Abstract
A series of pyridine-2-olates (pyO) and pyridine-2-thiolates (pyS) of silicon was studied in solid state and in solution. The crystal structures of Me3Si(pyO) (1a), Me3Si(pyS) (1b), Me2Si(pyO)2 (2a), Me2Si(pyS)2 (2b), Ph2Si(pyO)2 (3a) and Ph2Si(pyS)2 (3b) were determined by X-ray diffraction. For that purpose, crystals of the (at room temperature) liquid compounds 1a and 1b were grown in a capillary on the diffractometer. Compounds 1a, 1b, 2a, 2b and 3a feature tetracoordinate silicon atoms in the solid state, whereas 3b gave rise to a series of four crystal structures in which the Si atoms of this compound are hexacoordinate. Two isomers (3b1 with all-cis arrangement of the C2N2S2 donor atoms in P, and 3b2 with trans S-Si-S axis in P21/n) formed individual crystal batches, which allowed for their individual 29Si NMR spectroscopic study in the solid state (the determination of their chemical shift anisotropy tensors). Furthermore, the structures of a less stable modification of 3b2 (in C2/c) as well as a toluene solvate 3b2 (toluene) (in P) were determined. In CDCl3, the equimolar solutions of the corresponding pairs of pyO and pyS compounds (2a/2b and 3a/3b) showed substituent scrambling with the formation of the products Me2Si(pyO)(pyS) (2c) and Ph2Si(pyO)(pyS) (3c), respectively, as minor components in the respective substituent exchange equilibrium.
1. Introduction
The field of silicon coordination chemistry, especially the field of hypercoordinate silicon compounds, has been explored with a vast variety of Lewis bases, which may enhance the Si coordination number up to five or six in many cases [1,2,3]. Among many others, pyridine derivatives may serve as additional donor sites in the Si coordination sphere, e.g., in compounds such as (py)2SiCl4 [4], (3-Me-py)2SiH2Cl2 [5], I [6] and II [7] (Figure 1). 2-Substituted mono-anionic pyridine derivatives may serve as chelating (e.g, in compounds III [8], IV [9]) or as bridging multidentate ligands (e.g., in compounds V [10], VI [10] and VII [11]) in hypercoordinate Si compounds. The formation of Si-TM complexes (TM = transition metal) with the bridging coordination mode of an (O,N)- or (S,N)- bidentate ligand may be fostered by the oxidative addition of an Si-H moiety to TM (e.g., the reaction of Me(H)Si(pyO)2 and low-valent group 9 metal compounds, such as [Ir(μ-Cl)(COE)2]2 (COE = cyclooctene) [12,13,14,15] or reaction of Ph(H)Si(mt)2 and [Ru(COD)(COT)] (mt = methimazolyl, 1-methyl-2-mercaptoimidazol-3-yl, COD = 1,5-cyclooctadiene, COT = 1,3,5-cyclooctatriene) [16]. As reflected by compound V [10] and related μ-(S,N)-bridged complexes, such as [ClPd(μ-mt)4SiCl] [17], the lone pair acceptor features of Si may support complex formation with electron rich TM compounds even in the absence of Si-H bonds. This makes silanes with mono-anionic ambidentate substituents an interesting class of compounds with respect to transition metal coordination chemistry. The silanes themselves, however, exhibit interesting Si coordination chemistry, too. With respect to TM free non-ionic silicon compounds, for pyridine-2-olate (pyO−) and pyridine-2-thiolate (pyS−), the representatives Si(pyO)4 [10], MeSi(pyO)3 [10], [SiO(pyO)2]4 [18] and Si(pyS)4 [9] as well as Cl2Si(pyS)2 [8,9], MeClSi(pyS)2 and compounds of the types PhXSi(pyS)2 (X = F, Cl, Br, N3, NCO, NCS) [8] have been characterized crystallographically. Whereas the pyO-compounds feature tetracoordinate Si atoms (with exclusively Si-O bound pyO-moieties), the pyS-compounds feature hexacoordinate Si atoms and two chelating pyS-moieties. Compounds such as R2Si(8-oxyquinolinate)2 [19,20] clearly reveal that electron releasing substituents R (e.g., R2 = Me2, (CH2)6) lower the Si-atom’s tendency toward establishing higher coordination numbers. As the structural characterization (solid state structure) of the compounds of the types R3Si(pyX) and R2Si(pyX)2 (R = methyl or phenyl; X = O, S) is still missing in the literature, the following work addresses a systematic comparison of the Si-coordination chemistry of compounds of the types Me3Si(pyX) (1a, 1b), Me2Si(pyX)2 (2a, 2b) and Ph2Si(pyX)2 (3a, 3b) (X = O (a), S (b)), see Figure 1.
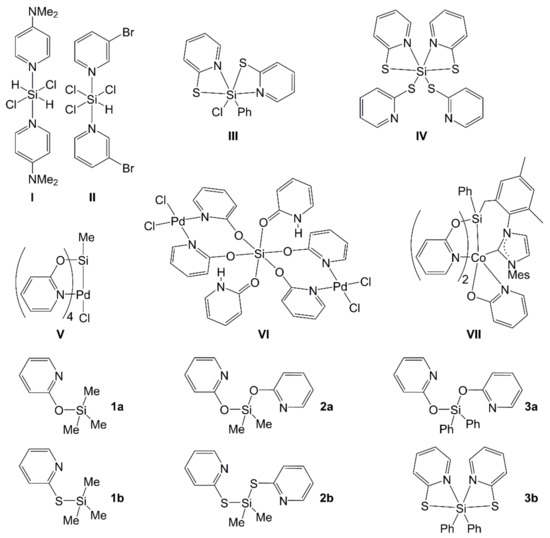
Figure 1.
Selected higher-coordinate silicon compounds with pyridine derivatives (I–VII) and compounds 1a, 1b, 2a, 2b, 3a and 3b studied in the current paper.
2. Materials and Methods
2.1. General Considerations
Starting materials 2-hydroxypyridine and 2-mercaptopyridine (ABCR, Karlsruhe, Germany, 98%) were used as received without further purification. Dichlorodimethylsilane and dichlorodiphenylsilane were distilled prior to use. Triethylamine was distilled from sodium/benzophenone and stored under argon atmosphere. THF, toluene and n-hexane were distilled from sodium/benzophenone prior to use. CDCl3 (Deutero, Kastellaun, Germany, 99.8%), toluene-d8 (ARMAR, Leipzig, Germany, 99.5%) and n-pentane (Th.Geyer, Renningen, Germany, >99%) were stored over activated molecular sieves (3 Å) for at least 7 days and used without further purification. All reactions were carried out under an atmosphere of dry argon utilizing standard Schlenk techniques. Solution NMR spectra (1H, 13C, 29Si) (cf. Figures S1–S12 in the supporting information) were recorded on Bruker Avance III 500 MHz and Bruker Nanobay 400 MHz spectrometers. 1H, 13C and 29Si chemical shifts are referenced to Me4Si (0 ppm) as internal reference. 1H and 13C NMR signals were assigned in accord with mutual coupling patterns (in case of 1H) and according to the shifts of corresponding 1H or 13C NMR signals in related compounds MeSi(pyO)3 [10], PhP(pyO)2 [21], P(pyS)3, PhSb(pyS)2 and Ph2Sb(pyS) [22]. The 29Si (CP/MAS) NMR spectra of compounds 3b1 and 3b2 (modification 1) (cf. Figures S13 and S14 in the supporting information) were recorded on a Bruker Avance 400 WB spectrometer using 7 mm zirconia (ZrO2) rotors with KelF-inserts at an MAS frequency of υspin = 900 Hz. The determination of the chemical shift anisotropy (CSA) tensor principal components from the spinning sideband spectra was carried out with the SOLA module contained in the Bruker software package TOPSPIN. Principal components δ11, δ22 and δ33 as well as span Ω and skew κ are reported according to the Herzfeld–Berger notation [23,24]. Elemental analyses were performed on an Elementar Vario MICRO cube. For single-crystal X-ray diffraction analyses, crystals were selected under an inert oil and mounted on a glass capillary (which was coated with silicone grease). In case of the (at room temperature) liquid compounds 1a and 1b, we adapted the method we had previously applied for the crystal structure determination of some liquid chlorosilanes [25]: Some drops of the compound were transferred (via syringe) into a glass capillary (diameter 1.0 mm), which was then flame sealed. This glass capillary was then mounted on a magnetic sample holder for Stoe goniometer heads and then placed under a stream of cold nitrogen at a fixed kappa angle of 45°. The sample was cooled until crystallization commenced, whereupon the sample was heated until its melting point to leave a tiny spot of crystalline material behind in the coldest part of the capillary. Thereafter the sample was slowly cooled to allow for the slow growth of larger single-crystalline domains. One of the domains adjacent to the initial crystallite was then moved towards the center of the cold stream, and heating/melting followed by slow cooling was repeated. After these two cycles the domains adjacent to the seed crystal were of sufficient quality for single-crystal X-ray diffraction. Thus, the upper domain was centered and used for data collection. Diffraction data were collected on a Stoe IPDS-2/2T diffractometer (STOE, Darmstadt, Germany) using Mo Kα-radiation. Data integration and absorption correction were performed with the STOE software XArea and XShape, respectively. The structures were solved by direct methods using SHELXS-97 or SHELXT and refined with the full-matrix least-squares methods of F2 against all reflections with SHELXL-2014/7 or SHELXL-2018/3 [26,27,28,29,30]. All non-hydrogen atoms were anisotropically refined. Hydrogen atoms were isotropically refined in idealized position (riding model). For the details of data collection and refinement (including the use of SQUEEZE in the refinement of the structure of 3b2 (toluene) see Appendix A. For the phase analyses of compounds 3b1 and 3b2 via X-ray powder diffraction at room temperature, a large single crystal of the respective compound (ca. 1.2 mm diameter) was mounted on a glass capillary and was embedded in a drop of epoxy resin. Upon unit cell determination (using some single-crystal X-ray diffraction frames), the powder diffraction diagram of this compound was simulated using a Gandolfi-scan (180° omega scan in combination with 6000 phi-rotations). For comparison, powdered bulk material of 3b1 and 3b2 (the samples which were used for solid state 29Si NMR investigations) was transferred into glass capillaries (diameter 1.0 mm), and their X-ray powder diffraction diagrams were recorded using the same Gandolfi-scan parameters on the same instrument (cf. Figure S17 in the supporting information). Powder diffraction data were converted using PowDLL converter [31]. The graphics of molecular structures were generated with ORTEP-3 [32,33] and POV-Ray 3.7 [34]. CCDC 2181113 (3a), 2181114 (2a), 2181115 (3b2 (toluene)), 2181116 (1a), 2181117 (1b), 2181118 (2b), 2181119 (3b2 (modif. 1)), 2181120 (3b2 (modif. 2)) and 2181121 (3b1) contain the supplementary crystal data for this article. These data can be obtained free of charge from the Cambridge Crystallographic Data Centre via https://www.ccdc.cam.ac.uk/structures/ (accessed on 22 June 2022).
The geometry optimizations were carried out with ORCA 5.0.3 [35] using the restricted PBE0 functional with a relativistically recontracted Karlsruhe basis set ZORA-def2-TZVPP [36,37] for all atoms, scalar relativistic ZORA Hamiltonian [38,39], atom-pairwise dispersion correction with the Becke–Johnson damping scheme (D3BJ) [40,41] and COSMO solvation (chloroform). Calculations were started from the molecular structures obtained by single-crystal X-ray diffraction analysis and isomers were generated therefrom. Numerical frequency calculations were performed to prove convergence at the local minimum after geometry optimization and to obtain the Gibbs free energy (293.15 K). Relaxed energy scans were performed in 10° steps by an increase of the Si-X-C2-N (X = O, S) torsion angle in the range from 0–180°. A transition state search was performed with nudged elastic band calculations with 11 steps on 1a. A numerical frequency calculation from the structure with the highest total energy level did confirm the transition state was reached. The transition state search for 1b was started from this geometry by changing oxygen to sulphur. A numerical frequency calculation was performed on this geometry and the result was used as initial Hessian for the transition state search. A numerical frequency calculation was performed on the final structure confirming the transition state was reached. Single-point calculations were performed with restricted B2T-PLYP functional [42] with a relativistically recontracted Karlsruhe basis set ZORA-def2-TZVPP [36,37] for all atoms and utilizing the AutoAux generation procedure [43], scalar relativistic ZORA Hamiltonian [38,39] and COSMO solvation (chloroform).
The H-atom positions of 3b1 and 3b2 (of the molecular structures obtained by single-crystal X-ray diffraction analyses) were optimized with ORCA 5.0.3. The geometry of tetrametylsilane (TMS) was optimized as described above. NMR chemical shifts were calculated with ADF-2019.303 [44] and referenced to TMS using an all-electron basis set TZ2P for all atoms [45] and scalar relativistic ZORA Hamiltonian [38,39]. As functionals PBE and PBE0 were used. Graphics were generated using Chemcraft [46].
2.2. Syntheses and Characterization
Samples of the trimethylsilyl derivatives of 2-hydroxypyridine (1a [47]) and of 2-mercaptopyridine (1b [48]) were prepared according to literature protocols and were available from previous investigations in our group.
