Behavior of B- and Z-DNA Crystals under High Hydrostatic Pressure
Abstract
:1. Introduction
2. Materials and Methods
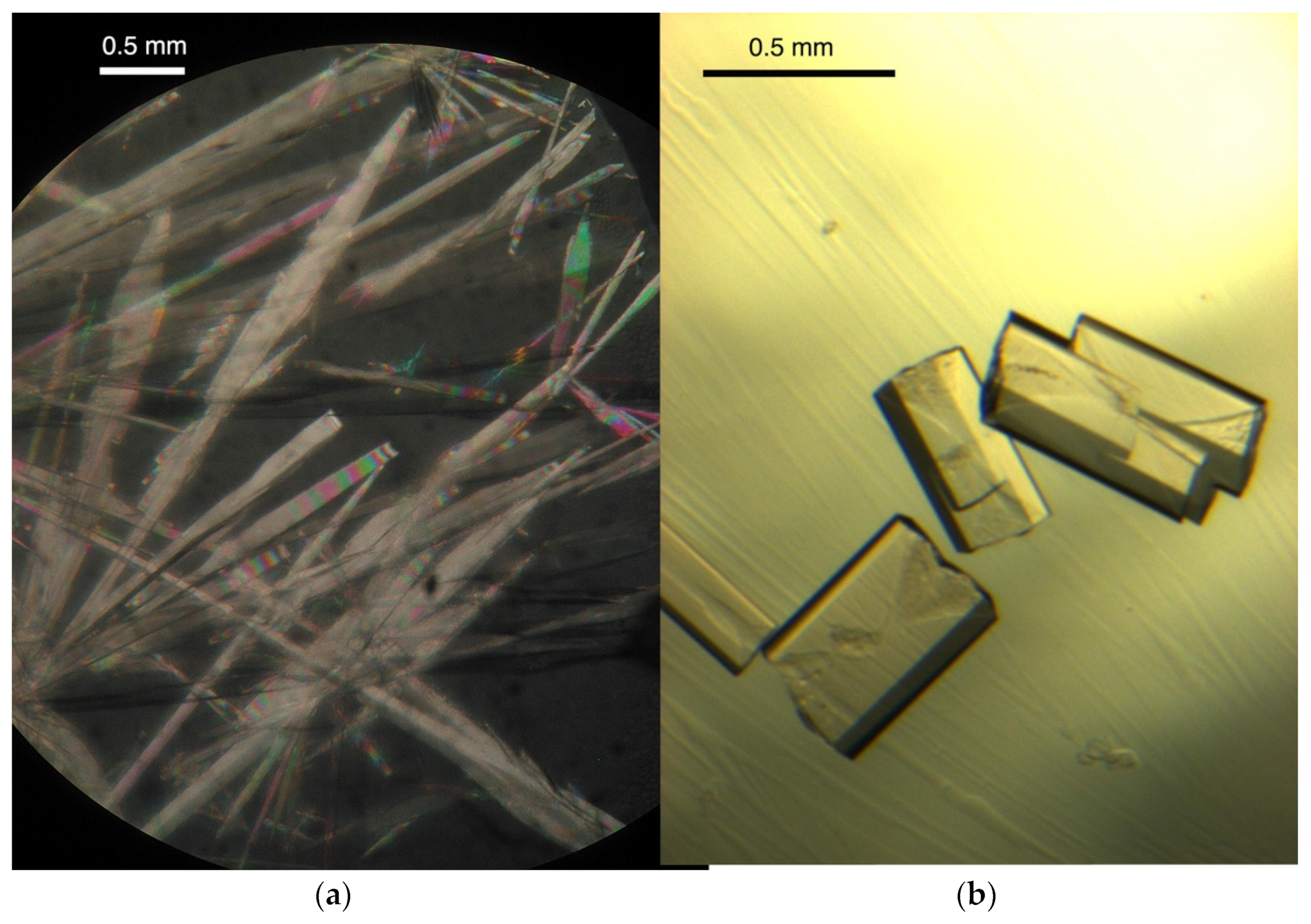
2.1. Crystalizations
2.2. Pressurization and Compression Monitorings
2.3. B-DNA and Z-DNA Data Recordings
- B-DNA: The crystals were relatively fragile and difficult to handle under pressure. Diffraction data were recorded on the ID27 beamline at the ESRF synchrotron (Grenoble, France) at a wavelength of 0.38 Å (Iodine K absorption edge). We used the DAC specially designed for protein crystallography based on previous developments [12,13,14] with an aperture of 55°, enough to record most of the Ewald sphere in a single rotation thanks to the high symmetry of the crystals. The detector was a MarResearch MarCCD165. The lifetime of the crystals and the geometry of the beamline were sufficient to record at ambient pressure at a resolution of 2 Å about 70 to 75% of the Ewald sphere (or the total unique reflections). Under pressure, the crystals decayed rapidly above 400–500 MPa. In these conditions, we were able to record a data set at 310 MPa before the crystal lost its diffraction ability.
- Z-DNA: Experiments were performed at ESRF (Grenoble) at the BM30A (FIP) beamline using a new DAC with a larger angular aperture (90°) [15]. The wavelength was set to 0.75003 Å. In these conditions, the maximum resolution expected at the edge of the detector (a Quantum ADSC Q315r CCD) was limited to about 1.4 Å, though the crystals usually diffracted to a better resolution. The X-ray beam was collimated to 50 × 50 μm2. Exposure times were 20 or 30 s, depending on the sample, for a rotation step of 1°. Several data sets were obtained at pressures of 300, 540, and 715 MPa (estimated ± 20 MPa) using one crystal per pressure step. Crystals are much more pressure-resistant than B-DNA crystals. Figure 2 shows the fate of a Z-DNA crystal following a compression cycle.
