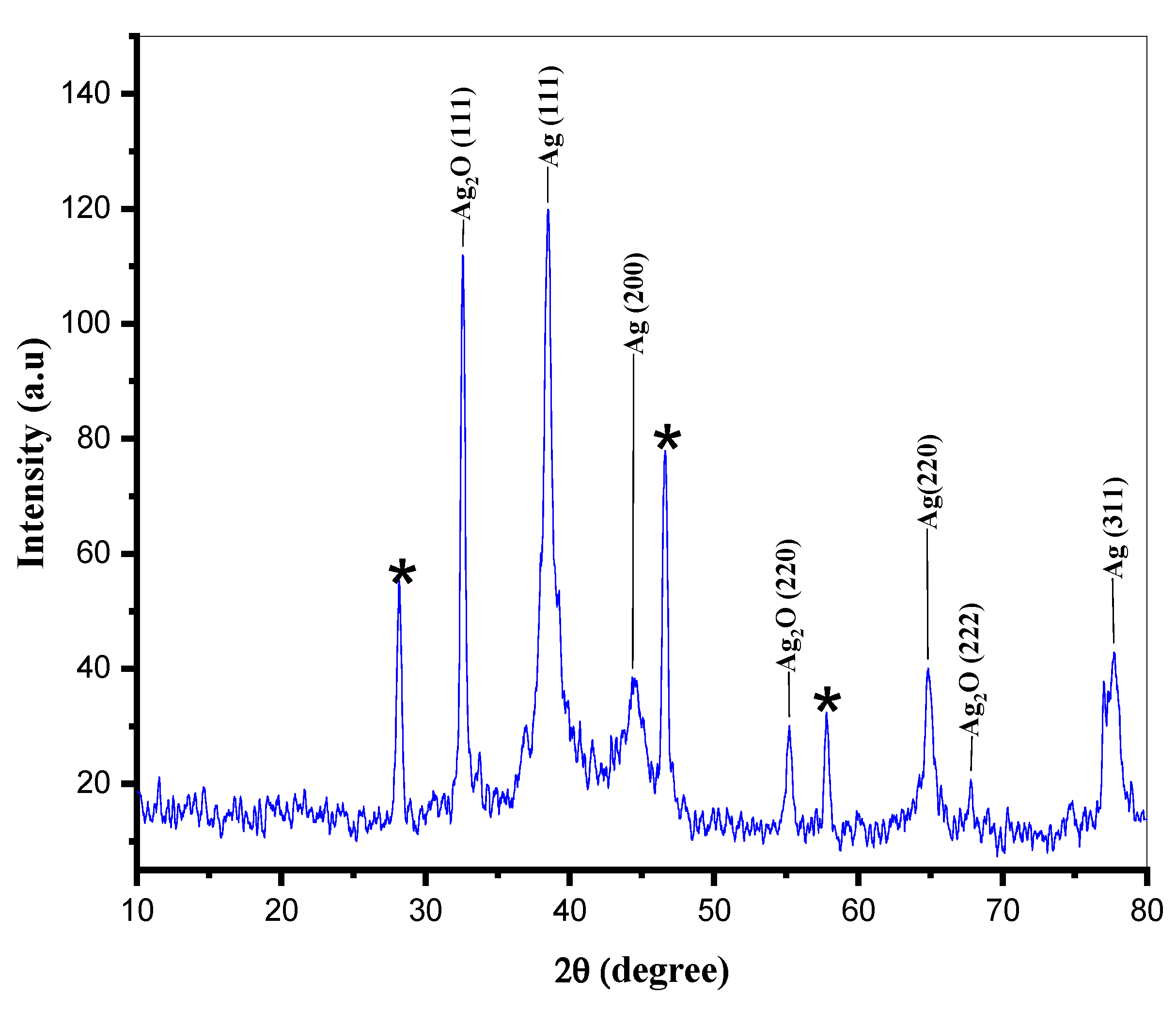
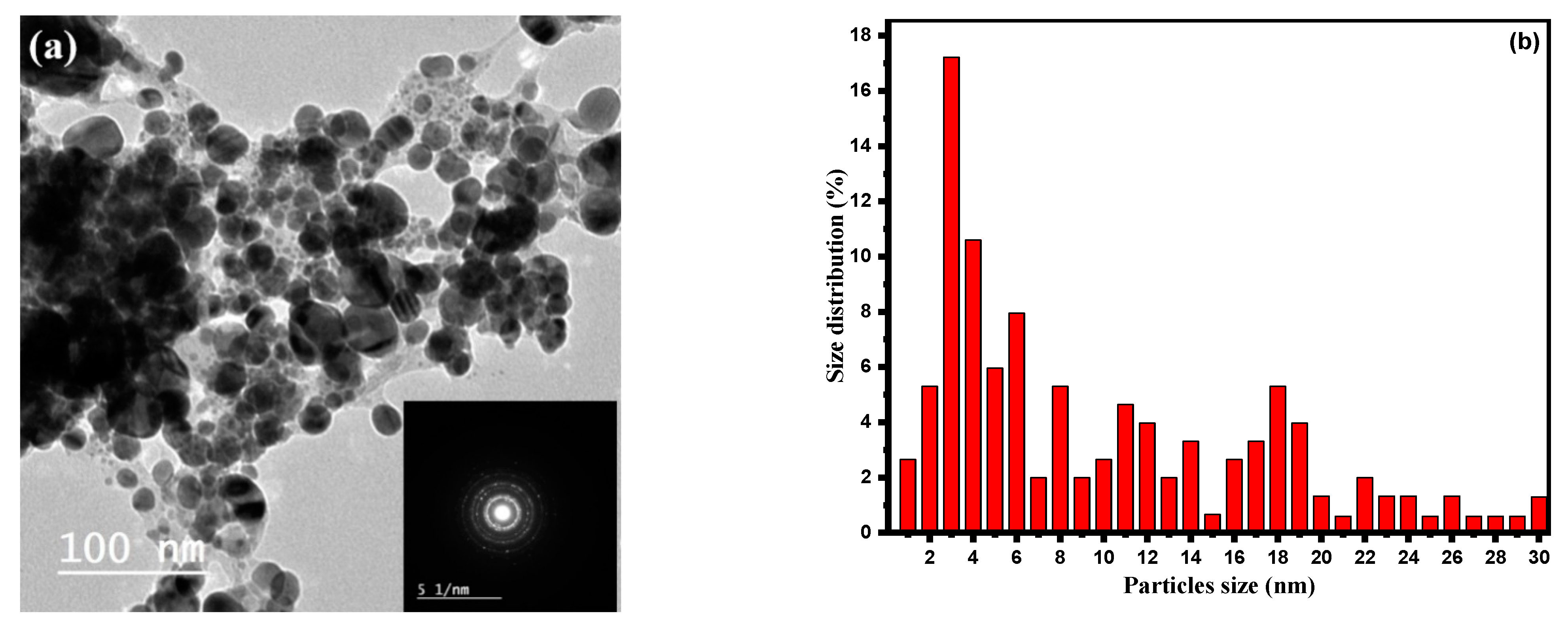



| Dye | Ionic Phase | Dye Conc. (mM) | Volume of Dye (μL) | Dye ppm in UV Cuvette | Volume of H2O (mL) | Volume of NaBH4 (5.0 mM) (mL) | Volume of CA-AgNPs(0.05%) (μL) | Reduction Time (min) |
|---|---|---|---|---|---|---|---|---|
| CR | Anionic | 1.0 | 300 | 63.15 | 1.700 | 0.990 | 10 | 36 |
| MB | Cationic | 1.0 | 100 | 10.25 | 1.900 | 0.980 | 20 | 32 |
| S. No. | Chemical Kinetic Model | Linear Equation Model | Dye | |||
|---|---|---|---|---|---|---|
| CR | MB | |||||
| Regression Coefficient (R2) Value | Rate Constant | Regression Coefficient (R2) Value | Rate Constant | |||
| 1 | Zero order | 0.9905 | 0.0311 (mol.L−1.min−1) | 0.9328 | 0.0431 (mol.L−1.min−1) | |
| 2 | First order | 0.8111 | 0.0715 (min−1) | 0.7491 | 0.1090 (min−1) | |
| 3 | Second order | 0.4446 | 0.2645 (L.mol−1.min−1) | 0.5167 | 0.5196 (L.mol−1.min−1) | |
| S. No | Plant Used for NPs Synthesis | Metal NPs/Electron Donor NaBH4 | Degradation % | Degradation Time | References |
|---|---|---|---|---|---|
| 1 | Terminalia arjuna | Ag | MB (93.60%), CR (92.20%), | 19 min for MB, 14 min for CR | [24] |
| 2 | Fruit extract of Gmelina arborea | Ag | MB (100%) | 30 min | [66] |
| 3 | Imperata cylindrica aqueous extract | Ag | MB (92.06%) | 14 min | [34] |
| 4 | Zingiber officinale aqueous extract | Ag | MB (99.9%) | 10 min | [67] |