Compound 2a (Me2Si(pyO)2, C12H14N2O2Si). A Schlenk flask was charged with magnetic stirring bar and 2-hydroxypyridine (3.00 g, 31.5 mmol), then evacuated and set under Ar atmosphere prior to adding THF (50 mL) and triethylamine (3.78 g, 37.4 mmol). The resultant mixture was stirred at room temperature, and dichlorodimethylsilane (2.06 g, 15.9 mmol) was added dropwise via syringe through a septum. Upon the completed addition of silane, stirring was continued for 30 min, whereupon the mixture was stored at 5 °C overnight. Thereafter, the triethylamine hydrochloride precipitate was removed by filtration and washed with THF (15 mL). From the combined filtrate and washings, the solvent was removed under reduced pressure (condensation into a cold trap) to afford a colorless oil. This crude product was dissolved into THF (4 mL), then n-pentane (4 mL) was added, and this solution was stored at 5 °C for 2 h. Within this time, some residual triethylamine hydrochloride precipitated, which was removed by filtration of the cold solution. From the clear filtrate, the volatiles were removed under reduced pressure, and the clear colorless oily residue was then dissolved into n-pentane (5 mL) and stored at −24 °C for one week to afford colorless crystals of 2a. The Schlenk flask was then stored in an ice/ethanol bath (ca. −10 °C), and from the solid product the supernatant solution was removed by decantation, followed by washing with n-pentane (2 × 2 mL) and drying in vacuum. Yield: 2.60 g (10.55 mmol, 67%). Elemental analysis for C12H14N2O2Si (246.34 g·mol−1): C, 58.51%; H, 5.73%; N, 11.37%; found C, 58.26%; H, 5.75%; N, 11.44%. 1H NMR (CDCl3): δ (ppm) 8.10 (br, 2H, H6), 7.56 (m, 2H, H4), 6.86 (br, 2H, H5), 6.80 (d, 2H, 8.2 Hz, H3), 0.64 (s, 6H, CH3); 13C{1H} NMR (CDCl3): δ (ppm) 161.6 (C2), 147.2 (C6), 139.2 (C4), 117.4 (C3), 112.9 (C5), −0.5 (CH3); 29Si{1H} NMR (CDCl3): δ (ppm) 0.8; 29Si{1H} NMR (toluene-d8): δ (ppm) 1.1.
Compound 2b (Me2Si(pyS)2, C12H14N2S2Si). A Schlenk flask was charged with magnetic stirring bar and 2-mercaptopyridine (4.10 g, 36.8 mmol), then evacuated and set under Ar atmosphere prior to adding THF (50 mL) and triethylamine (4.22 g, 41.6 mmol). The resultant mixture was stirred at room temperature, and dichlorodimethylsilane (2.44 g, 19.0 mmol) was added dropwise via syringe through a septum. Upon completed addition of silane, stirring was continued for 30 min, whereupon the mixture was stored at 5 °C overnight. Thereafter, the triethylamine hydrochloride precipitate was removed by filtration and washed with THF (20 mL). From the combined filtrate and washings, the solvent was removed under reduced pressure (condensation into a cold trap) to afford an orange oil. This crude product was dissolved into THF (4 mL), then n-pentane (4 mL) was added, and this solution was stored at 5 °C for 2 h. Within this time, some residual triethylamine hydrochloride precipitated, which was removed by filtration of the cold solution. From the clear filtrate, the volatiles were removed under reduced pressure, and the clear orange oily residue was then dissolved into toluene (1 mL) and n-pentane (4 mL) and stored at −24 °C for one week to afford orange crystals of 2b. The Schlenk flask was then stored in an ice/ethanol bath (ca. −10 °C), and from the solid product the supernatant solution was removed by decantation, followed by washing with n-pentane (2 × 2 mL) and drying in vacuum. Yield: 3.95 g (14.2 mmol, 77%). Elemental analysis for C12H14N2S2Si (278.47 g·mol−1): C, 51.76%; H, 5.07%; N, 10.06%; S, 23.03% found C, 51.45%; H, 4.96%; N, 10.37%; S, 23.09%. 1H NMR (CDCl3): δ (ppm) 8.35 (ddd, 2H, 1.0, 2.0, 5.0 Hz, H6), 7.46 (ddd, 2H, 2.0, 7.4, 8.0 Hz, H4), 7.34 (m, 2H, H3), 7.00 (ddd, 2H, 1.2, 5.0, 7.4 Hz, H5), 0.88 (s, 6H, CH3); 13C{1H} NMR (CDCl3): δ (ppm) 158.7 (C2), 148.8 (C6), 136.4 (C4), 126.7 (C3), 119.8 (C5), 3.7 (CH3); 29Si{1H} NMR (CDCl3): δ (ppm) 18.5; 29Si{1H} NMR (toluene-d8): δ (ppm) 17.9.
Compound 3a (Ph2Si(pyO)2, C22H18N2O2Si). A Schlenk flask was charged with magnetic stirring bar and 2-hydroxypyridine (2.50 g, 26.3 mmol), then evacuated and set under Ar atmosphere prior to adding THF (60 mL) and triethylamine (3.24 g, 32.2 mmol). The resultant mixture was stirred in an ice bath, and dichlorodiphenylsilane (3.46 g, 13.7 mmol) was added dropwise via syringe through a septum. Upon the completed addition of silane, stirring at 0 °C was continued for 60 min, whereupon the triethylamine hydrochloride precipitate was removed by filtration and washed with THF (20 mL). From the combined filtrate and washings, the solvent was removed under reduced pressure (condensation into a cold trap) to afford a white residue. This crude product was recrystallized in THF (1.5 mL). From the solid product the supernatant solution was removed by decantation, followed by washing with 8 mL of a mixture of n-pentane and THF (1:2 v/v) and drying in vacuum. Yield: 1.60 g (4.31 mmol, 31%). Elemental analysis for C22H18N2O2Si (370.47 g·mol−1): C, 71.32%; H, 4.90%; N, 7.56%; found C, 71.07%; H, 5.35%; N, 7.31%. 1H NMR (CDCl3): δ (ppm) 8.00 (m, 2H, H6), 7.91 (m, 4H, Ph-o), 7.50 (m, 2H, H4), 7.39 (m, 2H, Ph-p), 7.34 (m, 4H, Ph-m), 6.88 (d, 2H, 8.0 Hz, H3), 6.79 (dd, 2H, 5.3, 6.8 Hz, H5); 13C{1H} NMR (CDCl3): δ (ppm) 161.0 (C2), 147.4 (C6), 139.1 (C4), 135.4 (Ph-o), 131.8 (Ph-i), 130.5 (Ph-p), 127.6 (Ph-m), 117.9 (C3), 112.9 (C5); 29Si{1H} NMR (CDCl3): δ (ppm) −33.1.
Compound 3b (Ph2Si(pyS)2, C22H18N2S2Si). A Schlenk flask was charged with magnetic stirring bar and 2-mercaptopyridine (3.00 g, 27.0 mmol), then evacuated and set under Ar atmosphere prior to adding THF (45 mL) and triethylamine (2.95 g, 29.2 mmol). The resultant mixture was stirred at room temperature, and dichlorodiphenylsilane (3.37 g, 13.3 mmol) was added dropwise via syringe through a septum. Upon the completed addition of silane, stirring was continued for 60 min, whereupon the suspension was stored in an ice bath for 30 min prior to filtration of the triethylamine hydrochloride precipitate. The latter was washed with THF (10 mL). From the combined filtrate and washings, the solvent was removed under reduced pressure (condensation into a cold trap) to afford a yellow residue. This crude product was recrystallized in toluene (6 mL). From the solid product the supernatant solution was removed by decantation, followed by washing with 1 mL of toluene and drying in vacuum. Yield: 4.15 g (10.3 mmol, 77%). The coarse crystalline compound thus obtained consists of isomer 3b2 (modification 1) (this batch was employed for X-ray single-crystal and powder-diffraction analyses as well as the 29Si solid state NMR analyses of 3b2.). Elemental analysis for C22H18N2S2Si (402.61 g·mol−1): C, 65.63%; H, 4.51%; N, 6.96%; S, 15.93; found C, 65.47%; H, 4.66%; N, 7.09%; S, 15.63%. 1H NMR (CDCl3): δ (ppm) 8.13 (ddd, 2H, 0.9, 1.9, 5.1 Hz, H6), 7.75 (m, 4H, Ph-o), 7.19–7.31 (m, 10H, Ph-m/p, H3, H4), 6.82 (ddd, 2H, 1.2, 5.1, 7.4 Hz, H5); 13C{1H} NMR (CDCl3): δ (ppm) 158.1 (C2), 147.5 (C6), 136.6 (C4), 135.7 (Ph-i), 135.2 (Ph-o), 129.5 (Ph-p), 127.4 (Ph-m), 126.4 (C3), 119.7 (C5); 29Si{1H} NMR (CDCl3): δ (ppm) −19.0.
Isomer 3b1 was obtained following a similar protocol: A Schlenk flask was charged with magnetic stirring bar and 2-mercaptopyridine (2.00 g, 18.0 mmol), then evacuated and set under Ar atmosphere prior to adding THF (30 mL) and triethylamine (2.09 g, 20.7 mmol). The resultant mixture was stirred at room temperature, and dichlorodiphenylsilane (2.39 g, 9.40 mmol) was added dropwise via syringe through a septum. Upon the completed addition of silane, stirring was continued for 12 h, whereupon the suspension was stored at 5 °C for 4 h prior to filtration of the triethylamine hydrochloride precipitate. The latter was washed with THF (15 mL). From the combined filtrate and washings, the solvent was removed under reduced pressure (condensation into a cold trap) to afford a yellow residue. This crude product was recrystallized in toluene (3 mL). For crystallization, the solution was stored at 5 °C. To the solid product (with its supernatant) 5 mL of toluene were added, and then the solution was removed by filtration, followed by drying of the product in vacuum. Yield: 2.52 g (6.30 mmol, 70%). The product was identified as 3b1 by single-crystal X-ray diffraction. The filtrate was heated to ca. 60 °C, and n-hexane (2 mL) was added, whereupon this clear solution was placed in a fridge (5 °C) for further crystallization. Needles of the toluene solvate 3b2 (toluene) formed, which were suitable for single-crystal X-ray diffraction analysis. Within a few hours of storage (in the presence of the mother liquor), those crystals transformed into a fine crystalline powder.
The recrystallization of isomer 3b2 in THF afforded crystals of 3b1, too. A sample of 1.2 g of 3b2 (modification 1, as obtained from the abovementioned synthesis) was dissolved into 3.2 mL of hot THF to afford a clear yellow solution. Upon cooling to room temperature, n-pentane (2.5 mL) was added to the solution with stirring. The still clear yellow solution was placed in a fridge (5 °C), whereupon thin long plates of 3b2 (modification 2) formed. From this batch, a crystal was taken for single-crystal X-ray diffraction analysis of 3b2 (modification 2). Within few hours (still in the mother liquor and stored in the fridge), the thin plates 3b2 (modification 2) transformed into a fine-crystalline powder. From this product, the supernatant was removed with a syringe, and the solid residue was again recrystallized from THF. This time, THF was slowly added to the hot solution to avoid the complete dissolution of the starting material, to retain some seed crystals. During cooling to room temperature crystal growth commenced, and the crystalline material thus obtained was identified as 3b1 (this sample was employed for X-ray single-crystal and powder-diffraction analyses as well as the 29Si solid state NMR analyses of 3b1).
3. Results and Discussion
3.1. Compounds Me3Si(pyO) (1a) and Me3Si(pyS) (1b)
3.1.1. Crystallographic Analyses of 1a and 1b
Selected parameters of unit cell, data collection and structure refinement of 1a and 1b are summarized in Appendix A (Table A1). Compound 1a (Figure 2) crystallized in the orthorhombic space group Pbca with two molecules in the asymmetric unit. Compound 1b (Figure 2) crystallized in the monoclinic space group P21/m. The asymmetric unit comprises one half of the molecule because of its location on a crystallographic bisecting plane. In both compounds the pyridine N atom approaches the Si atom from far distance (ca. 3.0 Å), which is much longer than the sum of their covalent radii (1.82 Å [49]) but still shorter than the sum of van der Waals radii (3.65 Å [50]). Thus, it can be regarded as a remote [4+1]-coordination. The effects of Si-hypercoordination on the Si coordination sphere, however, are marginal; the lengthening of the N⋅⋅⋅Si trans-bond (with respect to the other Si-C bonds in the same molecule) lies between 3σ and 6σ, and the sum of idealized equatorial bond angles with respect to the N-Si-C axis (332.7° and 332.0° for the two molecules in the structure of 1a, 335.5° for 1b) still reflects the close to tetrahedral Si coordination sphere. Furthermore, the geometry parameter τ4′ [51] (as a measure for the distortion of the tetrahedral coordination sphere) is still very close to 1 (0.96 and 0.97 for the two independent molecules of 1a, 0.96 for 1b). Additionally, the C-X-Si (X = O, S) angles in compounds 1a and 1b are even wider than corresponding angles in aryl-X-SiMe3 compounds. Whereas in 1a, C-O-Si angles are 127.3(1)° and 128.4(1)°, 1,4-bis(trimethylsiloxy)benzene exhibits C-O-Si angles of 126.1(1)° and 126.7(1)° [52]. Whereas in 1b, the C-S-Si angle is 103.13(6)°, the azo-dye Et2N-C6H4-NN-C6H4-S-SiMe3 exhibits a C-S-Si angle of 98.46(6)° [53]. This can be explained by the torsion about the C-O or C-S bond in those aryl-X-SiMe3 compounds, which drives the Si-O or Si-S bond out of the aryl plane and thus lowers the steric demand (in terms of repulsion with SiMe3) of the aryl group.