2.4. Data Processings and Structure Refinements
3. Results
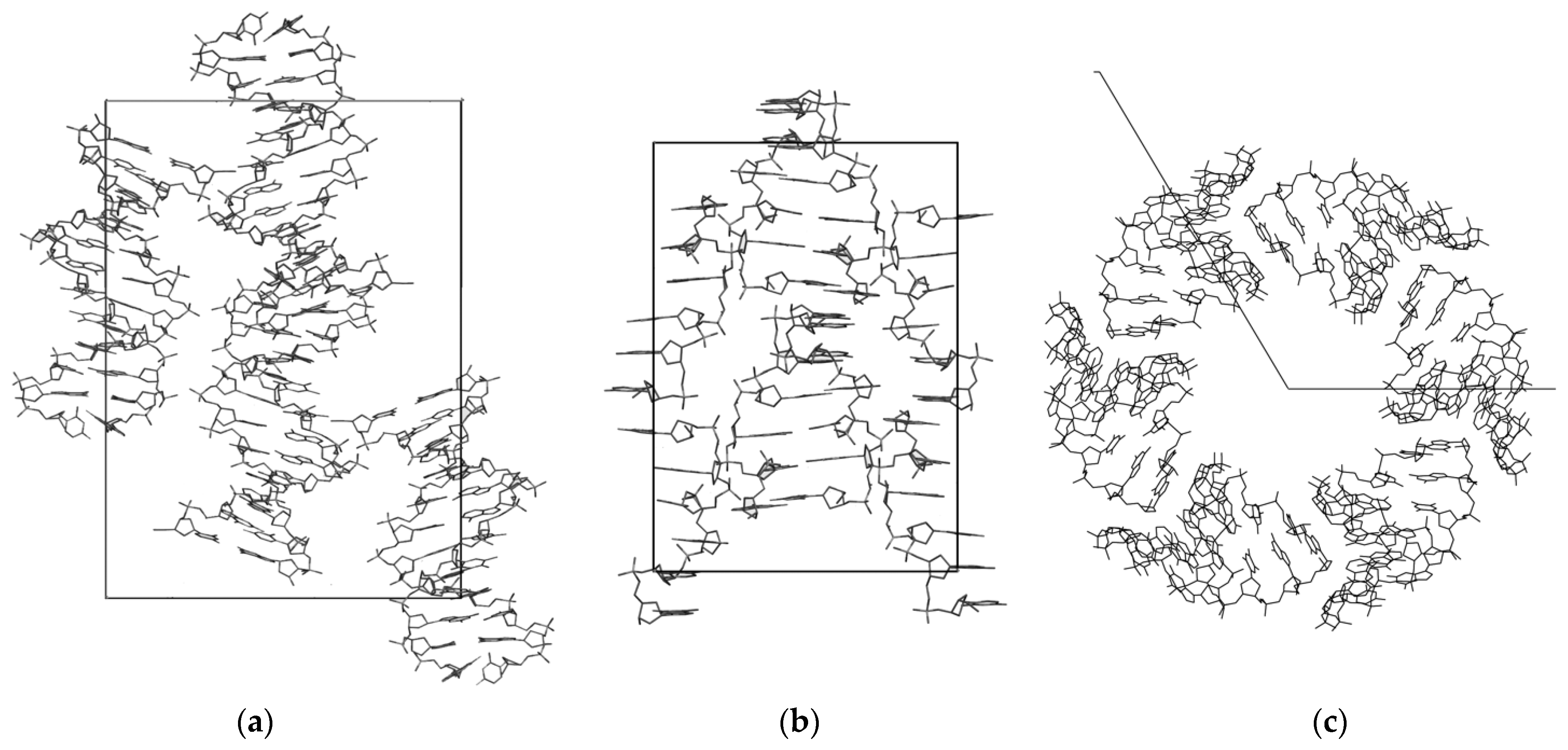
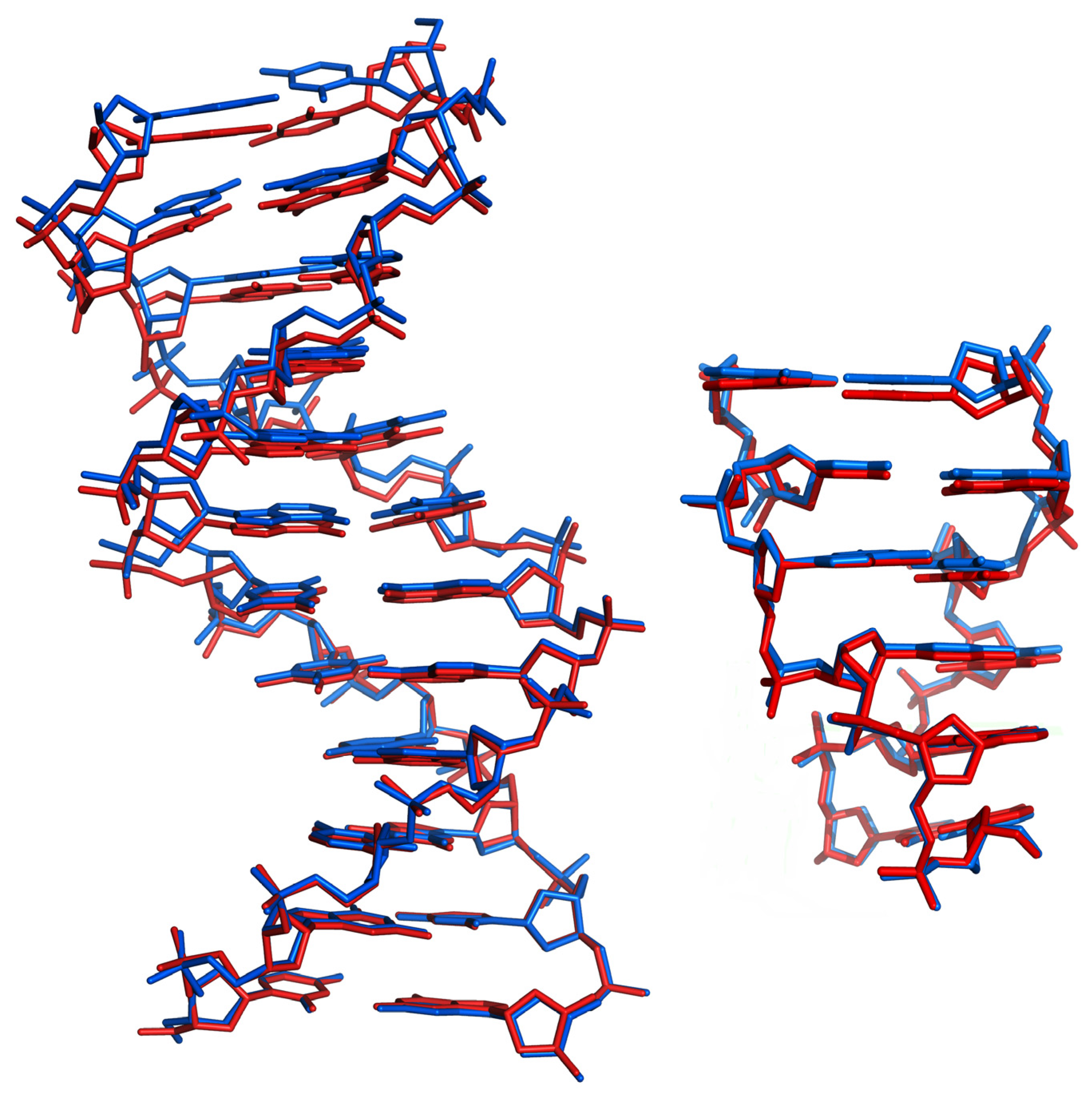
3.1. The DNA Crystal Packings
3.2. DNA Crystals’ Response to Hydrostatic Pressure—The “Reversible” Domain
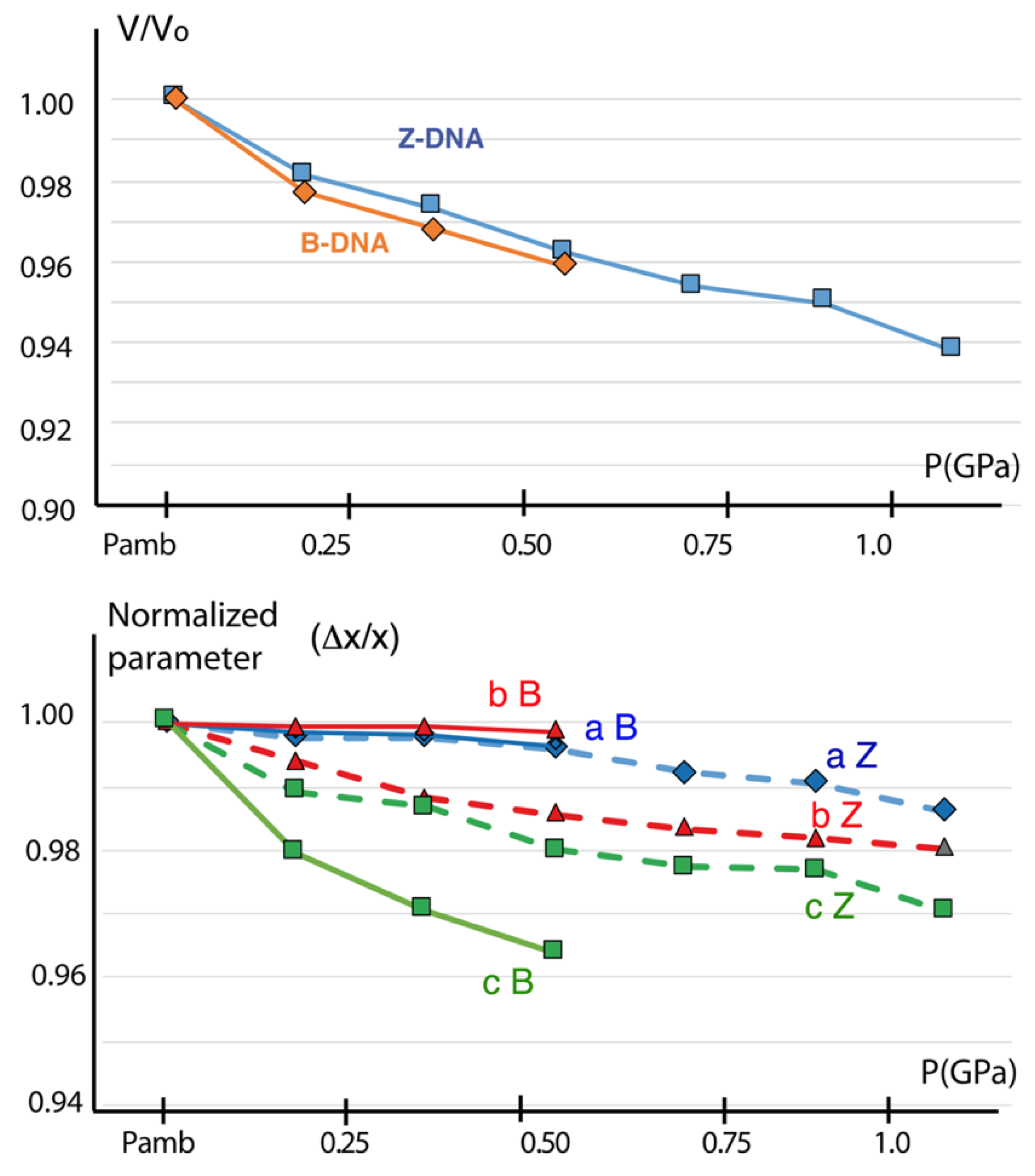
- Volumes: Figure 4 Top shows that the xV evolutions were similar in B and Z crystals. While the B-DNA crystals were destroyed above 400–500 GPa, the Z-DNA crystals entered a linear and constant compression domain up to their end of life estimated around 1.2–1.5 GPa.