| 5 | Phoenix Dactylifera L. a queous Leaves Extract | Ag/Ag2O | CR (80%), MB (84.60%) | 60 min for CR 50 min for MB | [68] |
| 6 | Caralluma acutangula | Ag | CR (95.24%) and MB (96.72%) | 36 min for CR 32 min for MB | This reported work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alamier, W.M.; Hasan, N.; Ali, S.K.; Oteef, M.D.Y. Biosynthesis of Ag Nanoparticles Using Caralluma acutangula Extract and Its Catalytic Functionality towards Degradation of Hazardous Dye Pollutants. Crystals 2022, 12, 1069. https://doi.org/10.3390/cryst12081069
Alamier WM, Hasan N, Ali SK, Oteef MDY. Biosynthesis of Ag Nanoparticles Using Caralluma acutangula Extract and Its Catalytic Functionality towards Degradation of Hazardous Dye Pollutants. Crystals. 2022; 12(8):1069. https://doi.org/10.3390/cryst12081069
Chicago/Turabian StyleAlamier, Waleed M., Nazim Hasan, Syed Kashif Ali, and Mohammed D. Y. Oteef. 2022. "Biosynthesis of Ag Nanoparticles Using Caralluma acutangula Extract and Its Catalytic Functionality towards Degradation of Hazardous Dye Pollutants" Crystals 12, no. 8: 1069. https://doi.org/10.3390/cryst12081069
APA StyleAlamier, W. M., Hasan, N., Ali, S. K., & Oteef, M. D. Y. (2022). Biosynthesis of Ag Nanoparticles Using Caralluma acutangula Extract and Its Catalytic Functionality towards Degradation of Hazardous Dye Pollutants. Crystals, 12(8), 1069. https://doi.org/10.3390/cryst12081069