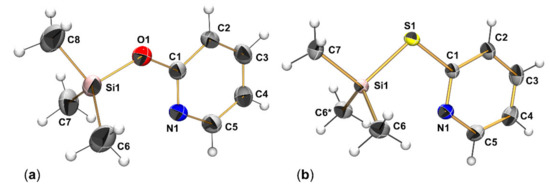
Figure 2.
Molecular structures of (a) Me3Si(pyO) (1a) and (b) Me3Si(pyS) (1b) in the crystal with thermal displacement ellipsoids at the 30% probability level and labels of non-hydrogen atoms. The asymmetric unit of 1a comprises two individual (but conformationally very similar) Me3Si(pyO) molecules, only one of them is depicted as a representative example. Selected interatomic distances (Å) and angles (deg.) of 1a: Si1-O1 1.6805(13), Si1-C6 1.843(2), Si1-C7 1.842(3), Si1-C8 1.853(2), Si1⋅⋅⋅N1 3.0036(15), Si1-O1-C1 127.25(11), N1-Si1-C8 150.10(9). The asymmetric unit of 1b comprises one half of the Me3Si(pyS) molecule, the atoms of which (except from methyl group C6) are located on a crystallographically imposed bisecting plane (symmetry operation *: x, 0.5−y, z). Selected interatomic distances (Å) and angles (deg.) of 1b: Si1-S1 2.1627(6), Si1-C6 1.8581(15), Si1-C7 1.867(2), Si1⋅⋅⋅N1 3.0454(17), Si1-S1-C1 103.13(6), N1-Si1-C7 160.21(8).
Our crystallographic analysis of 1b revises the original assumption of an Si-N bound thiopyridone group in this compound [9]. This assumption had been founded on the 29Si NMR shift of 1b, which was very similar to the 29Si NMR shift of trimethylsilylmethimazole, a compound which features an Si-N bound thiourea motif [54]. For insights into the energetic differences of isomers and conformers of compounds 1a and 1b, we performed quantum chemical calculations (cf. Section 3.1.2).
The different crystal packings of these (on the molecular level) structurally related compounds 1a and 1b can be attributed to the hard vs. soft nature of the Si-bound chalcogen atom. Whereas the closer proximity of O1 (and O2 of the second molecule in the asymmetric unit of 1a) is determined by C-H⋅⋅⋅O contacts to the pyridine rings of adjacent molecules, close contacts of S1 and CH3 groups are found in the packing of 1b.
3.1.2. Computational Analyses of Isomers of 1a and 1b
The molecular conformations of 1a and 1b in their crystal structures are posing two fundamental questions, which will be addressed here: (1) For the isomers with Si-X connectivity (X = O, S), as encountered in the crystals, is the conformation with remote N⋅⋅⋅Si coordination favorable for the isolated molecule or driven by crystal packing? (2) How favored is the Si-X over the Si-N binding mode, depending on X = O or S?
As in Me3Si-X-Aryl derivatives [52,53] the torsion of the Si-X (X = O, S) bond about the X-C bond out of the aryl plane indicates potentially favorable alternative molecular conformations for 1a and 1b, we performed potential energy surface (PES) scans, in which the Si-X-C-N dihedral angle was varied in steps of 10 degrees, followed by relaxation of the remaining bonds and angles of the respective molecule. Figure 3 illustrates the energetic relation of the conformers thus obtained. In both cases, the conformer with Si-X-C-N dihedral angle of 0 degrees was most favorable. Noteworthy, no local energetic minimum was encountered in the PES scans for dihedral angles around 90 degrees, only the decreasing slope of the graphs hints at some stabilizing effects through the lowered steric repulsion in those conformers. However, as the torsion approaches 180 degrees, a local minimum (for 1a) or at least a plateau (for 1b) was encountered. Regardless of the atom X (O vs. S), the 180 degree torsion about the X-C bond is accompanied by an energy difference of ca. 3 kcal·mol−1. Thus, for further investigation with the full optimization of the molecular conformation, two conformers of the Si-X-bound isomers were considered in addition to the respective isomer with Si-N bond.
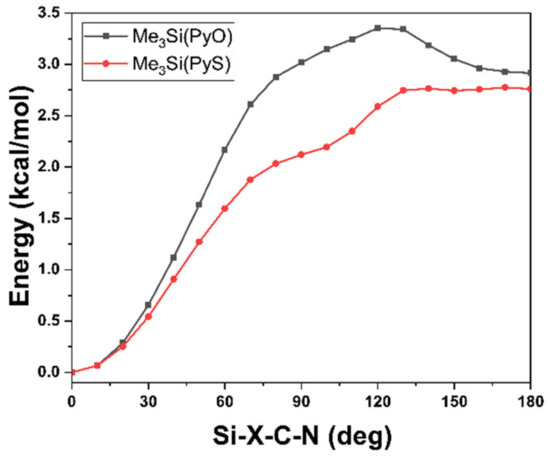
Figure 3.
Relative energy graphs of the potential energy surface (PES) scan for torsion about the O-C and S-C bond in 1a and 1b, respectively.
The full optimization of the two conformers of Si-X-bound compounds 1a and 1b essentially confirmed their energetic difference (in Gibbs free energy at 293 K) of ca. 3 kcal·mol−1. The alternative isomer of 1a and 1b with Si-N bound pyridine derivative was found to be less stable by 6.1 and 3.2 kcal·mol−1, respectively (Figure 4). Thus, in dynamic equilibria these isomers may still play a minor role. Especially in case of 1b, the transition state for the interconversion of the two isomers into one another (13.9 kcal·mol−1) is feasible at ambient temperature. The greater energetic difference in case of the isomers of 1a is in accord with the oxophilicity of silicon, and the higher energetic barrier posed by the transition state (19.9 kcal·mol−1) indicates a pronounced kinetic hindrance for the formation of isomer 1aN as well.
Figure 4.
Relative Gibbs free energy values (in kcal·mol−1) computed at the PBE0 D3BJ ZORA-def2-TZVPP level of theory for isomers of 1a and 1b. The superscript indices (from left to right) indicate the isomer/conformer with Si-X bond and ca. 180 degree torsion about Si-X-C-N (X180), with Si-X bond and ca. 0 degree torsion about Si-X-C-N (X0), with Si-N bond (N), and the transition state (TS) for the interconversion of isomers X0 and N into one another (cf. Figures S19–S26 and Tables S1–S8 in the supporting information).
3.2. Compounds Me2Si(pyO)2 (2a) and Me2Si(pyS)2 (2b)
3.2.1. Crystallographic Analyses of 2a and 2b
Selected parameters of unit cell, data collection and structure refinement of 2a and 2b are summarized in Appendix A (Table A1). Compounds 2a (in space group P21/c) and 2b (in space group P) both feature two crystallographically independent but conformationally very similar molecules in the asymmetric unit. For each of them, a representative example is shown in Figure 5. As found for 1a and 1b (cf. Section 3.1.1), compounds 2a and 2b feature essentially tetracoordinate Si atoms, the coordination spheres of which can be described as a remote [4+2]-coordination. Once again, Si⋅⋅⋅N contacts with interatomic separations around 3.0 Å complement the tetrahedral coordination sphere, and the Si-O-C- and Si-S-C angles in 2a and 2b, respectively, are similar to those in 1a and 1b, respectively. With respect to the geometry parameter τ4′ [51], the distortion of the almost tetrahedral coordination sphere of 2a (0.96 and 0.94 for the two independent molecules) is less pronounced than for 2b (0.91 for both of the two independent molecules).
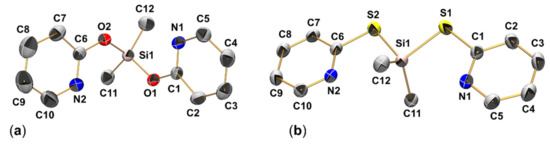
Figure 5.
Molecular structures of (a) Me2Si(pyO)2 (2a) and (b) Me2Si(pyS)2 (2b) in the crystal with thermal displacement ellipsoids at the 50% probability level and atom labels. For the sake of clarity, the H atoms are omitted from the graphic. In both cases, the asymmetric unit comprises two individual (but conformationally very similar) molecules. For each compound, only one of them is depicted as a representative example. Selected interatomic distances (Å) and angles (deg.) of 2a: Si1-O1 1.657(2), Si1-O2 1.659(2), Si1-C11 1.829(3), Si1-C12 1.837(4), Si1⋅⋅⋅N1 2.931(3), Si1⋅⋅⋅N2 3.031(3), Si1-O1-C1 125.32(18), Si1-O2-C6 128.70(19), O1-Si1-O2 111.41(12), C11-Si1-C12 111.72(18), N1-Si1-C11 155.63(12), N2-Si1-C12 152.29(14). Selected interatomic distances (Å) and angles (deg.) of 2b: Si1-S1 2.1708(5), Si1-S2 2.1696(4), Si1-C11 1.8582(13), Si1-C12 1.8593(14), Si1⋅⋅⋅N1 2.9560(11), Si1⋅⋅⋅N2 2.9734(11), Si1-S1-C1 100.26(4), Si1-S2-C6 101.00(4), S1-Si1-S2 92.601(18), C11-Si1-C12 116.80(7), N1-Si1-S2 153.12(3), N2-Si1-S1 152.41(3).
Besides those similarities, the presence of two pyX groups in compounds 2a and 2b gives rise to the formation of different conformers within the portfolio of [4+2]-coordination setups. Figure 6a illustrates the three conformations, which differ in the set of axial arrangements: Conformer 1 features two N⋅⋅⋅Si-C axes (i.e., the pyridine N atoms are capping tetrahedral faces opposite to an Si-C bond). This conformation is encountered in the solid state structure of 2a. Conformer 2 features two N⋅⋅⋅Si-X axes (i.e., the pyridine N atoms are capping tetrahedral faces opposite to the Si-X bonds, X being the anchor atom of an adjacent pyX moiety). This conformation is encountered in the solid state structure of 2b. Last but not least, conformation 3 with an N⋅⋅⋅Si-X and an N⋅⋅⋅Si-C axis should be feasible, but has not been encountered within these crystal structures, however. As the capping of tetrahedral faces trans to rather electronegative atoms X should be favorable because of N→Si lone pair donation into a lower lying σ*(Si-X) molecular orbital, the conformation 2 (as encountered with 2b) was rather expected, whereas the presence of conformer 1 in the structure of 2a was rather surprising. In dimethylsilicon carbamates Me2Si(-O(O=)C-NRR’)2 it is a common feature that the carbonyl O atoms are capping tetrahedral faces trans to Si-O bonds [55,56,57,58]. Therefore, we will have a closer look at the energetic differences between the conformers of compounds 2 in Section 3.2.3.
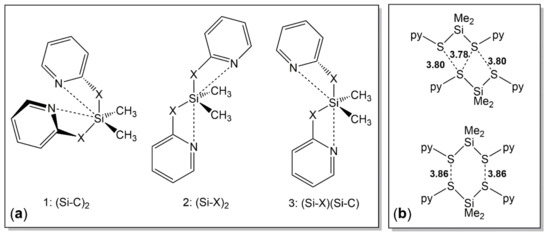
Figure 6.
(a) Different conformers of compounds 2 (i.e., of the type Me2Si(pyX)2 with X = O, S) with [4+2]-coordinate Si atom. The codes in parentheses indicate the trans bonds to the N→Si capped tetrahedral faces; (b) two different modes of weak intermolecular S⋅⋅⋅S contacts for molecule 1 (top) and molecule 2 (bottom) of 2b in the crystal packing with S⋅⋅⋅S separations in Å.
The different crystal packings of this set of (on the molecular level) structurally related compounds 2a and 2b can again be attributed to the hard vs. soft nature of the Si-bound chalcogen atom. Whereas the closer proximity of the O atoms of 2a is determined by C-H⋅⋅⋅O contacts to pyridine rings of adjacent molecules (such as in 1a), C-H⋅⋅⋅S contacts (also to pyridine rings of adjacent molecules) give rise to two different arrangements of sulfur atoms in the packing of 2b (as schematically represented in Figure 6b), in which the interatomic S⋅⋅⋅S-separations are in the upper range of the sum of the van der Waals radii [50].