- Cell parameters: (Figure 4 bottom). A strong anisotropy was evidenced for the B-DNA (xa and xb unaffected, xc strongly affected by pressure), whereas an isotropic compression was observed for the Z-DNA (the three x values were nearly equal).
4. Discussion
4.1. The DNA Scaffold Adaptation within the “Reversible Domain” of Pressure
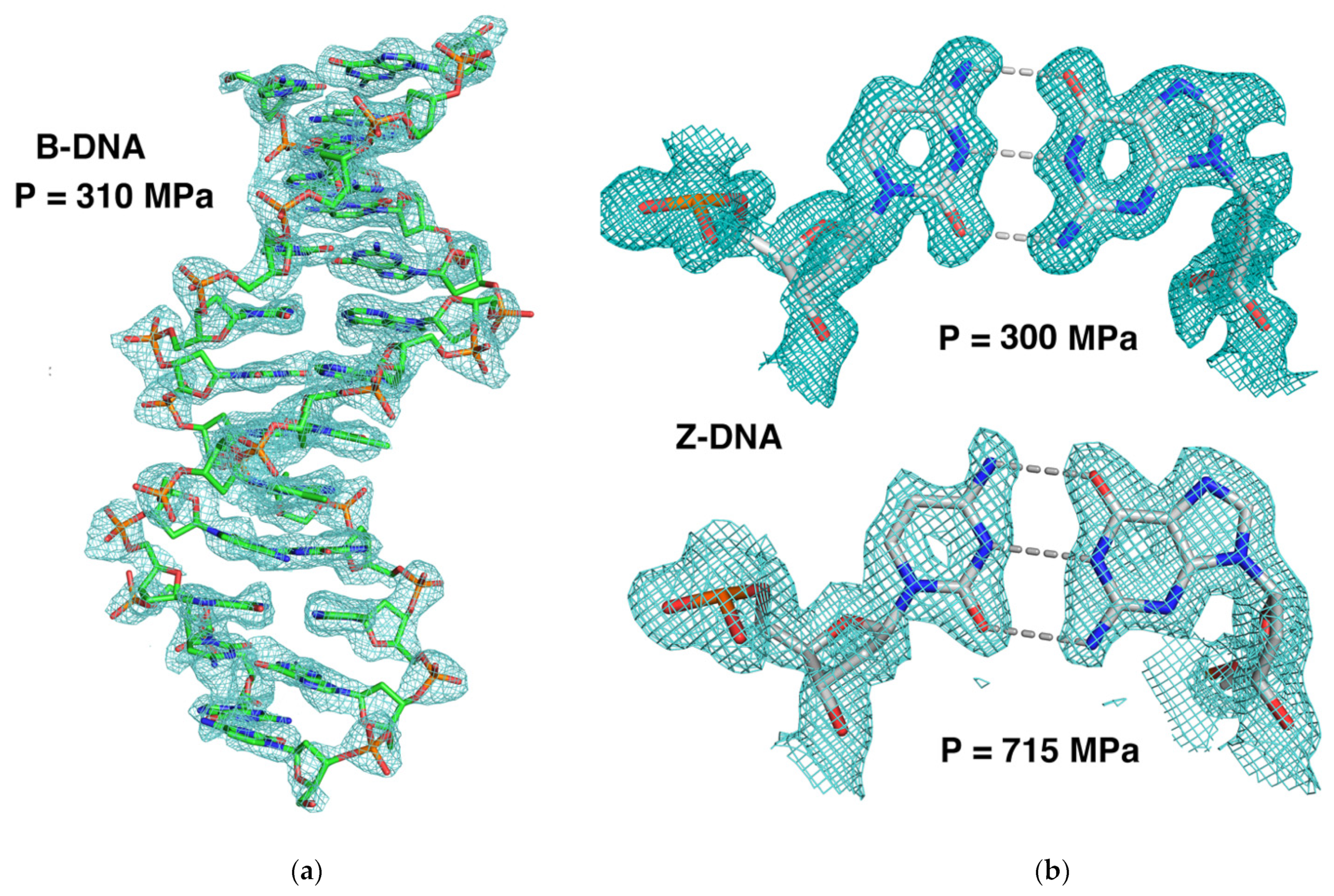
4.1.1. The Watson–Crick Base Pairings: The Transversal Compression
- B-DNA: The average C1′-C1′ distances were in favor of a small lateral compression at 310 MPa (in the range 0.1–0.2 Å for the two A-T and C-G base pairs). However, the base planes were found to be relatively rigid as the calculated shear, stretch, and stagger parameter changes were within the estimated standard variations (< 0.1 Å) at 310 MPa. The only significant perturbation we observed concerned the opening parameter of the four first C-G steps of the duplex, with a Δ-opening increase of 8° at the 5′ end of the dodecamer. This small change could also be deduced from the decrease of O2-N2 distances and the increase of N4/N6-O6 distances (Table 5). This tendency to rotate in the plane with respect to the central N1-N3 distances indicated that the major groove tends to become wider and the minor groove narrower. In addition, a packing effect was clearly present on the 3′-end regarding the propeller parameter of the last C-G base pair (Δ-propeller ~ −10°).