With respect to the poor tendency of forming hypercoordinate Si-compounds with SiMe2 moieties, the rather poor Lewis acidity of the Si atom (because of the rather electron releasing nature of alkyl groups) seems to play a partial role only, and the need for forming a (rather strained) four-membered chelate poses another obstacle. Using the N-oxide of the pyO ligand, which forms five-membered (O,O’)-chelates, Kraft and Brennessel have shown that hexacoordinate Si compounds of the type (O,O’)2Si(alkyl)2 are feasible [59].
3.2.2. NMR-Spectroscopic Study of Substituent Scrambling of Me2Si(pyO)2 (2a) and Me2Si(pyS)2 (2b) with the Formation of Me2Si(pyO)(pyS) (2c)
Even though the characterization of the molecular structures of the compounds of the type Me2Si(pyX)2 has been missing in the literature so far, compound Me2Si(pyO)2 (2a) had been mentioned in a patent already [60], but it was proposed to feature hexacoordinate Si with chelating pyO ligands. Our current study complements the knowledge of the small library of silicon compounds which carry either pyO or pyS groups. With view on their potential use as ligands in transition metal coordination chemistry, combinations of hard and soft donor atoms within the same molecule may become of interest in terms of ambiphilic ligand properties. Thus, we also address the fundamental question as to the accessibility of compounds of the type R2Si(pyO)(pyS), in this particular case compound Me2Si(pyO)(pyS) (2c).
As the computational analyses in Section 3.1.2 indicated a rather high mobility of Si-bound pyS groups (including Si-N/Si-S bond exchange), we expected easy substituent scrambling in the (pyO)(pyS)-silane system, similar to the substituent scrambling encountered with mixtures of methimazolylsilanes and chlorosilanes [54]. In an NMR-scale experiment, the equimolar solutions of 2a in CDCl3 (0.88 M) and 2b in CDCl3 (0.88 M) were prepared, and the 29Si NMR spectra of these stock solution as well as of a mixture of equal volume fractions of these solutions were recorded (Figure 7). Whereas the individual stock solutions of 2a and 2b gave rise to only one 29Si NMR signal (at 0.7 ppm and 18.6 ppm, respectively), the solution of the mixture gave rise to three signals of similar intensity (at 0.7 ppm, 4.3 ppm and 18.6 ppm). The additional signal at 4.3 ppm can be assigned to the product of substituent scrambling (2c). In contrast to a statistical distribution and intensity ratio 2a:2b:2c of 1:1:2 (according to the equilibrium shown in Figure 7), compound 2c is underrepresented, the equilibrium side of starting materials 2a and 2b favored. (See also Figure S15 in the supporting information.) This finding will be underlined by results presented in the computational Section 3.2.3, and it points to potential problems associated with attempts of synthesizing organosilicon compounds, which bear both pyO and pyS groups within one molecule. They may be likely to undergo substituent scrambling in solution in a dynamic equilibrium, thus hampering the isolation of the desired product.
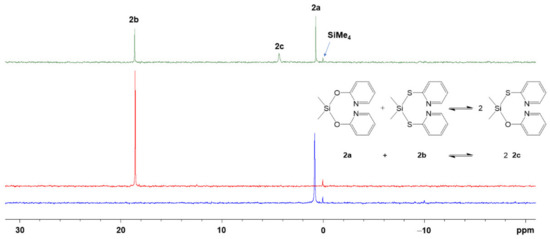
Figure 7.
Stack plot of 29Si{1H} NMR spectra of (from bottom to top) a 0.88 M solution of 2a in CDCl3 (blue trace), a 0.88 M solution of 2b in CDCl3 (red trace) and of an equimolar mixture of these stock solutions (green trace) as well as a reaction scheme and signal assignment. We attribute the signal broadening for 2c to dynamic effects in the Si coordination sphere (cf. Section 3.2.3, computational analyses of conformers and isomers of compound 2c).
3.2.3. Computational Analyses of Conformers of 2a, 2b and 2c
The molecular conformations of compounds 2a, 2b and 2c, which correspond to the conformations shown in Figure 6, were optimized at the PBE0 D3BJ ZORA-def2-TZVPP level of theory, and their relative Gibbs free energies were estimated for 293 K (Figure 8). In case of compound 2c, two different conformers with (Si-X)(Si-C) trans-capping had to be considered because of the mixed substitution pattern, which gives rise to conformers with (Si-S)(Si-C) and (Si-O)(Si-C) trans-capping.
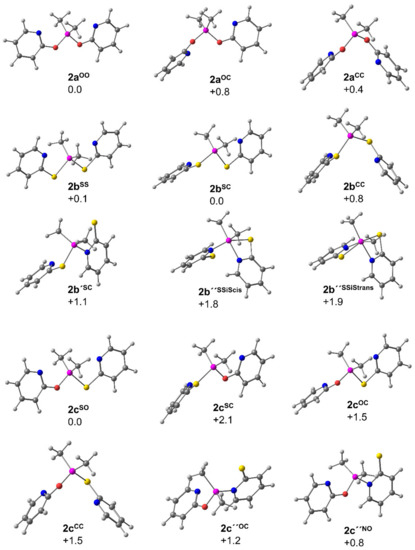
Figure 8.
Relative Gibbs free energy values (in kcal·mol−1) computed at the PBE0 D3BJ ZORA-def2-TZVPP level of theory for conformers of 2a, 2b and 2c and selected isomers with Si-N-bound substituent(s) 2b’, 2b’’ and 2c’’. For 2a, 2b and 2c, the superscript indices indicate the conformer in which the N atoms are capping tetrahedral faces trans to the superscript indexed atoms. Labels of the other isomers are based on related capping or on the relative S-Si-S arrangement within an octahedral coordination sphere (cf. Figures S27–S59 and Tables S9–S41 in the supporting information).
For compound 2a, the three conformers are very similar in Gibbs free energy, thus explaining as to why conformer 1 (2aCC) could crystallize, as it is only 0.4 kcal·mol−1 less stable than the energetic minimum conformer (2aOO). In case of compound 2b, the conformers 2bSS and 2bSC (with an energetic difference of only 0.1 kcal·mol−1) represent the minima, well in accord with the experimentally encountered conformation 2 (2bSS) in the solid state. In case of compound 2c, conformer 2 (2cSO) should dominate as the alternative configurations are at least 1.5 kcal·mol−1 higher in energy. The comparison of the total energies (in Hartree) associated with the Gibbs free energy values of the couples of energetic minimum conformers 2aOO + 2bSC vs. 2 2cSO implies that the formation of two molecules of 2c out of 2a+2b would be exergonic (with −0.45 kcal·mol−1). The accessibility of further conformers low in energy (2aCC and 2bSS), however, may explain as to why formation 2c is less favored in the dynamic equilibrium observed by NMR spectroscopy (cf. Figure 7).
As the computational analysis of compound 1b revealed that the isomerization of the pyS ligand into its Si-N bound form (cf. Section 3.1.2) may be associated with rather low energy difference, we considered isomers with Si-N bound pyS and/or pyO group(s) in the computational analyses of compounds 2 as well. The set of molecular structures derived therefrom, as well as their energies, can be found in the supporting information. For compound 2a the isomers with at least one Si-N bound pyO group are noticeably less stable (relative Gibbs free energy +5.6 kcal·mol−1 or more with respect to 2aOO). For compound 2b, three further isomers relatively low in energy were found upon changing one (2b’SC +1.1 kcal·mol−1) or two pyS groups into Si-N binding mode (2b’’SSiScis +1.8 kcal·mol−1 and 2b’’SSiStrans +1.9 kcal·mol−1). The Si coordination sphere of the former resembles a trigonal bipyramid with S-Si-S axis, the latter two are related to two isomers with rather octahedral Si coordination sphere and cis-disposed methyl groups (with cis-disposed S-Si-S bonds in one and trans-situation in the other). These isomers indicate that, in a dynamic equilibrium, Si hypercoordination may play a role in solutions of compound 2b. In fact, with respect to 2a (Δδ = +0.3 ppm) the 29Si NMR shift of compound 2b is more responsive to change of solvent and exhibits a slight upfield shift of the signal when using toluene-d8 instead of CDCl3 as NMR solvent (Δδ = −0.6 ppm). However, this effect may also originate from varying contributions of other conformers and is not unequivocally speaking for the influence of Si hypercoordination. For compound 2c, two isomers low in energy with Si-N bound pyS group were encountered (2c’’OC +1.2 kcal·mol−1 and 2b’’NO +0.8 kcal·mol−1), both exhibiting a rather trigonal-bipyramidal Si coordination sphere with O-Si-S axis.
3.3. Compounds Ph2Si(pyO)2 (3a) and Ph2Si(pyS)2 (3b)
3.3.1. Crystallographic Analyses of 3a and 3b
Selected parameters of unit cell, data collection and structure refinement of 3a are summarized in Appendix A (Table A1). Compound 3a (Figure 9) crystallized in the triclinic space group P1, and the asymmetric unit consists of three individual molecules of 3a, which adopt the same conformation. That is, their Si atoms are tetracoordinate and the N atoms cap the tetrahedral faces trans to the Si-C bonds (as in 2a). Additionally, on all three sites the same motif of disorder is present (cf. Figure 9b). That is, the formal site exchange of one pyO group and one phenyl group, while the remaining pair of pyO and Ph groups essentially remains on their places and just responds to the slightly different spatial demand of the neighboring groups. Even though this disorder lowers the precision of bond lengths and angles, the rather high occupancies of the predominant parts allowed for their refinement without restraints (restraints were applied to the low occupancy parts only) and the data obtained give a reasonable impression of the molecular conformations. Selected data of molecule 1 will be discussed as a representative example. Even though the geometry parameter τ4′ [51] hints at rather weak distortion (0.94) as it accounts for the deviations of angles from the tetrahedral angle regardless of atom type, the distortion of the tetrahedral Si coordination sphere is both slightly more pronounced than in compound 2a and, moreover, the deviations from tetrahedral angle are counter-VSEPR; i.e., the C11-Si1-C17 angle, which should be somewhat wider than 109.5° according to VSEPR, is 108.88(13)°, whereas the O1-Si1-O2 (113.31(11)°) angle is widened. This kind of deformation can be rationalized by the type of capping of tetrahedral faces: as N1 approaches Si1 trans to C17, the tetrahedral face Si1(O1,O2,C11) will be widened, thus widening the O1-Si1-O2 angle and compressing the C11-Si1-C17 angle. Analogously, as N2 approaches Si1 trans to C11, the tetrahedral face Si1(O1,O2,C17) will be widened, thus widening the O1-Si1-O2 angle again and compressing the C11-Si1-C17 angle again. The intramolecular Si⋅⋅⋅N contacts (separations Si1⋅⋅⋅N1 2.966(3) Å, Si1⋅⋅⋅N2 2.936(4) Å) are similar to those in 2a, however. Thus, the lowered steric demand of the phenyl groups (with respect to the methyl groups in 2a), which arises from their ability to adapt to steric requirements by torsion about the Si-C bond, is expected to be the origin of the pronounced distortion of the tetrahedral Si coordination sphere in 3a. The Si1-O1-C1 and Si1-O2-C6 angles (126.7(2)° and 125.6(3)°, respectively) are also similar to the corresponding angles in 2a. Thus, the remote N→Si coordination in the diphenylsilicon compound 3a is a similar to the situation in the methylsilicon compounds 1a, 2a and MeSi(pyO)3 [10] as well as in compound Si(pyO)4 [10].
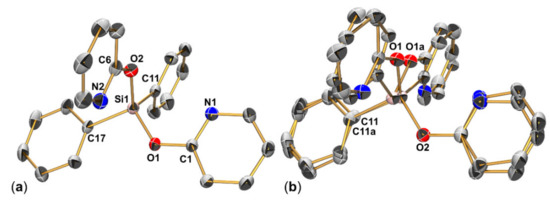
Figure 9.
(a) Molecular structure of one of the three independent (but conformationally very similar) molecules of the asymmetric unit of 3a (thermal displacement ellipsoids are drawn at the 30% probability level, selected atoms are labeled, H atoms and disorder parts are omitted for clarity). (b) The disorder encountered in this structure corresponds to a site mismatch of one set of Ph and pyO groups. The same disorder motif is encountered with the other two molecules in the asymmetric unit as well. The site occupancies of disorder parts 1 and 2 are 0.930(2):0.070(2) for molecule 1 (the one shown in this graphic) and 0.881(2):0.119(2) for the related disorders in the remaining two molecules in the asymmetric unit.
The crystal structure of 3a exhibits an interesting feature in its molecular packing. As shown in Figure 10, the orientations of the three individual molecules of the asymmetric unit of this packing are almost related to one another by local (not crystallographic) symmetry elements. While molecules 2 and 3 reflect inversion symmetry, the relative positions of molecules 1 and 3 indicate a glide plane perpendicular to the longest axis. Therefore, a test structure solution in the centrosymmetric space group P delivered a model with two molecules in the unit cell (which correspond to the pair of molecules 2 and 3), while data integration in a monoclinic setting (which was possible because of two angles of the unit cell being close to 90°) and a test structure solution in the non-centrosymmetric space group Pc delivered another model with two molecules in the unit cell (which correspond to the pair of molecules 1 and 3). In both cases (models in P and in Pc), the models failed to describe the missing molecule 1 and 2, respectively.