- Z-DNA: The base-pair geometries remained remarkably constant during the pressurization up to 715 MPa. All variations of the six base-pair parameters varied less than 0.1 Å (Δ-shear, Δ-stretch, Δ-stagger) and less than 1° (Δ-buckle, Δ-propeller, and Δ-opening) between ambient pressure and 715 MPa. This illustrated the rigidity and stability of the Z-duplex under high pressure.
4.1.2. The Base Stackings: The Longitudinal Compression
- Z-DNA:
- B-DNA:
4.2. The Water Mediation
4.3. The Irreversible Zone of Pressure: A Phase Transition as a Response to Pressure?
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Levy, M.; Miller, S.L. The Stability of the RNA Bases: Implications for the Origin of Life. Proc. Natl. Acad. Sci. USA 1998, 95, 7933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindahl, T. Instability and Decay of the Primary Structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Chmela, V.; Green, N.J.; Russell, D.A.; Janicki, M.J.; Góra, R.W.; Szabla, R.; Bond, A.D.; Sutherland, J.D. Selective Prebiotic Formation of RNA Pyrimidine and DNA Purine Nucleosides. Nature 2020, 582, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Marguet, E.; Forterre, P. DNA Stability at Temperatures Typical for Hyperthermophiles. Nucleic Acids Res. 1994, 22, 1681–1686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karni, M.; Zidon, D.; Polak, P.; Zalevsky, Z.; Shefi, O. Thermal Degradation of DNA. DNA Cell Biol. 2013, 32, 298–301. [Google Scholar] [CrossRef] [Green Version]
- Dubins, D.N.; Lee, A.; Macgregor, R.B.; Chalikian, T.V. On the Stability of Double Stranded Nucleic Acids. J. Am. Chem. Soc. 2001, 123, 9254–9259. [Google Scholar] [CrossRef]
- Macgregor, R.B. Effect of Hydrostatic Pressure on Nucleic Acids. Biopolymers 1998, 48, 253. [Google Scholar] [CrossRef]
- Green, R.E.; Malaspinas, A.-S.; Krause, J.; Briggs, A.W.; Johnson, P.L.F.; Uhler, C.; Meyer, M.; Good, J.M.; Maricic, T.; Stenzel, U.; et al. A Complete Neandertal Mitochondrial Genome Sequence Determined by High-Throughput Sequencing. Cell 2008, 134, 416–426. [Google Scholar] [CrossRef] [Green Version]
- Stiller, M.; Fulton, T.L. Multiplex PCR Amplification of Ancient DNA. In Ancient DNA; Shapiro, B., Hofreiter, M., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2012; Volume 840, pp. 133–141. ISBN 978-1-61779-515-2. [Google Scholar]
- Girard, E.; Prangé, T.; Dhaussy, A.-C.; Migianu-Griffoni, E.; Lecouvey, M.; Chervin, J.-C.; Mezouar, M.; Kahn, R.; Fourme, R. Adaptation of the Base-Paired Double-Helix Molecular Architecture to Extreme Pressure. Nucleic Acids Res. 2007, 35, 4800–4808. [Google Scholar] [CrossRef] [Green Version]
- Wing, R.; Drew, H.; Takano, T.; Broka, C.; Tanaka, S.; Itakura, K.; Dickerson, R.E. Crystal Structure Analysis of a Complete Turn of B-DNA. Nature 1980, 287, 755–758. [Google Scholar] [CrossRef]
- Wang, A.H.-J.; Quigley, G.J.; Kolpak, F.J.; Crawford, J.L.; van Boom, J.H.; van der Marel, G.; Rich, A. Molecular Structure of a Left-Handed Double Helical DNA Fragment at Atomic Resolution. Nature 1979, 282, 680–686. [Google Scholar] [CrossRef]
- Roy, S.; Caruthers, M. Synthesis of DNA/RNA and Their Analogs via Phosphoramidite and H-Phosphonate Chemistries. Molecules 2013, 18, 14268–14284. [Google Scholar] [CrossRef] [Green Version]
- Timsit, Y.; Moras, D. Crystallization of DNA. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1992; Volume 211, pp. 409–429. ISBN 978-0-12-182112-8. [Google Scholar]
- Chervin, J.C.; Canny, B.; Besson, J.M.; Pruzan, P. A Diamond Anvil Cell for IR Microspectroscopy. Rev. Sci. Instrum. 1995, 66, 2595–2598. [Google Scholar] [CrossRef]
- Fourme, R.; Kahn, R.; Mezouar, M.; Girard, E.; Hoerentrup, C.; Prangé, T.; Ascone, I. High-Pressure Protein Crystallography (HPPX): Instrumentation, Methodology and Results on Lysozyme Crystals. J. Synchrotron Radiat. 2001, 8, 1149–1156. [Google Scholar] [CrossRef] [Green Version]
- Fourme, R.; Ascone, I.; Kahn, R.; Mezouar, M.; Bouvier, P.; Girard, E.; Lin, T.; Johnson, J.E. Opening the High-Pressure Domain beyond 2 Kbar to Protein and Virus Crystallography—Technical Advance. Structure 2002, 10, 1409–1414. [Google Scholar] [CrossRef]
- Girard, E.; Dhaussy, A.-C.; Couzinet, B.; Chervin, J.-C.; Mezouar, M.; Kahn, R.; Ascone, I.; Fourme, R. Toward Fully Fledged High-Pressure Macromolecular Crystallography. J. Appl. Crystallogr. 2007, 40, 912–918. [Google Scholar] [CrossRef]
- Shen, G.; Wang, Y.; Dewaele, A.; Wu, C.; Fratanduono, D.E.; Eggert, J.; Klotz, S.; Dziubek, K.F.; Loubeyre, P.; Fat’yanov, O.V.; et al. Toward an International Practical Pressure Scale: A Proposal for an IPPS Ruby Gauge (IPPS-Ruby2020). High Press. Res. 2020, 40, 299–314. [Google Scholar] [CrossRef]
- Dauter, Z. Data-Collection Strategies. Acta Crystallogr. D Biol. Crystallogr. 1999, 55, 1703–1717. [Google Scholar] [CrossRef] [Green Version]
- Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [Green Version]
- Evans, P.R. An Introduction to Data Reduction: Space-Group Determination, Scaling and Intensity Statistics. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 282–292. [Google Scholar] [CrossRef] [Green Version]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.W.; McCoy, A.; et al. Overview of the CCP4 Suite and Current Developments. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Berman, H.M. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Murshudov, G.N.; Vagin, A.A.; Dodson, E.J. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallogr. D Biol. Crystallogr. 1997, 53, 240–255. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [Green Version]
- Doucet, J.; Benoit, J.-P.; Cruse, W.B.T.; Prangé, T.; Kennard, O. Coexistence of A- and B-Form DNA in a Single Crystal Lattice. Nature 1989, 337, 190–192. [Google Scholar] [CrossRef]
- Chen, Y.Z.; Prohofsky, E.W. Theory of Pressure-Dependent Melting of the DNA Double Helix: Role of Strained Hydrogen Bonds. Phys. Rev. E 1993, 47, 2100–2108. [Google Scholar] [CrossRef]
- Knop, J.-M.; Mukherjee, S.K.; Gault, S.; Cockell, C.S.; Winter, R. Structural Responses of Nucleic Acids to Mars-Relevant Salts at Deep Subsurface Conditions. Life 2022, 12, 677. [Google Scholar] [CrossRef]
- Hervé, G.; Tobé, S.; Heams, T.; Vergne, J.; Maurel, M.-C. Hydrostatic and Osmotic Pressure Study of the Hairpin Ribozyme. Biochim. Biophys. Acta BBA-Proteins Proteomics 2006, 1764, 573–577. [Google Scholar] [CrossRef] [Green Version]
- Giel-Pietraszuk, M.; Barciszewski, J. A Nature of Conformational Changes of Yeast TRNAPhe. Int. J. Biol. Macromol. 2005, 37, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Fourme, R.; Girard, E.; Akasaka, K. High-Pressure Macromolecular Crystallography and NMR: Status, Achievements and Prospects. Curr. Opin. Struct. Biol. 2012, 22, 636–642. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Sugimoto, N. Effect of Pressure on Thermal Stability of G-Quadruplex DNA and Double-Stranded DNA Structures. Molecules 2013, 18, 13297–13319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagae, T.; Kawamura, T.; Chavas, L.M.G.; Niwa, K.; Hasegawa, M.; Kato, C.; Watanabe, N. High-Pressure-Induced Water Penetration into 3-Isopropylmalate Dehydrogenase. Acta Crystallogr. D Biol. Crystallogr. 2012, 68, 300–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ascone, I.; Kahn, R.; Girard, E.; Prangé, T.; Dhaussy, A.-C.; Mezouar, M.; Ponikwicki, N.; Fourme, R. Isothermal Compressibility of Macromolecular Crystals and Macromolecules Derived from High-Pressure X-Ray Crystallography. J. Appl. Crystallogr. 2010, 43, 407–416. [Google Scholar] [CrossRef]
- Wlodawer, A. Stereochemistry and Validation of Macromolecular Structures. In Protein Crystallography; Wlodawer, A., Dauter, Z., Jaskolski, M., Eds.; Methods in Molecular Biology; Springer New York: New York, NY, 2017; Volume 1607, pp. 595–610. ISBN 978-1-4939-6998-2. [Google Scholar]