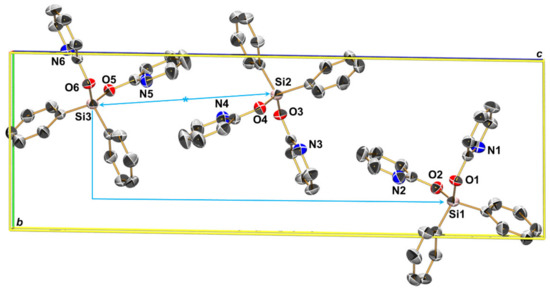
Figure 10.
Unit cell packing (view along the a axis) of the three independent molecules of the asymmetric unit of 3a (thermal displacement ellipsoids are drawn at the 50% probability level, selected atoms are labeled, H atoms and disorder parts are omitted for clarity). The light blue arrows indicate pseudo-symmetry relations: whereas molecules 3 and 2 are roughly related to one another by a local center of inversion, the relation between molecules 3 and 1 indicates a local b-glide symmetry (with respect to this unit cell setting).
In case of the analogous pyridine-2-thiolato compounds, the switch from SiMe2 (in 2b) to SiPh2 (in 3b) gave rise to noticeable changes in the coordination chemistry. While the Si atom of 2b is tetracoordinate even in the solid state, compound 3b exhibits hexacoordinate Si atoms in the solid. Several attempts at crystallizing compound 3b delivered (so far) four different crystalline solids, all of which feature 3b as hexacoordinate Si complex. An interesting aspect about the different crystal structures determined therefrom is the individual characterization of two different coordination isomers (3b1 and 3b2). Figure 11 shows the molecular structures of 3b1 and 3b2 (the latter in the solvent free modification 1, in space group P21/n). As the other two crystal structures comprise isomer 3b2, too (in a solvate 3b2 (toluene) and in a less stable solvent-free modification 3b2 in C2/c), the latter two structures are contained in this publication as Supplementary Materials. The molecular conformation of 3b2 in its three different crystal structures is essentially alike, and bond lengths and also angles are very similar. For example, the S-Si-S angles range from 156.82(3)° (in modification 1) to 157.89(6)° (in modification 2), C-Si-C angles from 99.43(8)° (in modification 1) to 100.64(9)° (in toluene solvate), N-Si-N angles from 81.02(7)° (in toluene solvate) to 82.31(6)° (in modification 1). Therefore, we just point out that the crystallographically determined density of 3b2 in modification 2 in C2/c (1.29 g⋅cm−1 at 200 K) is noticeable lower than in modification 1 in P21/n (1.36 g⋅cm−1 at 180 K), which is in accord with the stabilization of the modification 1 by closer packing.
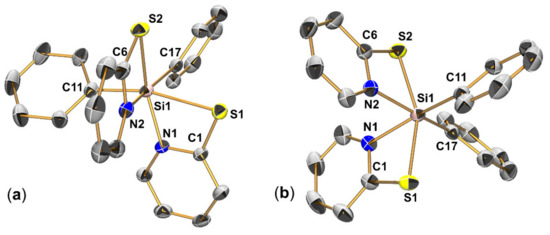
Figure 11.
Molecular structures of isomers of Ph2Si(pyS)2 (3b): (a) isomer 3b1 and (b) isomer 3b2 (in modification 1, space group P21/n) in the crystal with thermal displacement ellipsoids at the 50% probability level and selected atom labels. For the sake of clarity, the H atoms are omitted from the graphic. Selected interatomic distances (Å) and angles (degrees) of 3b1: Si1-S1 2.5683(5), Si1-S2 2.3335(5), Si1-N1 1.9058(11), Si1-N2 1.9694(12), Si1-C11 1.9181(14), Si1-C17 1.9162(14), S1-Si1-S2 91.868(18), N1-Si1-N2 90.00(5), C11-Si1-C17 101.37(6), S1-Si1-C11 165.91(5), S2-Si1-N1 154.73(4), N2-Si1-C17 163.44(6). Selected interatomic distances (Å) and angles (degrees) of 3b2 (modification 1): Si1-S1 2.3280(7), Si1-S2 2.3372(7), Si1-N1 2.0103(16), Si1-N2 2.0127(16), Si1-C11 1.9108(19), Si1-C17 1.9164(19), S1-Si1-S2 156.82(3), N1-Si1-N2 82.31(6), C11-Si1-C17 99.43(8), N1-Si1-C11 165.82(8), N2-Si1-C17 165.68(8).
The molecular conformation of 3b1 resembles all-cis arrangement of the C,C-, N,N- and S,S-pairs of donor atoms, whereas in 3b2 only C,C and N,N are mutually cis and the S atoms are trans to one another. Thus, the two conformations correspond to those which had been found previously with other hexacoordinate pyridinethiolato-Si-complexes [8,9]. Tacke et al. even reported the presence of two corresponding isomers of one of their compounds (PhClSi(pyS)2) within one crystal structure in a disordered manner (while for the other compounds one of the isomers was found (e.g., all-cis for Ph(N3)Si(pyS)2 [8], trans-S2 for Cl2Si(pyS)2 [8,9]). With computational methods they have shown that, within the portfolio of PhXSi(pyS)2 complexes with two cis-disposed monodentate substituents (Ph and X, X = halide, N3, NCO, NCS), these two diastereomeric forms (all-cis isomer with trans-X-Si-S axis and trans-S-Si-S isomer) are more stable than the alternative diastereomers (all-cis isomer with trans-C-Si-S axis and trans-N-Si-N isomer). In this regard, encountering diastereomers 3b1 and 3b2 is in accord with the expectation derived from those previous calculations. Compound 3b, however, allows for the individual characterization of two such isomers, which will also be addressed in Section 3.3.3 (analysis of 29Si chemical shift anisotropy).
The molecular structure of isomer 3b2 resembles C2 symmetry. Accordingly, the Si-S bonds are similar to one another (ca. 2.33 Å), and the same holds true for the Si-N and Si-C bonds (with ca. 2.01 Å and 1.91 Å, respectively). In isomer 3b1, however, each pair of donor atoms is facing two different trans situations. Resulting therefrom, bond Si1-S1 (which is trans to Si-C) is ca. 0.23 Å longer than bond Si1-S2 (which is trans to Si-N). In case of the previously published all-cis featured complexes of the type RXSi(pyS)2 (R = Ph, Me; X = F, Cl, N3, NCS, NCO) [8] the bond length differences between the two Si-S bonds within a molecule are less pronounced. The Si-S bonds trans to a pyridine N atom are still in the range 2.30–2.34 Å, but the Si-S bonds trans to one of the monodentate ligands are always trans to the rather electronegative (pseudo)halide and thus significantly shorter than the corresponding bond in 3b1 (in the range 2.39–2.45 Å). Significant bond length difference in 3b1 is also observed for the different Si-N bonds. Si1-N1 (trans to Si-S) is 0.06 Å shorter than Si1-N2 (trans to Si-C). The Si-C bonds (of both 3b1 and 3b2), however, exhibit the same length within the boundaries of experimental error. In both compounds, the small (four-membered) chelate rings allow for wider angles between the monodentate substituents. Thus, in both compounds the C-Si-C angles are close to 100°. These effects in the Si coordination spheres of 3b1 and 3b2 (small chelate bite angles, wider angles between monodentate substituents, donor atoms DA of different size and thus Si-DA bond length) result in the heavy distortion of the idealized octahedral coordination spheres (see Figure S18 in the supporting information). With respect to the structure of dimethylsilicon compound 2b, in which the pyS groups are essentially monodentate, the chelation of the Si atoms in 3b1 and 3b2 causes the bending of the pyridine rings toward Si by adapting smaller N-C-S angles. Whereas in 2b these angles range between 117.68(9)° and 118.37(9)°, they are S1-C1-N1 112.59(9)° and S2-C6-N2 109.38(10)° in 3b1, S1-C1-N1 108.98(14)° and S2-C6-N2 109.47(14)° in 3b2 (modification 1).
3.3.2. NMR-Spectroscopic Study of Substituent Scrambling of Ph2Si(pyO)2 (3a) and Ph2Si(pyS)2 (3b) with Formation of Ph2Si(pyO)(pyS) (3c)
As ligand scrambling between 2a and 2b in CDCl3 solution gave rise to the formation of compound 2c, we also studied the system of diphenylsilicon compounds 3a and 3b in CDCl3 solution in an analogous manner (cf. Figure 12). As expected, the solution of compound 3a gives rise to a 29Si NMR signal at −33.2 ppm, characteristic of a tetracoordinate Si compound. In CDCl3 solution, the Si atoms of compounds 3b are also tetracoordinate, the 29Si NMR signal (at −18.2 ppm) is in the typical region of tetracoordinate Si compounds, too. The equimolar mixture of these stock solutions also produces a third signal (at −13.9 ppm), which can be assigned to compound 3c, the product of substituent scrambling (see also Figure S16 in the supporting information.) In case of the equilibrium mixture of the diphenylsilicon compounds, the product of substituent scrambling (3c) is even more underrepresented than in the case of the corresponding system of dimethylsilicon compounds. We attribute the further shift of the equilibrium to the side of the starting materials 3a+3b to the stabilization of 3b by the chelation of its Si atom. Even though the 29Si solution NMR signal of 3b is speaking for predominant silicon tetracoordination, the isomers with hexacoordinate Si atom should still serve a small contribution within a dynamic coordination equilibrium.
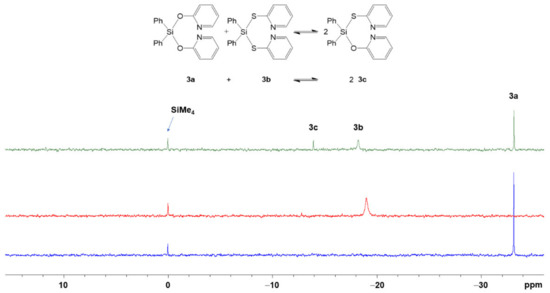
Figure 12.
Stack plot of 29Si{1H} NMR spectra of (from bottom to top) a 0.38 M solution of 3a in CDCl3 (blue trace), a 0.38 M solution of 3b in CDCl3 (red trace) and of an equimolar mixture of these stock solutions (green trace) as well as a reaction scheme and signal assignment. We attribute the signal broadening for 3b to dynamic effects in the Si coordination sphere (cf. Section 3.2.3, the computational analyses of conformers and isomers of compound 2b).
3.3.3. NMR Spectroscopic and Computational Analyses of 29Si Chemical Shift Anisotropy Tensors of 3b1 and 3b2 (Two Isomers of Ph2Si(pyS)2 (3b))
The crystallization of the individual isomers 3b1 and 3b2, both of which feature hexacoordinate Si atoms, allowed to study their 29Si NMR chemical shift anisotropy (CSA) by solid state NMR spectroscopy. Their individual CSA tensors should be influenced by the different donor atom arrangements in the Si coordination sphere. Therefore, two batches of 3b (one of which contained isomer 3b1 and the other one contained 3b2) were checked for the clear predominance of the isomer under investigation by (1) homogeneous appearance of crystal shapes, (2) multiple crystal picking and unit cell determination and (3) the grinding of a representative sample for X-ray powder diffraction. Even though we cannot guarantee the absolute absence of the respective other isomer, the samples are “phase pure” for the individual isomer of 3b within the boundaries of the powder XRD diagrams recorded, and thus, the solid state NMR spectra should represent the CSA features of the respective isomer.
The two samples produced 29Si CP/MAS NMR spectra with essentially the same isotropic chemical shift (δiso = −127 ppm). Using high spinning rates, thus recording only the isotropic shift signal, one would not be able to distinguish between the two isomers in this case. Spinning side band spectra, however, reveal the different nature of the two individual samples (Figure 13). To confirm the assignment of the different CSA tensors to the respective isomers of 3b, their CSA tensors were calculated (based on the atomic coordinates of a single molecule of the respective crystal structure, i.e., 3b1 and modification 1 of 3b2). The computational and experimental results are collated in Table 1. In principle, the computational data reflect the features of the experimentally determined CSA tensors in a satisfactory manner. (In addition to general experimental errors, both in data extraction and analysis from CP/MAS NMR experiments as well as deviations caused by the computational method applied, the computational analysis of a single molecule vs. experimental analysis of molecules within the crystal packing can be expected to give rise to some deviations).
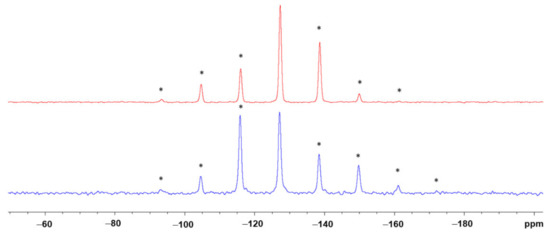
Figure 13.
29Si CP/MAS NMR spectra of isomers 3b1 (bottom, blue trace) and 3b2 (top, red trace) recorded at a spinning frequency of 900 Hz. The isotropic chemical shifts are almost identical (δiso = −127.2 and −127.3 ppm, respectively). The different chemical shift anisotropy features of the two isomers are obvious from the spinning sidebands (asterisked peaks).