- Blanc, E.; Roversi, P.; Vonrhein, C.; Flensburg, C.; Lea, S.M.; Bricogne, G. Refinement of Severely Incomplete Structures with Maximum Likelihood in BUSTER–TNT. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2210–2221. [Google Scholar] [CrossRef] [Green Version]
- Vaguine, A.A.; Richelle, J.; Wodak, S.J. SFCHECK: A Unified Set of Procedures for Evaluating the Quality of Macromolecular Structure-Factor Data and Their Agreement with the Atomic Model. Acta Crystallogr. D Biol. Crystallogr. 1999, 55, 191–205. [Google Scholar] [CrossRef]
- Fourme, R.; Ascone, I.; Kahn, R.; Girard, E.; Mezouar, M.; Lin, T.; Johnson, J.E. New Trends in Macromolecular Crystallography at High Hydrostatic Pressure. In Proceedings of the Advances in High Pressure Bioscience and Biotechnology II; Winter, R., Ed.; Springer: Berlin/Heidelberg, Germany, 2003; pp. 161–170. [Google Scholar]
- Li, H.; Yamada, H.; Akasaka, K. Effect of Pressure on Individual Hydrogen Bonds in Proteins. Basic Pancreatic Trypsin Inhibitor. Biochemistry 1998, 37, 1167–1173. [Google Scholar] [CrossRef]
- Lu, X.-J. 3DNA: A Software Package for the Analysis, Rebuilding and Visualization of Three-Dimensional Nucleic Acid Structures. Nucleic Acids Res. 2003, 31, 5108–5121. [Google Scholar] [CrossRef] [Green Version]
- Sinden, R.R. DNA Structure and Function; Academic Press: San Diego, CA, USA, 1994; ISBN 978-0-12-645750-6. [Google Scholar]
- Westhof, E.; Prangé, T.; Chevrier, B.; Moras, D. Solvent Distribution in Crystals of B- and Z-Oligomers. Biochimie 1985, 67, 811–817. [Google Scholar] [CrossRef]
- Westhof, E. Water: An Integral Part of Nucleic Acid Structure. Annu. Rev. Biophys. Biophys. Chem. 1988, 17, 125–144. [Google Scholar] [CrossRef]
- Berman, H.M. Hydration of DNA. Curr. Opin. Struct. Biol. 1991, 1, 423–427. [Google Scholar] [CrossRef]
- Jeffrey, G.A.; Saenger, W. Hydrogen Bonding in Biological Structures; Springer: Berlin/Heidelberg, Germany, 1991; ISBN 978-3-642-85135-3. [Google Scholar]
- Schneider, B.; Cohen, D.; Berman, H.M. Hydration of DNA Bases: Analysis of Crystallographic Data. Biopolymers 1992, 32, 725–750. [Google Scholar] [CrossRef]
- Likhtenshtein, G.I. Nucleic Acids Hydration. In Biological Water; Soft and Biological Matter; Springer International Publishing: Cham, Switzerland, 2021; ISBN 978-3-030-82502-7. [Google Scholar]
- Saenger, W. Water and Nucleic Acids. In Principles of Nucleic Acid Structure; Springer Advanced Texts in Chemistry; Springer: New York, NY, USA, 1984; ISBN 978-0-387-90761-1. [Google Scholar]
- Barciszewski, J.; Jurczak, J.; Porowski, S.; Specht, T.; Erdmann, V.A. The Role of Water Structure in Conformational Changes of Nucleic Acids in Ambient and High-Pressure Conditions: Nucleic Acids at High Pressure. Eur. J. Biochem. 2001, 260, 293–307. [Google Scholar] [CrossRef]
- Krzyźaniak, A.; Salański, P.; Jurczak, J.; Barciszewski, J. B-Z DNA Reversible Conformation Changes Effected by High Pressure. FEBS Lett. 1991, 279, 1–4. [Google Scholar] [CrossRef] [Green Version]


| DNA 1—Pressure (MPa) | B–310 | Z–300 | Z–540 | Z–715 |
|---|---|---|---|---|
| Resolution all (Å) | 34–2.55 | 25.1–1.41 | 25.1–1.41 | 25.1–1.41 |
| Resolution high (Å) | 2.6–2.55 | 1.44–1.41 | 1.44–1.41 | 1.44–1.41 |
| Cell dimensions | ||||
| a (Å) | 25.23 | 17.90 | 17.87 | 17.80 |
| b (Å) | 40.66 | 31.04 | 30.96 | 30.89 |
| c (Å) | 65.20 | 44.19 | 43.89 | 43.77 |
| Cell volume (Å3) | 66 885 | 24 554 | 24 282 | 24 066 |
| Completeness (%) | 78 (70) | 91.2 (79.1) | 77.4 (75.1) | 69.9 (66.6) |
| Redundancy | 4.1 (3.3) | 5.2 (4.9) | 5.8 (5.7) | 5.6 (5.7) |
| Unique reflections | 2412 (277) | 4712 (291) | 3922 (271) | 3464 (255) |
| Rmerge (%) | 11.2 (35.3) | 14.6 (20.6) | 11.2 (18.9) | 10.4 (42.5) |
| Rpim (%) | 5.4 (23.3) | 7.8 (17.7) | 4.7 (17.9) | 6.6 (30.1) |
| I/σ (I) | 8.1 (1.9) | 9.6 (4.9) | 7.8 (2.1) | 10.6 (2.4) |
| DNA 1–Pressure (MPa) | B−310 | Z−300 | Z−540 | Z−715 |
|---|---|---|---|---|
| PDB ID’s | 7zql | 7zqm | 7zqn | 7zqo |
| Rwork (%) | 18.0 | 19.1 | 19.0 | 20.7 |
| Rfree (%) | 23.5 | 29.5 | 22.5 | 26.8 |
| Estimated coords error (Å) | 0.250 | 0.053 | 0.060 | 0.062 |
| Mean standard deviations from ideality | ||||
| 1–2 distances (Å) | 0.005 | 0.009 | 0.008 | 0.006 |
| 1–2 Angles (°) | 1.947 | 2.493 | 2.422 | 1.967 |
| Planes (Å) | 0.018 | 0.023 | 0.022 | 0.022 |
| No of waters | 19 | 62 | 52 | 47 |
| Average B factors (Å2) | ||||
| Overall | 22.2 | 9.9 | 10.8 | 10.9 |
| Bases (A-T, C-G) | 17.8 | 6.4 | 7.5 | 9.2 |
| Deoxy-riboses | 25.8 | 7.8 | 8.7 | 9.5 |
| Phosphates | 30.3 | 11.4 | 12.6 | 12.0 |
| Water | 22.7 | 23.4 | 22.5 | 20.4 |
| B-DNA | Z-DNA | A-DNA 1 | |
|---|---|---|---|
| compound | d(CGCGAATTCGCG)2 | d(CGCGCG)2 | d(GGTATACC)2 |
| System/space group | Orthorhombic P212121 | Orthorhombic P212121 | Hexagonal P61 |
| Z = | 4 | 4 | 6 |
| Cell parameters (Å) | |||
| a = | 25.22 | 17.94 | 45.034 |
| b = | 40.66 | 31.41 | 45.034 |
| c = | 67.19 | 44.78 | 41.747 |
| Vo (Å3) | 68,899.7 | 25,233.3 | 73,322.4 |
| Resolution (Å) | 2.4 | 1.4 2 | 1.6 |
| Compactness(Å3/resid.) | 717 | 525 | 763 |
| Pmax (GPa) | 0.4–0.5 | 1.3–1.5 | >1.9 |
| B-DNA Dodecamer | Z-DNA Hexamer | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Pressure | a | b | c | V | xV | Pressure | a | b | c | V | xV |
| Ambient | 25.22 | 40.66 | 67.19 | 68,899 | Ambient | 17.94 | 31.41 | 44.78 | 25,233 | ||
| 0.21 | 25.24 | 40.66 | 65.80 | 67,528 | 0.08 | 0.22 | 17.90 | 31.22 | 44.30 | 24,757 | 0.10 |
| 0.31 | 25.23 | 40.66 | 65.20 | 66,885 | 0.09 | 0.30 | 17.90 | 31.04 | 44.19 | 24,553 | 0.08 |
| 0.45 | 25.19 | 40.64 | 64.75 | 66,286 | 0.09 | 0.54 | 17.87 | 30.96 | 43.89 | 24,282 | 0.05 |
| 0.71 | 17.80 | 30.89 | 43.77 | 24,067 | 0.05 | ||||||
| 0.80 | 17.77 | 30.84 | 43.74 | 23,971 | 0.04 | ||||||
| 1.1 | 17.69 | 30.79 | 43.45 | 23,666 | 0.04 | ||||||
| B-DNA Dodecamer 1 | Z-DNA Hexamer 1 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Base pair | A-T | C-G | C-G | |||||||||
| Pressure | N1-N3 | N6-O4 | C1′-C1′ | N4-O6 | N3-N1 | O2-N2 | C1′-C1′ | Pressure | N4-O6 | N3-N1 | O2-N2 | C1′-C1′ |
| Ambient | 2.81 | 3.01 | 10.53 | 2.85 | 2.89 | 2.86 | 10.71 | Ambient | 2.90 | 2.96 | 2.88 | 10.86 |
| 310 | 2.81 | 3.13 | 10.37 | 2.87 | 2.87 | 2.81 | 10.63 | 300 | 2.86 | 2.90 | 2.86 | 10.72 |
| 540 | 2.87 | 2.92 | 2.85 | 10.76 | ||||||||
| 715 | 2.86 | 2.94 | 2.90 | 10.78 | ||||||||
| B-DNA | Z-DNA | ||||||
|---|---|---|---|---|---|---|---|
| Pressure (MPa) | Height (Å) | Base Stack (Å) | ∂x/∂p (Å·GPa−1) | Pressure (MPa) | Height (Å) | Base Stack (Å) | ∂x/∂p (Å·GPa−1) |
| Ambient | 36.60 | 3.39 | - | Ambient | 18.0 | 3.58 | - |
| 210 | - | 3.30 1 | 0.43 1 | 300 | 17.63 | 3.53 | 0.23 |
| 310 | 35.45 | 3.26 | 0.40 | 540 | 17.40 | 3.48 | 0.21 |
| 450 | - | 3.20 1 | 0.43 1 | 715 | 17.24 | 3.47 | 0.17 |
| 1100 1 | 17.10 | 3.42 | 0.13 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prangé, T.; Colloc’h, N.; Dhaussy, A.-C.; Lecouvey, M.; Migianu-Griffoni, E.; Girard, E. Behavior of B- and Z-DNA Crystals under High Hydrostatic Pressure. Crystals 2022, 12, 871. https://doi.org/10.3390/cryst12060871
Prangé T, Colloc’h N, Dhaussy A-C, Lecouvey M, Migianu-Griffoni E, Girard E. Behavior of B- and Z-DNA Crystals under High Hydrostatic Pressure. Crystals. 2022; 12(6):871. https://doi.org/10.3390/cryst12060871
Chicago/Turabian StylePrangé, Thierry, Nathalie Colloc’h, Anne-Claire Dhaussy, Marc Lecouvey, Evelyne Migianu-Griffoni, and Eric Girard. 2022. "Behavior of B- and Z-DNA Crystals under High Hydrostatic Pressure" Crystals 12, no. 6: 871. https://doi.org/10.3390/cryst12060871
APA StylePrangé, T., Colloc’h, N., Dhaussy, A.-C., Lecouvey, M., Migianu-Griffoni, E., & Girard, E. (2022). Behavior of B- and Z-DNA Crystals under High Hydrostatic Pressure. Crystals, 12(6), 871. https://doi.org/10.3390/cryst12060871