Table 1.
Comparison of 29Si CSA tensor characteristics (isotropic chemical shift δiso, principal components δ11, δ22, δ33, span Ω and skew κ) of 3b1 and of modification 1 of 3b2 determined from solid state NMR spectra (“exp”, cf. Figure 13) and calculated at the PBE TZ2P and PBE0 TZ2P level of theory.
Interestingly, in compound 3b1 one principal component is particularly well shielded (positive skew of the CSA tensor), a feature which has also been found with hexacoordinate diorganosilicon complexes with a salen-type ligand (with SiO2N2C2 coordination sphere and trans-C-Si-C axis) [61] and with paddlewheel-shaped complexes with SiN4ClE (E = Ge, Sn) coordination spheres (N4 equatorial plane and Cl-Si-E axis) [62]. In contrast, the CSA tensor of 3b2 features one considerably less shielded component (i.e., negative skew). This feature has been encountered with the pyridine and bipyridine complexes of dichlorosilane [6,63]. Figure 14 illustrates the directions of the principal components of the 29Si CSA tensor with respect to the Si coordination sphere for compounds 3b1 and 3b2. As the Si coordination spheres are highly distorted, far from octahedral, they exhibit hardly any idealized symmetry axes, which should coincide with a particular principal axis of the CSA tensor. In compound 3b1, the particularly well shielded direction (33) is close to perpendicular to the idealized Si,N,N- plane of the molecule, thus (with some offset) pointing along the direction of a C-Si-S axis. The two principal components of lower shielding are located close to within this plane. In compound 3b2, one component (11) is particularly less shielded. The molecule nearly exhibits C2 symmetry (the idealized two-fold axis passing through and bisecting the N-N-, S-S- and C-C-axes), and from the principal components the direction of component (11) is closest to this idealized axis, but nonetheless exhibits a noticeable offset therefrom.
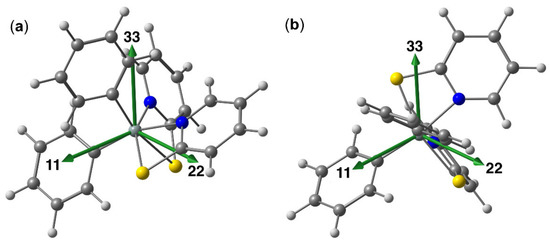
Figure 14.
Graphical representation of the orientation of the 29Si CSA tensor principal components in compounds 3b1 (a) and 3b2 (b) obtained at the PBE0 TZ2P level of theory. The respective orientation of the CSA tensors obtained at the PBE TZ2P level (not depicted) is very similar.
4. Conclusions
With this contribution, we give some first insights into the Si coordination chemistry of tri- and diorganosilicon compounds with pyridine-2-(thi)olato substituents. The combination of the latter (which are potentially four-membered chelators) with rather electron releasing hydrocarbyl substituents at silicon (Me) lead to compounds with tetracoordinate Si atom even in the solid state. In all cases (Me3Si(pyO) (1a), Me3Si(pyS) (1b), Me2Si(pyO)2 (2a), Me2Si(pyS)2 (2b)) the pyridine derivative is Si-O or Si-S bound, and the pyridine nitrogen atoms establish Si⋅⋅⋅N contacts with separations of ca. 3 Å. Whereas this Si tetracoordination motif can be found for other silicon pyridine-2-olates (such as Ph2Si(pyO)2 (3a), as reported herein), the corresponding thiolate (Ph2Si(pyS)2 (3b)) features Si hexacoordination in the solid state and Si tetracoordination in chloroform solution. It thus represents a link between tetracoordinate Si compounds, such as 2b, and hexacoordinate Si compounds, such as the previously reported complex MeClSi(pyS)2 [8], which retains Si hexacoordination in solution. Isomerization in solution (in dynamic equilibria) gave rise to the crystallization of two individual isomers of 3b (3b1 and 3b2), which allowed the study of their individual molecular features by X-ray crystallography and 29Si solid state NMR spectroscopy.
The solutions to the combinations of the corresponding silicon pyridine-2-olates and -thiolates (2a + 2b, 3a + 3b) undergo substituent scrambling. In those two systems, the products with mixed substituent patterns (R2Si(pyO)(pyS), 2c R = Me, 3c R = Ph) are underrepresented. As shown by computational analyses, the mixed substitution pattern does not provide any significant energetic sink to foster their formation. In turn, compounds of the type R2Si(pyO)(pyS) can be expected to undergo substituent scrambling with the formation of the respective homoleptic compounds (with respect to PyO and pyS). As a practical consequence thereof, the use of such compounds in transition metal coordination chemistry might be highly limited. Transition metal compounds of pyridine-2-olato silanes are currently under investigation with respect to their use as catalysts [12,13,14,15]. Catalyst tuning by the partial replacement of pyO by pyS bridges, if relevant for future studies, may thus be highly challenging.
Supplementary Materials
The following Supporting Information can be downloaded at: https://www.mdpi.com/article/10.3390/cryst12081054/s1. Crystallographic data for the compounds reported in this paper (in CIF format) and a document containing graphics of the 1H, 13C and 29Si NMR spectra of compounds 2a, 2b, 3a and 3b; X-ray powder diffraction diagrams of 3b1 and 3b2 (modification 1); graphical representations of the Si coordination polyhedra of compounds 3b1 and 3b2; data sets (consisting of molecular graphic, atomic coordinates and total energies) of the optimized molecular structures of isomers/conformers of 1a, 1b, 2a, 2b and 2c as well as of the transition state for the Si-O/Si-N and Si-S/Si-N isomerization of 1a and 1b, respectively. Figure S1: 1H NMR spectrum of 2a in CDCl3 (full spectrum and magnified inset of group of signals); Figure S2: 13C{1H} NMR spectrum of 2a in CDCl3; Figure S3: 29Si{1H} NMR spectrum of 2a in CDCl3; Figure S4: 1H NMR spectrum of 2b in CDCl3 (full spectrum and magnified insets of group of signals); Figure S5: 13C{1H} NMR spectrum of 2b in CDCl3; Figure S6: 29Si{1H} NMR spectrum of 2b in CDCl3; Figure S7: 1H NMR spectrum of 3a in CDCl3 (full spectrum and magnified insets of group of signals); Figure S8: 13C{1H} NMR spectrum of 3a in CDCl3; Figure S9: 29Si{1H} NMR spectrum of 3a in CDCl3; Figure S10: 1H NMR spectrum of 3b in CDCl3 (full spectrum and magnified inset of group of signals); Figure S11: 13C{1H} NMR spectrum of 3b in CDCl3; Figure S12: 29Si{1H} NMR spectrum of 3b in CDCl3; Figure S13: 29Si CP/MAS NMR spectrum of 3b1 (MAS frequency 900 Hz, spinning side bands are asterisked) showing the experimental spectrum (top, blue trace) and the spinning side band spectrum simulated from the CSA tensor derived therefrom (bottom, red trace); Figure S14: 29Si CP/MAS NMR spectrum of 3b2 (MAS frequency 900 Hz, spinning side bands are asterisked) showing the experimental spectrum (top, blue trace) and the spinning side band spectrum simulated from the CSA tensor derived therefrom (bottom, red trace); Figure S15: 29Si{1H} NMR spectrum of an equimolar mixture of 2a and 2b in CDCl3 (with integrals of the signal intensities of 2a, 2b and 2c); Figure S16: 29Si{1H} NMR spectrum of an equimolar mixture of 3a and 3b in CDCl3 (with integrals of the signal intensities of 3a, 3b and 3c); Figure S17: X-ray powder diffraction patterns of compounds 3b1 and 3b2 (modification 1) recorded with Mo Kα radiation using Gandolfi scans: (a) Diffraction pattern recorded from a powdered bulk sample (blue trace) and from a single crystal (red trace) of compound 3b1: (b) Diffraction pattern recorded from a powdered bulk sample (blue trace) and from a single crystal (red trace) of modification 1 of compound 3b2: (c) Superposition (low-angle fraction) of the diffraction patterns recorded from powdered bulk samples of compound 3b1 (blue trace) and of modification 1 of compound 3b2 (red trace); Figure S18: Graphical representations of the Si coordination polyhedra of compounds 3b1 (a) and 3b2 (modification 1) (b) with atom labels (bottom) and with some additional atoms attached for a better image of the polyhedron within the molecule (top): Thermal displacement ellipsoids are shown at the 50% probability level; Figure S19: Optimized molecular structure of 1aO0; Figure S20: Optimized molecular structure of 1aN; Figure S21: Optimized molecular structure of 1aO180; Figure S22: Optimized molecular structure of 1aTS; Figure S23: Optimized molecular structure of 1bS0; Figure S24: Optimized molecular structure of 1bN; Figure S25: Optimized molecular structure of 1bS180; Figure S26: Optimized molecular structure of 1bTS; Figure S27: Optimized molecular structure of 2aOO; Figure S28: Optimized molecular structure of 2aOC; Figure S29: Optimized molecular structure of 2aCC; Figure S30: Optimized molecular structure of 2a´NO; Figure S31: Optimized molecular structure of 2a´OC; Figure S32: Optimized molecular structure of 2a´CC; Figure S33: Optimized molecular structure of 2a´NC; Figure S34: Optimized molecular structure of 2a´´NN; Figure S35: Optimized molecular structure of 2bSS; Figure S36: Optimized molecular structure of 2bSC; Figure S37: Optimized molecular structure of 2bCC; Figure S38: Optimized molecular structure of 2b´NS; Figure S39: Optimized molecular structure of 2b´SC; Figure S40: Optimized molecular structure of 2b´CC; Figure S41: Optimized molecular structure of 2b´NC; Figure S42: Optimized molecular structure of 2b´´NN; Figure S43: Optimized molecular structure of 2b´´SSiScis; Figure S44: Optimized molecular structure of 2b´´SSiStrans; Figure S45: Optimized molecular structure of 2cSO; Figure S46: Optimized molecular structure of 2cOC; Figure S47: Optimized molecular structure of 2cSC; Figure S48: Optimized molecular structure of 2cCC; Figure S49: Optimized molecular structure of 2c´NS; Figure S50: Optimized molecular structure of 2c´SC; Figure S51: Optimized molecular structure of 2c´CC; Figure S52: Optimized molecular structure of 2c´NC; Figure S53: Optimized molecular structure of 2c´´NO; Figure S54: Optimized molecular structure of 2c´´OC; Figure S55: Optimized molecular structure of 2c´´CC; Figure S56: Optimized molecular structure of 2c´´NC; Figure S57: Optimized molecular structure of 2c´´´NN; Figure S58: Optimized molecular structure of 2c´´´OSiCtrans; Figure S59: Optimized molecular structure of 2c´´´SSiCtrans; Table S1: Atomic coordinates for optimized structure of 1aO0; Table S2: Atomic coordinates for optimized structure of 1aN; Table S3: Atomic coordinates for optimized structure of 1a O180; Table S4: Atomic coordinates for optimized structure of 1aTS; Table S5: Atomic coordinates for optimized structure of 1bS0; Table S6: Atomic coordinates for optimized structure of 1bN; Table S7: Atomic coordinates for optimized structure of 1b180; Table S8: Atomic coordinates for optimized structure of 1bTS; Table S9: Atomic coordinates for optimized structure of 2aOO; Table S10: Atomic coordinates for optimized structure of 2aOC; Table S11: Atomic coordinates for optimized structure of 2aCC; Table S12: Atomic coordinates for optimized structure of 2a´NO; Table S13: Atomic coordinates for optimized structure of 2a´OC; Table S14: Atomic coordinates for optimized structure of 2a´CC; Table S15: Atomic coordinates for optimized structure of 2a´NC; Table S16: Atomic coordinates for optimized structure of 2a´´NN; Table S17: Atomic coordinates for optimized structure of 2bSS; Table S18: Atomic coordinates for optimized structure of 2bSC; Table S19: Atomic coordinates for optimized structure of 2bCC; Table S20: Atomic coordinates for optimized structure of 2b´NS; Table S21: Atomic coordinates for optimized structure of 2b´SC; Table S22: Atomic coordinates for optimized structure of 2b´CC; Table S23: Atomic coordinates for optimized structure of 2b´NC; Table S24: Atomic coordinates for optimized structure of 2b´´NN; Table S25: Atomic coordinates for optimized structure of 2b´´SSiScis: Table S26: Atomic coordinates for optimized structure of 2b´´SSiStrans; Table S27: Atomic coordinates for optimized structure of 2cSO; Table S28: Atomic coordinates for optimized structure of 2cOC; Table S29: Atomic coordinates for optimized structure of 2cSC; Table S30: Atomic coordinates for optimized structure of 2cCC; Table S31: Atomic coordinates for optimized structure of 2c´NS; Table S32: Atomic coordinates for optimized structure of 2c´SC; Table S33: Atomic coordinates for optimized structure of 2c´CC; Table S34: Atomic coordinates for optimized structure of 2c´NC; Table S35: Atomic coordinates for optimized structure of 2c´´NO; Table S36: Atomic coordinates for optimized structure of 2c´´OC; Table S37: Atomic coordinates for optimized structure of 2c´´CC; Table S38: Atomic coordinates for optimized structure of 2c´´NC; Table S39: Atomic coordinates for optimized structure of 2c´´´NN; Table S40: Atomic coordinates for optimized structure of 2c´´´OSiCtrans; Table S41: Atomic coordinates for optimized structure of 2c´´´SSiCtrans.
Author Contributions
Conceptualization, J.W.; investigation, A.S., M.W., L.E., R.G., E.B. and J.W.; writing—original draft preparation, J.W.; writing—review and editing, R.G. and J.W.; visualization, R.G. and J.W.; supervision, J.W.; All authors have read and agreed to the published version of the manuscript.
Funding
The authors are grateful for computing time at the High-Performance Computing Cluster at TU Bergakademie Freiberg, which was funded by Deutsche Forschungsgemeinschaft (DFG)—397252409.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors are grateful to Marcel Swart (Institut de Química Computacional i Catàlisi, Facultat de Ciències, Universitat de Girona, Spain) for his support with computational analyses (with the ADF package), to Beate Kutzner and Franziska Gründler (TU Bergakademie Freiberg, Institut für Anorganische Chemie) for solution NMR and elemental microanalysis service, respectively, and Anika Rogoll (TU Bergakademie Freiberg, Institut für Analytische Chemie) for preparing a sample of compound 1a.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A

Table A1.
Crystallographic data from data collection and refinement for 1a, 1b, 2a, 2b and 3a.
Table A1.
Crystallographic data from data collection and refinement for 1a, 1b, 2a, 2b and 3a.
| Parameter | 1a | 1b | 2a 1 | 2b | 3a 2 |
|---|---|---|---|---|---|
| Formula | C8H13NOSi | C8H13NSSi | C12H14N2O2Si | C12H14N2S2Si | C22H18N2O2Si |
| Mr | 167.28 | 183.34 | 246.34 | 278.46 | 370.47 |
| T(K) | 220(2) | 220(2) | 200(2) | 200(2) | 200(2) |
| λ(Å) | 0.71073 | 0.71073 | 0.71073 | 0.71073 | 0.71073 |
| Crystal system | orthorhombic | monoclinic | monoclinic | triclinic | triclinic |
| Space group | Pbca | P21/m | P21/c | P1 | |
| a(Å) | 11.6876(1) | 6.6079(7) | 12.7881(6) | 9.7145(4) | 6.1626(2) |
| b(Å) | 24.898(3) | 7.2078(6) | 10.7211(6) | 11.5896(5) | 8.8822(3) |
| c(Å) | 13.833(2) | 11.1025(12) | 18.6634(8) | 12.9509(6) | 26.4639(9) |
| α(°) | 90 | 90 | 90 | 73.601(3) | 88.692(3) |
| β(°) | 90 | 97.954(9) | 90.247(4) | 89.903(3) | 88.561(3) |
| γ(°) | 90 | 90 | 90 | 85.998(3) | 81.453(3) |
| V(Å3) | 4025.4(9) | 523.71(9) | 2558.8(2) | 1395.13(11) | 1431.76(8) |
| Z | 16 | 2 | 8 | 4 | 3 |
| ρcalc(g⋅cm−1) | 1.10 | 1.16 | 1.28 | 1.33 | 1.29 |
| μMoKα (mm−1) | 0.2 | 0.4 | 0.2 | 0.4 | 0.1 |
| F(000) | 1440 | 196 | 1040 | 584 | 582 |
| θmax(°), Rint | 25.0, 0.0463 | 28.0, 0.0573 | 25.0, 0.0761 | 28.0, 0.0181 | 28.0, 0.0197 |
| Completeness | 99.4% | 99.9% | 99.3% | 99.6% | 99.8% |
| Reflns collected | 26,377 | 7376 | 19,701 | 35,958 | 48,444 |
| Reflns unique | 3522 | 1255 | 4515 | 6716 | 13251 |
| Restraints | 0 | 0 | 0 | 0 | 36 |
| Parameters | 205 | 67 | 313 | 311 | 846 |
| GoF | 1.074 | 1.106 | 1.045 | 1.058 | 1.057 |
| R1, wR2 [I > 2σ(I)] | 0.0419, 0.1136 | 0.0330, 0.0914 | 0.0460, 0.0965 | 0.0293, 0.0779 | 0.0389, 0.1002 |
| R1, wR2 (all data) | 0.0478, 0.1205 | 0.0354, 0.0948 | 0.0849, 0.1112 | 0.0310, 0.0793 | 0.0443, 0.1061 |
| Largest peak/hole (e⋅Å−3) | 0.28, −0.25 | 0.25, −0.24 | 0.19, −0.24 | 0.50, −0.37 | 0.40, −0.28 |
1 The structure of compound 2a was refined as a twin (instruction TWIN 1 0 0 0 1 0 0 0 −1 using a HKLF4 format data set). The batch scale factor (twin population) refined to 0.211(1). 2 The absolute structure parameter of this structure refined to χFlack = 0.10(2).

Table A2.
Crystallographic data from data collection and refinement for 3b1, 3b2 (two modifications) and 3b2 (toluene).
Table A2.
Crystallographic data from data collection and refinement for 3b1, 3b2 (two modifications) and 3b2 (toluene).
| Parameter | 3b1 | 3b2 (Modif. 1) | 3b2 (Modif. 2) | 3b2⋅(Toluene) 1 |
|---|---|---|---|---|
| Formula | C22H18N2S2Si | C22H18N2S2Si | C22H18N2S2Si | C29H26N2S2Si |
| Mr | 402.59 | 402.59 | 402.59 | 494.73 |
| T(K) | 200(2) | 180(2) | 200(2) | 200(2) |
| λ(Å) | 0.71073 | 0.71073 | 0.71073 | 0.71073 |
| Crystal system | triclinic | monoclinic | monoclinic | triclinic |
| Space group | P21/n | C2/c | ||
| a(Å) | 9.4763(8) | 12.3581(4) | 10.9877(5) | 9.7034(9) |
| b(Å) | 10.2415(8) | 12.7040(4) | 19.5766(9) | 11.2023(9) |
| c(Å) | 11.1044(8) | 12.5693(4) | 9.7361(5) | 13.2853(13) |
| α(°) | 96.698(6) | 90 | 90 | 73.607(7) |
| β(°) | 114.442(6) | 90.262(2) | 97.501(4) | 72.232(7) |
| γ(°) | 90.107(6) | 90 | 90 | 82.511(7) |
| V(Å3) | 972.92(14) | 1973.33(11) | 2076.33(17) | 1317.8(2) |
| Z | 2 | 4 | 4 | 2 |
| ρcalc(g⋅cm−1) | 1.37 | 1.36 | 1.29 | 1.25 |
| μMoKα (mm−1) | 0.3 | 0.3 | 0.3 | 0.3 |
| F(000) | 420 | 840 | 840 | 520 |
| θmax(°), Rint | 28.0, 0.0182 | 27.0, 0.0470 | 25.0, 0.0755 | 25.0, 0.0316 |
| Completeness | 99.8% | 100% | 100% | 99.8% |
| Reflns collected | 14558 | 20583 | 12049 | 12384 |
| Reflns unique | 4698 | 4302 | 1829 | 4641 |
| Restraints | 0 | 0 | 0 | 59 |
| Parameters | 244 | 244 | 124 | 296 |
| GoF | 1.102 | 1.042 | 1.081 | 1.043 |
| R1, wR2 [I > 2σ(I)] | 0.0313, 0.0794 | 0.0389, 0.0847 | 0.0429, 0.0913 | 0.0365, 0.0861 |
| R1, wR2 (all data) | 0.0367, 0.0837 | 0.0533, 0.0905 | 0.0690, 0.1001 | 0.0563, 0.0929 |
| Largest peak/hole (e⋅Å−3) | 0.36, −0.24 | 0.30, −0.31 | 0.22, −0.25 | 0.33, −0.32 |
1 The asymmetric unit of the structure of compound 3b2 (toluene) comprises one molecule of complex 3b2 and two half molecules of toluene (which are located in close proximity of crystallographic centers of inversion and therefore disordered by symmetry). One of the solvent molecules was refined with an idealized hexagon for the phenyl group (using ShelXL instruction AFIX 66). The second solvent molecule was, in addition to the symmetry-related disorder, severely disordered over many sites. Therefore, this part of the solvent was not refined but treated with SQUEEZE as implemented in PLATON [64,65,66]. This procedure detected, per unit cell, solvent accessible volume of 231 Å3 and contributions of 50 electrons therein (well in accordance with 50 electrons for the toluene molecule per unit cell, which has been omitted from refinement).
References
- Chuit, C.; Corriu, R.J.P.; Reye, C.; Young, J.C. Reactivity of penta- and hexacoordinate silicon compounds and their role as reaction intermediates. Chem. Rev. 1993, 93, 1371–1448. [Google Scholar] [CrossRef]
- Wagler, J.; Böhme, U.; Kroke, E. Higher-Coordinated Molecular Silicon Compounds. Struct. Bond. 2013, 155, 29–105. [Google Scholar] [CrossRef]
- Lemière, G.; Millanvois, A.; Ollivier, C.; Fensterbank, L. A Parisian Vision of the Chemistry of Hypercoordinated Silicon Derivatives. Chem. Rec. 2021, 21, 1119–1129. [Google Scholar] [CrossRef] [PubMed]
- Bechstein, S.O.; Ziemer, B.; Hass, D.; Trojanov, S.I.; Rybakov, V.B.; Maso, G.N. Halogen Exchange on Silicon Halides. XIII. Structure and Reactivity of Silicon Halide-Pyridine Compounds. Z. Anorg. Allg. Chem. 1990, 582, 211–216. [Google Scholar] [CrossRef]
- Hensen, K.; Stumpf, T.; Bolte, M.; Näther, A.C.; Fleischer, H. Experimental Investigations and ab Initio Studies on Hexacoordinated Complexes of Dichlorosilane. J. Am. Chem. Soc. 1998, 120, 10402–10408. [Google Scholar] [CrossRef]
- Fester, G.W.; Wagler, J.; Brendler, E.; Böhme, U.; Roewer, G.; Kroke, E. Octahedral Adducts of Dichlorosilane with Substituted Pyridines: Synthesis, Reactivity and a Comparison of Their Structures and29Si NMR Chemical Shifts. Chem. Eur. J. 2008, 14, 3164–3176. [Google Scholar] [CrossRef]
- Fester, G.W.; Wagler, J.; Brendler, E.; Kroke, E. Stable Trichlorosilane–Pyridine Adducts. Eur. J. Inorg. Chem. 2008, 2008, 5020–5023. [Google Scholar] [CrossRef]
- Baus, J.A.; Burschka, C.; Bertermann, R.; Guerra, C.F.; Bickelhaupt, F.M.; Tacke, R. Neutral Six-Coordinate and Cationic Five-Coordinate Silicon(IV) Complexes with Two Bidentate MonoanionicN,S-Pyridine-2-thiolato(−) Ligands. Inorg. Chem. 2013, 52, 10664–10676. [Google Scholar] [CrossRef]
- Wächtler, E.; Gericke, R.; Kutter, S.; Brendler, E.; Wagler, J. Molecular structures of pyridinethiolato complexes of Sn(II), Sn(IV), Ge(IV), and Si(IV). Main Group Met. Chem. 2013, 36, 181–191. [Google Scholar] [CrossRef]
- Ehrlich, L.; Gericke, R.; Brendler, E.; Wagler, J. (2-Pyridyloxy)silanes as Ligands in Transition Metal Coordination Chemistry. Inorganics 2018, 6, 119. [Google Scholar] [CrossRef]
- Sun, J.; Ou, C.; Wang, C.; Uchiyama, M.; Deng, L. Silane-Functionalized N-Heterocyclic Carbene–Cobalt Complexes Containing a Five-Coordinate Silicon with a Covalent Co–Si Bond. Organometallics 2015, 34, 1546–1551. [Google Scholar] [CrossRef]
- Lalrempuia, R.; Iglesias, M.; Polo, V.; Miguel, P.J.S.; Fernández-Alvarez, F.J.; Pérez-Torrente, J.J.; Oro, L.A. Effective Fixation of CO2 by Iridium-Catalyzed Hydrosilylation. Angew. Chem. Int. 2012, 51, 12824–12827. [Google Scholar] [CrossRef]
- Garcés, K.; Lalrempuia, R.; Polo, V.; Fernández-Alvarez, F.J.; García-Orduña, P.; Lahoz, F.J.; Pérez-Torrente, J.J.; Oro, L.A. Rhodium-Catalyzed Dehydrogenative Silylation of Acetophenone Derivatives: Formation of Silyl Enol Ethers versus Silyl Ethers. Chem. Eur. J. 2016, 22, 14717–14729. [Google Scholar] [CrossRef]
- Julián, A.; Guzmán, J.; Jaseer, E.A.; Fernández-Alvarez, F.J.; Royo, R.; Polo, V.; Orduña, M.P.G.; Lahoz, F.J.; Oro, L.A. Mechanistic Insights on the Reduction of CO2 to Silylformates Catalyzed by Ir-NSiN Species. Chem. Eur. J. 2017, 23, 11898–11907. [Google Scholar] [CrossRef]
- Julián, A.; Garcés, K.; Lalrempuia, R.; Jaseer, E.A.; García-Orduña, P.; Fernández-Alvarez, F.J.; Lahoz, F.J.; Oro, L.A. Reactivity of Ir-NSiN Complexes: Ir-Catalyzed Dehydrogenative Silylation of Carboxylic Acids. ChemCatChem 2018, 10, 1027–1034. [Google Scholar] [CrossRef]
- Hill, A.F.; Neumann, H.; Wagler, J. Bis(methimazolyl)silyl Complexes of Ruthenium. Organometallics 2010, 29, 1026–1031. [Google Scholar] [CrossRef]
- Wagler, J.; Brendler, E. Metallasilatranes: Palladium(II) and Platinum(II) as Lone-Pair Donors to Silicon(IV). Angew. Chem. Int. Ed. 2010, 49, 624–627. [Google Scholar] [CrossRef]
- Kazimierczuk, K.; Dołęga, A. Synthesis and structural characterization of new cyclic siloxane with functionalized organic substituents. Phosphorus Sulfur Silicon Relat. Elem. 2017, 192, 1140–1143. [Google Scholar] [CrossRef]
- Wagler, J.; Schley, M.; Gerlach, D.; Böhme, U.; Brendler, E.; Roewer, G. Surprising Insights in the Various Molecular Structures of Hypercoordinate Bis(oxinato)silicon Complexes. Z. Naturforsch. B 2005, 60, 1054–1064. [Google Scholar] [CrossRef]
- Brendler, E.; Wächtler, E.; Wagler, J. Hypercoordinate Silacycloalkanes: Step-by-Step Tuning of N→Si Interactions. Organometallics 2009, 28, 5459–5465. [Google Scholar] [CrossRef]
- Gericke, R.; Wagler, J. Ruthenium complexes of phosphino derivatives of carboxylic amides: Synthesis and characterization of tridentate P,E2 and tetradentate P,E3 (E = N,O) ligands and their reactivity towards [RuCl2(PPh3)3]. Polyhedron 2017, 125, 57–67. [Google Scholar] [CrossRef]
- Wächtler, E.; Gericke, R.; Block, T.; Gerke, B.; Pöttgen, R.; Wagler, J. Compounds of the types Pn(pyS)3 (Pn = P, As, Bi; pyS: Pyridine-2-thiolate) and Sb(pyS) x Ph3–x (x = 3–1); molecular structures and electronic situations of the Pn atoms. Z. Naturforsch. B 2021, 76, 103–118. [Google Scholar] [CrossRef]
- Herzfeld, J.; Berger, A.E. Sideband intensities in NMR spectra of samples spinning at the magic angle. J. Chem. Phys. 1980, 73, 6021–6030. [Google Scholar] [CrossRef]
- Mason, J. Conventions for the reporting of nuclear magnetic shielding (or shift) tensors suggested by participants in the NATO ARW on NMR shielding constants at the University of Maryland, College Park, July 1992. Solid State Nucl. Magn. Reson. 1993, 2, 285–288. [Google Scholar] [CrossRef]
- Wagler, J.; Gericke, R. Molecular structures of various alkyldichlorosilanes in the solid state. Dalton Trans. 2017, 46, 8875–8882. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Program for the Solution of Crystal Structures; SHELXS-97; University of Göttingen: Göttingen, Germany, 1997. [Google Scholar]
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. A Found. Adv. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Program for the Refinement of Crystal Structures; SHELXL-2014/7; University of Göttingen: Göttingen, Germany, 2014. [Google Scholar]
- Sheldrick, G.M. Program for the Refinement of Crystal Structures; SHELXL-2018/3; University of Göttingen: Göttingen, Germany, 2018. [Google Scholar]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. Sect. A 2008, A64, 112–122. [Google Scholar] [CrossRef]
- Kourkoumelis, N. A Program for the Interconversion Procedure between Variable Formats of Powder X-ray Files. PowDLL (Version 2.991.0.0). Available online: http://users.uoi.gr/nkourkou/powdll/#download (accessed on 14 June 2022).
- Farrugia, L.J. ORTEP-3 for Windows—A version ofORTEP-III with a Graphical User Interface (GUI). J. Appl. Crystallogr. 1997, 30, 565. [Google Scholar] [CrossRef]
- Farrugia, L.J. WinGXandORTEP for Windows: An update. J. Appl. Crystallogr. 2012, 45, 849–854. [Google Scholar] [CrossRef]
- POV-RAY (Version 3.7), Trademark of Persistence of Vision Raytracer Pty. Ltd., Williamstown, Victoria (Australia). Copyright Hallam Oaks Pty. Ltd., 1994–2004. Available online: http://www.povray.org/download/ (accessed on 28 June 2021).
- Neese, F. Software update: The ORCA program system—Version 5.0. WIREs Comput. Mol. Sci. 2022, 8, e1606. [Google Scholar] [CrossRef]
- Weigend, F.; Ahlrichs, R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: Design and assessment of accuracy. Phys. Chem. Chem. Phys. 2005, 7, 3297–3305. [Google Scholar] [CrossRef] [PubMed]
- Pantazis, D.A.; Neese, F. All-electron basis sets for heavy elements. WIREs Comput. Mol. Sci. 2014, 4, 363–374. [Google Scholar] [CrossRef]
- van Lenthe, E.; Baerends, E.J.; Snijders, J.G. Relativistic regular two-component Hamiltonians. J. Chem. Phys. 1993, 99, 4597–4610. [Google Scholar] [CrossRef]
- Van Wüllen, C. Molecular density functional calculations in the regular relativistic approximation: Method, application to coinage metal diatomics, hydrides, fluorides and chlorides, and comparison with first-order relativistic calculations. J. Chem. Phys. 1998, 109, 392–399. [Google Scholar] [CrossRef]
- Grimme, S.; Ehrlich, S.; Goerigk, L. Effect of the damping function in dispersion corrected density functional theory. J. Comput. Chem. 2011, 32, 1456. [Google Scholar] [CrossRef]
- Grimme, S.; Antony, J.; Ehrlich, S.; Krieg, H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 2010, 132, 154104–154119. [Google Scholar] [CrossRef]
- Karton, A.; Tarnopolsky, A.; Lamère, J.-F.; Schatz, G.C.; Martin, J.M.L. Highly Accurate First-Principles Benchmark Data Sets for the Parametrization and Validation of Density Functional and Other Approximate Methods. Derivation of a Robust, Generally Applicable, Double-Hybrid Functional for Thermochemistry and Thermochemical Kinetics. J. Phys. Chem. A 2008, 112, 12868–12886. [Google Scholar] [CrossRef]
- Stoychev, G.L.; Auer, A.A.; Neese, F. Automatic Generation of Auxiliary Basis Sets. J. Chem. Theory Comput. 2017, 13, 554–562. [Google Scholar] [CrossRef]
- Te Velde, G.; Bickelhaupt, F.M.; Baerends, E.J.; Fonseca Guerra, C.; van Gisbergen, S.J.A.; Snijders, J.G.; Ziegler, T. Chemistry with ADF. J. Comput. Chem. 2001, 22, 931–967. [Google Scholar] [CrossRef]
- Van Lenthe, E.; Baerends, E.J. Optimized Slater-type basis sets for the elements 1-118. J. Comput. Chem. 2003, 24, 1142–1156. [Google Scholar] [CrossRef]
- Chemcraft, Version 1.8 (Build 164). 2016. Available online: http://www.chemcraftprog.com/ (accessed on 19 September 2015).
- Motherwell, W.B.; Storey, L.J. Some studies on nucleophilic trifluoromethylation using the shelf-stable trifluoromethylacetophenone-N,N-dimethyltrimethylsilylamine adduct. J. Fluor. Chem. 2005, 126, 489–496. [Google Scholar] [CrossRef]
- Bassindale, A.R.; Parker, D.J.; Taylor, P.G. Modelling nucleophilic substitution at silicon in solution using hypervalent silicon compounds based on 2-thiopyridones. J. Chem. Soc. Perkin Trans. 2000, 2, 1059–1066. [Google Scholar] [CrossRef]
- Cordero, B.; Gómez, V.; Platero-Prats, A.E.; Revés, M.; Echeverría, J.; Cremades, E.; Barragán, F.; Alvarez, S. Covalent radii revisited. Dalton Trans. 2008, 2832–2838. [Google Scholar] [CrossRef]
- Mantina, M.; Chamberlin, A.C.; Valero, R.; Cramer, C.J.; Truhlar, D.G. Consistent van der Waals Radii for the Whole Main Group. J. Phys. Chem. A 2009, 113, 5806–5812. [Google Scholar] [CrossRef]
- Okuniewski, A.; Rosiak, D.; Chojnacki, J.; Becker, B. Coordination polymers and molecular structures among complexes of mercury(II) halides with selected 1-benzoylthioureas. Polyhedron 2015, 90, 47–57. [Google Scholar] [CrossRef]
- Bock, H.; Meuret, J.; Bats, J.W.; Havlas, Z. Structures of Sterically Overcrowded Molecules 39 [1,2] Single Crystal and Gas Phase Structures of l,4-Bis(trimethylsiloxy)benzene. Z. Naturforsch. B 1994, 49, 288–296. [Google Scholar] [CrossRef][Green Version]
- Fuhr, O.; Fenske, D. Sulfur Bridged Copper Complexes with dye Ligands. Z. Anorg. Allg. Chem. 2004, 630, 1607–1612. [Google Scholar] [CrossRef]
- Wagler, J.; Heine, T.; Hill, A.F. Poly(methimazolyl)silanes: Syntheses and Molecular Structures. Organometallics 2010, 29, 5607–5613. [Google Scholar] [CrossRef]
- Wiltzsch, C.; Kraushaar, K.; Schwarzer, A.; Kroke, E. CO2 as an Oxygen Source for Polysiloxanes—Preparation, Crystal Structure and Thermal Decomposition of Two Novel Silylcarbamates. Z. Naturforsch. B 2011, 66, 917–922. [Google Scholar] [CrossRef]
- Kraushaar, K.; Wiltzsch, C.; Wagler, J.; Böhme, U.; Schwarzer, A.; Roewer, G.; Kroke, E. From CO2 to Polysiloxanes: Di(carbamoyloxy)silanes Me2Si[(OCO)NRR′]2 as Precursors for PDMS. Organometallics 2012, 31, 4779–4785. [Google Scholar] [CrossRef]
- Herbig, M.; Böhme, U.; Kroke, E. Insertion of CO2 and related heteroallenes into the Si–N-bond of methyl(N-morpholino)silanes. Inorg. Chim. Acta 2018, 473, 20–28. [Google Scholar] [CrossRef]
- Gründler, F.; Scholz, H.; Herbig, M.; Schwarzer, S.; Wagler, J.; Kroke, E. Formation of Aromatic O-Silylcarbamates from Aminosilanes and Their Subsequent Thermal Decomposition with Formation of Isocyanates. Eur. J. Inorg. Chem. 2021, 2021, 2211–2224. [Google Scholar] [CrossRef]
- Kraft, B.M.; Brennessel, W.W. Chelation and Stereodynamic Equilibria in Neutral Hypercoordinate Organosilicon Complexes of 1-Hydroxy-2-pyridinone. Organometallics 2013, 33, 158–171. [Google Scholar] [CrossRef]
- Ismail, R.M. Metal 2-Pyridone. Complexes. Patent FR1475896, 7 July 1964. [Google Scholar]
- Wagler, J.; Böhme, U.; Brendler, E.; Blaurock, S.; Roewer, G. Novel Hexacoordinate Diorganosilanes with Salen-Type Ligands: Molecular Structure versus 29Si NMR Chemical Shifts. Z. Anorg. Allg. Chem. 2005, 631, 2907–2913. [Google Scholar] [CrossRef]
- Wagler, J.; Brendler, E.; Langer, D.-C.T.; Pöttgen, R.; Heine, T.; Zhechkov, L. Ylenes in the MII→SiIV (M=Si, Ge, Sn) Coordination Mode. Chem. A Eur. J. 2010, 16, 13429–13434. [Google Scholar] [CrossRef]
- Fester, G.W.; Eckstein, J.; Gerlach, D.; Wagler, J.; Brendler, E.; Kroke, E. Reactions of Hydridochlorosilanes with 2,2′-Bipyridine and 1,10-Phenanthroline: Complexation versus Dismutation and Metal-Catalyst-Free 1,4-Hydrosilylation. Inorg. Chem. 2010, 49, 2667–2673. [Google Scholar] [CrossRef]
- Spek, A.L. Single-crystal structure validation with the program PLATON. J. Appl. Crystallogr. 2003, 36, 7–13. [Google Scholar] [CrossRef]
- Spek, A.L. Structure validation in chemical crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009, 65, 148–155. [Google Scholar] [CrossRef]
- Spek, A.L. PLATON SQUEEZE: A tool for the calculation of the disordered solvent contribution to the calculated structure factors. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 9–18. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).